Quantification of Pneumocystis jirovecii: Cross-Platform Comparison of One qPCR Assay with Leading Platforms and Six Master Mixes
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Preparation
2.2. Evaluation of Different Master Mixes and Thermocyclers
2.3. Statistical Analysis
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Gigliotti, F.; Wright, T.W. Pneumocystis: Where Does It Live? PLoS Pathogens 2012, 8, e1003025. [Google Scholar] [CrossRef] [PubMed]
- Thomas, C.F.; Limper, A.H. Pneumocystis pneumonia. N. Engl. J. Med. 2004, 350, 2487–2498. [Google Scholar] [CrossRef] [PubMed]
- Roux, A.; Canet, E.; Valade, S.; Gangneux-Robert, F.; Hamane, S.; Lafabrie, A.; Maubon, D.; Debourgogne, A.; Le Gal, S.; Dalle, F.; et al. Pneumocystis jirovecii Pneumonia in Patients with or without AIDS, France. Emerg. Infect. Dis. 2014, 20, 1490–1497. [Google Scholar] [CrossRef] [PubMed]
- Salzer, H.J.F.; Schäfer, G.; Hoenigl, M.; Günther, G.; Hoffmann, C.; Kalsdorf, B.; Alanio, A.; Lange, C. Clinical, Diagnostic, and Treatment Disparities between HIV-Infected and Non-HIV-Infected Immunocompromised Patients with Pneumocystis jirovecii Pneumonia. Respiration 2018, 96, 52–65. [Google Scholar] [CrossRef] [PubMed]
- Alanio, A.; Hauser, P.M.; Lagrou, K.; Melchers, W.J.G.; Helweg-Larsen, J.; Matos, O.; Cesaro, S.; Maschmeyer, G.; Einsele, H.; Donnelly, J.P.; et al. ECIL guidelines for the diagnosis of Pneumocystis jirovecii pneumonia in patients with haematological malignancies and stem cell transplant recipients. J. Antimicrob. Chemother. 2016, 71, 2386–2396. [Google Scholar] [CrossRef] [PubMed]
- Dellière, S.; Gits-Muselli, M.; Bretagne, S.; Alanio, A. Outbreak causing Fungi: Pneumocystis jirovecii. Mycopathologia 2019, in press. [Google Scholar]
- Cordonnier, C.; Cesaro, S.; Maschmeyer, G.; Einsele, H.; Donnelly, J.P.; Alanio, A.; Hauser, P.M.; Lagrou, K.; Melchers, W.J.G.; Helweg-Larsen, J.; et al. Pneumocystis jirovecii pneumonia: Still a concern in patients with haematological malignancies and stem cell transplant recipients. J. Antimicrob. Chemother. 2016, 71, 2379–2385. [Google Scholar] [CrossRef] [PubMed]
- Fan, L.-C.; Lu, H.-W.; Cheng, K.-B.; Li, H.-P.; Xu, J.-F. Evaluation of PCR in Bronchoalveolar Lavage Fluid for Diagnosis of Pneumocystis jirovecii Pneumonia: A Bivariate Meta-Analysis and Systematic Review. PLoS ONE 2013, 8, e73099. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.; Ling, G.; Qiang, C.; Ming, Q.; Wu, C.; Wang, K.; Ying, Z. PCR Diagnosis of Pneumocystis Pneumonia: A Bivariate Meta-Analysis. J. Clin. Microbiol. 2011, 49, 4361–4363. [Google Scholar] [CrossRef] [PubMed]
- Alanio, A.; Bretagne, S. Difficulties with molecular diagnostic tests for mould and yeast infections: Where do we stand? Clin. Microbiol. Infect. 2014, 20, 36–41. [Google Scholar] [CrossRef] [PubMed]
- Bustin, S.A.; Benes, V.; Garson, J.A.; Hellemans, J.; Huggett, J.; Kubista, M.; Mueller, R.; Nolan, T.; Pfaffl, M.W.; Shipley, G.L.; et al. The MIQE guidelines: Minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 2009, 55, 611–622. [Google Scholar] [CrossRef] [PubMed]
- Alanio, A.; Bretagne, S. Pneumocystis jirovecii detection in asymptomatic patients: What does its natural history tell us? F1000Research 2017, 6, 739. [Google Scholar] [CrossRef] [PubMed]
- Robin, C.; Alanio, A.; Gits-Muselli, M.; la Martire, G.; Schlemmer, F.; Botterel, F.; Angebault, C.; Leclerc, M.; Beckerich, F.; Redjoul, R.; et al. Molecular Demonstration of a Pneumocystis Outbreak in Stem Cell Transplant Patients: Evidence for Transmission in the Daycare Center. Front. Microbiol. 2017, 8, 1531–1538. [Google Scholar] [CrossRef] [PubMed]
- Mori, S.; Cho, I.; Sugimoto, M. A follow-up study of asymptomatic carriers of Pneumocystis jirovecii during immunosuppressive therapy for rheumatoid arthritis. J. Rheumatol. 2009, 36, 1600–1605. [Google Scholar] [CrossRef] [PubMed]
- Gits-Muselli, M.; White, L.P.; Mengoli, C.; Chen, S.; Crowley, B.; Dingemans, G.; Fréalle, E.; Gorton, R.; Guiver, M.; Hagen, F.; et al. The Fungal PCR Initiative’s evaluation of in-house and commercial Pneumocystis jirovecii qPCR assays: Towards a standard for a diagnostics assay. Med. Mycol. 2019. [Google Scholar] [CrossRef] [PubMed]
- White, P.L.; Barton, R.; Guiver, M.; Linton, C.J.; Wilson, S.; Smith, M.; Gómez, B.L.; Carr, M.J.; Kimmitt, P.T.; Seaton, S.; et al. A Consensus on Fungal Polymerase Chain Reaction Diagnosis? J. Mol. Diagn. 2006, 8, 376–384. [Google Scholar] [CrossRef] [PubMed]
- Gits-Muselli, M.; Peraldi, M.-N.; de Castro, N.; Delcey, V.; Menotti, J.; Guigue, N.; Hamane, S.; Raffoux, E.; Bergeron, A.; Valade, S.; et al. New Short Tandem Repeat-Based Molecular Typing Method for Pneumocystis jirovecii Reveals Intrahospital Transmission between Patients from Different Wards. PLoS ONE 2015, 10, e0125763. [Google Scholar] [CrossRef] [PubMed]
- Valero, C.; Buitrago, M.J.; Gits-Muselli, M.; Benazra, M.; Sturny-Leclere, A.; Hamane, S.; Guigue, N.; Bretagne, S.; Alanio, A. Copy Number Variation of Mitochondrial DNA Genes in Pneumocystis jirovecii According to the Fungal Load in BAL Specimens. Front. Microbiol. 2016, 7, 1413. [Google Scholar] [CrossRef] [PubMed]
- Javorski-Miller, S.; Delgado, I. Real-Time PCR Instrumentation: An Instrument Selection Guide. In PCR Troubleshooting and Optimization; Kennedy, S., Oswald, N., Eds.; Caister Academic Press: Poole, UK, 2011. [Google Scholar]
- Dalpke, A.H.; Hofko, M.; Zimmermann, S. Development and Evaluation of a Real-Time PCR Assay for Detection of Pneumocystis jirovecii on the Fully Automated BD MAX Platform. J. Clin. Microbiol. 2013, 51, 2337–2343. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Linssen, C.F.M.; Jacobs, J.A.; Beckers, P.; Templeton, K.E.; Bakkers, J.; Kuijper, E.J.; Melchers, W.J.G.; Drent, M.; Vink, C. Inter-laboratory comparison of three different real-time PCR assays for the detection of Pneumocystis jiroveci in bronchoalveolar lavage fluid samples. J. Med. Microbiol. 2006, 55, 1229–1235. [Google Scholar] [CrossRef] [PubMed][Green Version]
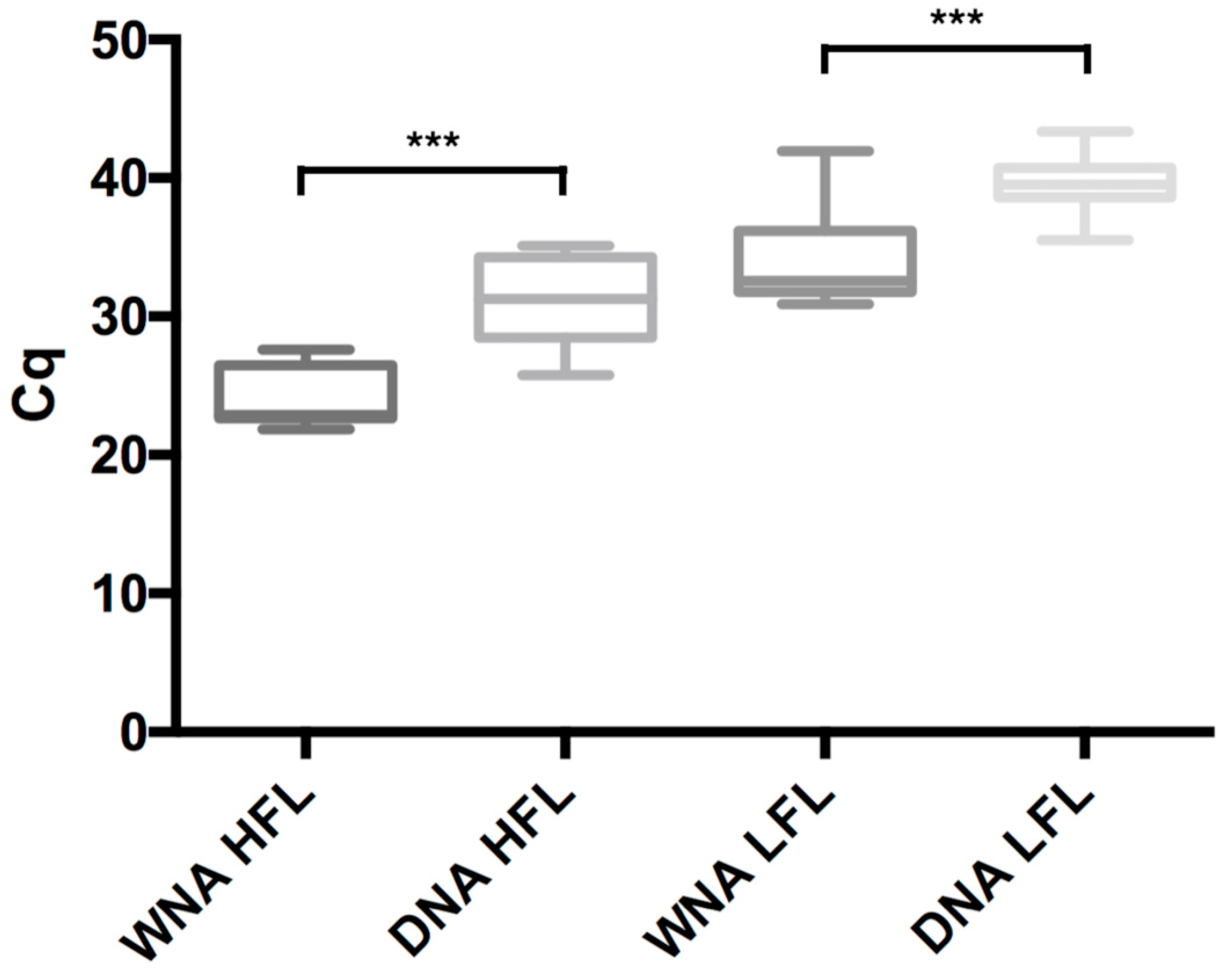
| Mix | Mean Cq HFL (±SD) | p | Mean Cq LFL (±SD) | p | Mean PCR Efficiency in % (±SD) | |
|---|---|---|---|---|---|---|
| DNA detection | Mix D1 | 28.8 (±2.7) | 0.009 | 37.2 (±2.1) | 0.008 | nd |
| Mix D2 | 31.6 (±3.4) | 40.0 (±2.2) | nd | |||
| Mix D3 | 30.7 (±4.7) | 39.5 (±2.4) | nd | |||
| WNA detection | Mix W4 | 24.0 (±2.2) | NS | 33.3 (±2.8) | NS | 104.5 (±7.4) |
| Mix W5 | 23.5 (±2.4) | 33.4 (±3.0) | 100.5 (±2.9) | |||
| Mix W6 | 24.4 (±2.2) | 33.7 (±3.6) | 103.8 (±9.3) | |||
| Mix | ABI7500 | QuantStudio | Rotor-Gene Q | LC480 | |
|---|---|---|---|---|---|
| Fit Point | Fit Point | Fit Point | Fit Point | Derivative | |
| Mix W4 | 99.8 | 100.1 | 115.4 | 102.5 | 110.0 |
| Mix W5 | 96.2 | 102.4 | 101.3 | 102.1 | 152.7 |
| Mix W6 | 92.1 | 113.5 | 101.1 | 108.6 | 149.3 |
| Mean gap from perfect efficiency (i.e., 100%) (±SD) | 4.0 (±3.8) | 5.3 (±7.1) | 5.9 (±8.1) | 4.4 (±3.6) | 37.3 (±23.7) |
| Target Load | Platform 1 | Platform 2 | Contrast | Std. Err. | p | Better |
|---|---|---|---|---|---|---|
| 1:1000 | none | none | - | - | - | - |
| 1:100 | Quantstudio | ABI 7500 | −5.537 | 2.858 | 0.053 | first |
| Rotor-Gene Q | ABI 7500 | −4.080 | 1.853 | 0.028 | first | |
| 1:1 | Light Cycler 480 | Quantstudio | 4.523 | 2.490 | 0.069 | second |
| Rotor-Gene Q | Light Cycler 480 | −3.378 | 1.525 | 0.027 | first |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dellière, S.; Gits-Muselli, M.; White, P.L.; Mengoli, C.; Bretagne, S.; Alanio, A. Quantification of Pneumocystis jirovecii: Cross-Platform Comparison of One qPCR Assay with Leading Platforms and Six Master Mixes. J. Fungi 2020, 6, 9. https://doi.org/10.3390/jof6010009
Dellière S, Gits-Muselli M, White PL, Mengoli C, Bretagne S, Alanio A. Quantification of Pneumocystis jirovecii: Cross-Platform Comparison of One qPCR Assay with Leading Platforms and Six Master Mixes. Journal of Fungi. 2020; 6(1):9. https://doi.org/10.3390/jof6010009
Chicago/Turabian StyleDellière, Sarah, Maud Gits-Muselli, P. Lewis White, Carlo Mengoli, Stéphane Bretagne, and Alexandre Alanio. 2020. "Quantification of Pneumocystis jirovecii: Cross-Platform Comparison of One qPCR Assay with Leading Platforms and Six Master Mixes" Journal of Fungi 6, no. 1: 9. https://doi.org/10.3390/jof6010009
APA StyleDellière, S., Gits-Muselli, M., White, P. L., Mengoli, C., Bretagne, S., & Alanio, A. (2020). Quantification of Pneumocystis jirovecii: Cross-Platform Comparison of One qPCR Assay with Leading Platforms and Six Master Mixes. Journal of Fungi, 6(1), 9. https://doi.org/10.3390/jof6010009







