A Re-Evaluation of the Relationship between Morphology and Pathogenicity in Candida Species
Abstract
1. Introduction
2. Morphogenesis in Candida Species
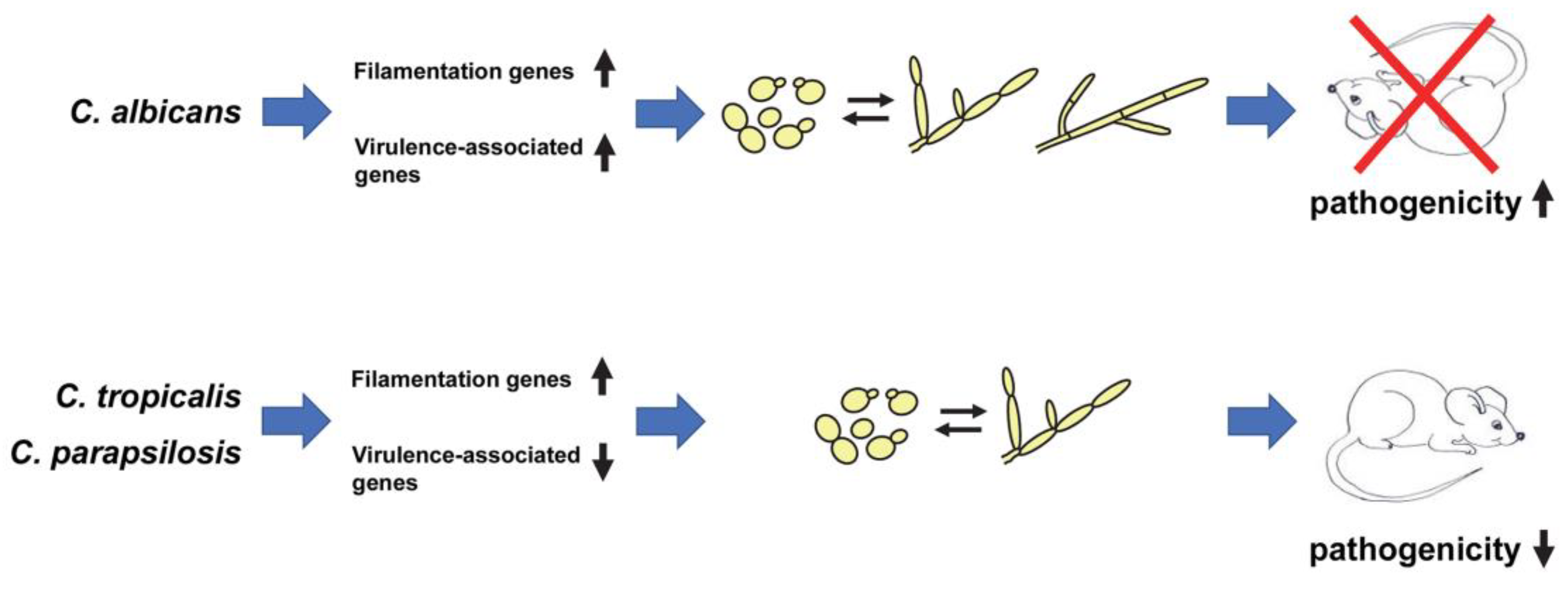
3. Relationship between Morphology and Pathogenicity in C. albicans
4. Relationship between Morphology and Pathogenicity in Non-albicans Candida Species
5. Evolutionary Differences in the Relationship between Morphology and Pathogenicity in C. albicans vs. Non-albicans Candida Species
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Edmond, M.B.; Wallace, S.E.; McClish, D.K.; Pfaller, M.A.; Jones, R.N.; Wenzel, R.P. Nosocomial bloodstream infections in United States hospitals: A three-year analysis. Clin. Infect. Dis. 1999, 29, 239–244. [Google Scholar] [CrossRef] [PubMed]
- Wisplinghoff, H.; Bischoff, T.; Tallent, S.M.; Seifert, H.; Wenzel, R.P.; Edmond, M.B. Nosocomial bloodstream infections in US hospitals: Analysis of 24,179 cases from a prospective nationwide surveillance study. Clin. Infect. Dis. 2004, 39, 309–317. [Google Scholar] [CrossRef] [PubMed]
- Odds, F.C. Candida species and virulence. ASM News 1994, 60, 313–318. [Google Scholar]
- Odds, F.C. Candida and Candidosis, 2nd ed.; Baillière Tindall: London, UK, 1988; p. 468. [Google Scholar]
- Odds, F.C. Pathogenesis of Candida infections. J. Am. Acad. Dermatol. 1994, 31, S2–S5. [Google Scholar] [CrossRef]
- Dupont, P.F. Candida albicans, the opportunist. A cellular and molecular perspective. J. Am. Podiatr. Med. Assoc. 1995, 85, 104–115. [Google Scholar] [CrossRef]
- Weig, M.; Gross, U.; Muhlschlegel, F. Clinical aspects and pathogenesis of Candida infection. Trends Microbiol. 1998, 6, 468–470. [Google Scholar] [CrossRef]
- Perlroth, J.; Choi, B.; Spellberg, B. Nosocomial fungal infections: Epidemiology, diagnosis, and treatment. Med. Mycol. 2007, 45, 321–346. [Google Scholar] [CrossRef]
- Gudlaugsson, O.; Gillespie, S.; Lee, K.; Vande Berg, J.; Hu, J.; Messer, S.; Herwaldt, L.; Pfaller, M.; Diekema, D. Attributable mortality of nosocomial candidemia, revisited. Clin. Infect. Dis. 2003, 37, 1172–1177. [Google Scholar] [CrossRef]
- Picazo, J.J.; Gonzalez-Romo, F.; Candel, F.J. Candidemia in the critically ill patient. Int. J. Antimicrob. Agents 2008, 32 (Suppl. S2), S83–S85. [Google Scholar] [CrossRef]
- Miller, L.G.; Hajjeh, R.A.; Edwards, J.E., Jr. Estimating the cost of nosocomial candidemia in the United States. Clin. Infect. Dis. 2001, 32, 1110. [Google Scholar] [CrossRef]
- McCarty, T.P.; Pappas, P.G. Invasive Candidiasis. Infect. Dis. Clin. N. Am. 2016, 30, 103–124. [Google Scholar] [CrossRef] [PubMed]
- Caceres, D.H.; Forsberg, K.; Welsh, R.M.; Sexton, D.J.; Lockhart, S.R.; Jackson, B.R.; Chiller, T. Candida auris: A review of recommendations for detection and control in healthcare settings. J. Fungi 2019, 5, 111. [Google Scholar] [CrossRef] [PubMed]
- Pfaller, M.A.; Diekema, D.J. Epidemiology of invasive candidiasis: A persistent public health problem. Clin. Microbiol. Rev. 2007, 20, 133–163. [Google Scholar] [CrossRef]
- Patel, P.K.; Erlandsen, J.E.; Kirkpatrick, W.R.; Berg, D.K.; Westbrook, S.D.; Louden, C.; Cornell, J.E.; Thompson, G.R.; Vallor, A.C.; Wickes, B.L.; et al. The changing epidemiology of oropharyngeal candidiasis in patients with HIV/AIDS in the era of antiretroviral therapy. AIDS Res. Treat. 2012, 2012, 262471. [Google Scholar] [CrossRef] [PubMed]
- Moran, G.P.; Sullivan, D.J.; Coleman, D.C. Emergence of non-Candida albicans Candida species as pathogens. In Candida and Candidiasis; Calderone, R.A., Ed.; ASM Press: Washinton, DC, USA, 2002; pp. 37–54. [Google Scholar]
- Calderone, R.A.; Fonzi, W.A. Virulence factors of Candida albicans. Trends Microbiol. 2001, 9, 327–335. [Google Scholar] [CrossRef]
- Kumamoto, C.A.; Vinces, M.D. Contributions of hyphae and hypha-co-regulated genes to Candida albicans virulence. Cell. Microbiol. 2005, 7, 1546–1554. [Google Scholar] [CrossRef]
- Sudbery, P.; Gow, N.; Berman, J. The distinct morphogenic states of Candida albicans. Trends Microbiol. 2004, 12, 317–324. [Google Scholar] [CrossRef]
- Thompson, D.S.; Carlisle, P.L.; Kadosh, D. Coevolution of morphology and virulence in Candida species. Eukaryot. Cell 2011, 10, 1173–1182. [Google Scholar] [CrossRef]
- Gow, N.A.; Henderson, G.; Gooday, G.W. Cytological interrelationships between the cell cycle and duplication cycle of Candida albicans. Microbios 1986, 47, 97–105. [Google Scholar]
- Sudbery, P.E. The germ tubes of Candida albicans hyphae and pseudohyphae show different patterns of septin ring localization. Mol. Microbiol. 2001, 41, 19–31. [Google Scholar] [CrossRef]
- Warenda, A.J.; Konopka, J.B. Septin function in Candida albicans morphogenesis. Mol. Biol. Cell 2002, 13, 2732–2746. [Google Scholar] [CrossRef]
- Sutton, D.A.; Fothergill, A.W.; Rinaldi, M.G. Guide to Clinically Significant Fungi, 1st ed.; Williams and Wilkins: Baltimore, MD, USA, 1998. [Google Scholar]
- Staib, P.; Morschhauser, J. Chlamydospore formation in Candida albicans and Candida dubliniensis—An enigmatic developmental programme. Mycoses 2007, 50, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Chabasse, D.; Bouchara, J.P.; de Gentile, L.; Chennebault, J.M. Candida albicans chlamydospores observed in vivo in a patient with AIDS. Ann. Biol. Clin. 1988, 46, 817–818. [Google Scholar]
- Bartie, K.L.; Williams, D.W.; Wilson, M.J.; Potts, A.J.; Lewis, M.A. Differential invasion of Candida albicans isolates in an in vitro model of oral candidosis. Oral Microbiol. Immunol. 2004, 19, 293–296. [Google Scholar] [CrossRef] [PubMed]
- Lo, H.J.; Kohler, J.R.; DiDomenico, B.; Loebenberg, D.; Cacciapuoti, A.; Fink, G.R. Nonfilamentous C. albicans mutants are avirulent. Cell 1997, 90, 939–949. [Google Scholar] [CrossRef]
- Braun, B.R.; Head, W.S.; Wang, M.X.; Johnson, A.D. Identification and characterization of TUP1-regulated genes in Candida albicans. Genetics 2000, 156, 31–44. [Google Scholar]
- Braun, B.R.; Johnson, A.D. Control of filament formation in Candida albicans by the transcriptional repressor TUP1. Science 1997, 277, 105–109. [Google Scholar] [CrossRef]
- Saville, S.P.; Lazzell, A.L.; Monteagudo, C.; Lopez-Ribot, J.L. Engineered control of cell morphology in vivo reveals distinct roles for yeast and filamentous forms of Candida albicans during infection. Eukaryot. Cell 2003, 2, 1053–1060. [Google Scholar] [CrossRef]
- Carlisle, P.L.; Banerjee, M.; Lazzell, A.; Monteagudo, C.; Lopez-Ribot, J.L.; Kadosh, D. Expression levels of a filament-specific transcriptional regulator are sufficient to determine Candida albicans morphology and virulence. Proc. Natl. Acad. Sci. USA 2009, 106, 599–604. [Google Scholar] [CrossRef]
- Banerjee, M.; Uppuluri, P.; Zhao, X.R.; Carlisle, P.L.; Vipulanandan, G.; Villar, C.C.; Lopez-Ribot, J.L.; Kadosh, D. Expression of UME6, a key regulator of Candida albicans hyphal development, enhances biofilm formation via Hgc1- and Sun41-dependent mechanisms. Eukaryot. Cell 2013, 12, 224–232. [Google Scholar] [CrossRef]
- Zheng, X.; Wang, Y.; Wang, Y. Hgc1, a novel hypha-specific G1 cyclin-related protein regulates Candida albicans hyphal morphogenesis. EMBO J. 2004, 23, 1845–1856. [Google Scholar] [CrossRef] [PubMed]
- O’Meara, T.R.; Veri, A.O.; Ketela, T.; Jiang, B.; Roemer, T.; Cowen, L.E. Global analysis of fungal morphology exposes mechanisms of host cell escape. Nat. Commun. 2015, 6, 6741. [Google Scholar] [CrossRef] [PubMed]
- Noble, S.M.; French, S.; Kohn, L.A.; Chen, V.; Johnson, A.D. Systematic screens of a Candida albicans homozygous deletion library decouple morphogenetic switching and pathogenicity. Nat. Genet. 2010, 42, 590–598. [Google Scholar] [CrossRef]
- Priest, S.J.; Lorenz, M.C. Characterization of virulence-related phenotypes in Candida species of the CUG clade. Eukaryot. Cell 2015, 14, 931–940. [Google Scholar] [CrossRef]
- Bramley, T.A.; Menzies, G.S.; Williams, R.J.; Kinsman, O.S.; Adams, D.J. Binding sites for LH in Candida albicans: Comparison with the mammalian corpus luteum LH receptor. J. Endocrinol. 1991, 130, 177–190. [Google Scholar] [CrossRef]
- Caticha, O.; Grover, S.; Winge, D.; Odell, W.D. Stimulation of Candida albicans transition by human chorionic gonadotrophin and a bacterial protein. Endocr. Res. 1992, 18, 133–143. [Google Scholar] [CrossRef]
- Kinsman, O.S.; Pitblado, K.; Coulson, C.J. Effect of mammalian steroid hormones and luteinizing hormone on the germination of Candida albicans and implications for vaginal candidosis. Mycoses 1988, 31, 617–626. [Google Scholar] [CrossRef]
- Merson-Davies, L.A.; Odds, F.C. A morphology index for characterization of cell shape in Candida albicans. J. Gen. Microbiol. 1989, 135, 3143–3152. [Google Scholar] [CrossRef]
- Brown, A.J. Expression of growth form-specific factors during morphogenesis in Candida albicans. In Candida and Candidiasis; Calderone, R.A., Ed.; ASM Press: Washington, DC, USA, 2002; pp. 87–93. [Google Scholar]
- Lackey, E.; Vipulanandan, G.; Childers, D.S.; Kadosh, D. Comparative evolution of morphological regulatory functions in Candida species. Eukaryot. Cell 2013, 12, 1356–1368. [Google Scholar] [CrossRef]
- Yu, S.; Li, W.; Liu, X.; Che, J.; Wu, Y.; Lu, J. Distinct expression levels of ALS, LIP, and SAP genes in Candida tropicalis with diverse virulent activities. Front. Microbiol. 2016, 7, 1175. [Google Scholar] [CrossRef]
- Silva, S.; Hooper, S.J.; Henriques, M.; Oliveira, R.; Azeredo, J.; Williams, D.W. The role of secreted aspartyl proteinases in Candida tropicalis invasion and damage of oral mucosa. Clin. Microbiol. Infect. 2011, 17, 264–272. [Google Scholar] [CrossRef]
- Jiang, C.; Li, Z.; Zhang, L.; Tian, Y.; Dong, D.; Peng, Y. Significance of hyphae formation in virulence of Candida tropicalis and transcriptomic analysis of hyphal cells. Microbiol. Res. 2016, 192, 65–72. [Google Scholar] [CrossRef]
- Silva, S.; Henriques, M.; Oliveira, R.; Azeredo, J.; Malic, S.; Hooper, S.J.; Williams, D.W. Characterization of Candida parapsilosis infection of an in vitro reconstituted human oral epithelium. Eur. J. Oral Sci. 2009, 117, 669–675. [Google Scholar] [CrossRef]
- Toth, R.; Cabral, V.; Thuer, E.; Bohner, F.; Nemeth, T.; Papp, C.; Nimrichter, L.; Molnar, G.; Vagvolgyi, C.; Gabaldon, T.; et al. Investigation of Candida parapsilosis virulence regulatory factors during host-pathogen interaction. Sci. Rep. 2018, 8, 1346. [Google Scholar] [CrossRef] [PubMed]
- Banerjee, M.; Lazzell, A.L.; Romo, J.A.; Lopez-Ribot, J.L.; Kadosh, D. Filamentation is associated with reduced pathogenicity of multiple non-albicans Candida species. mSphere 2019, 4. [Google Scholar] [CrossRef]
- Kadosh, D. Control of Candida albicans morphology and pathogenicity by post-transcriptional mechanisms. Cell. Mol. Life Sci. 2016, 73, 4265–4278. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.H.; Iyer, K.R.; Pardeshi, L.; Munoz, J.F.; Robbins, N.; Cuomo, C.A.; Wong, K.H.; Cowen, L.E. Genetic analysis of Candida auris implicates Hsp90 in morphogenesis and azole tolerance and Cdr1 in azole resistance. mBio 2019, 10. [Google Scholar] [CrossRef] [PubMed]
- Yue, H.; Bing, J.; Zheng, Q.; Zhang, Y.; Hu, T.; Du, H.; Wang, H.; Huang, G. Filamentation in Candida auris, an emerging fungal pathogen of humans: Passage through the mammalian body induces a heritable phenotypic switch. Emerg. Microbes Infect. 2018, 7, 188. [Google Scholar] [CrossRef]
- Kadosh, D.; Johnson, A.D. Induction of the Candida albicans filamentous growth program by relief of transcriptional repression: A genome-wide analysis. Mol. Biol. Cell 2005, 16, 2903–2912. [Google Scholar] [CrossRef]
- Nantel, A.; Dignard, D.; Bachewich, C.; Harcus, D.; Marcil, A.; Bouin, A.P.; Sensen, C.W.; Hogues, H.; van het Hoog, M.; Gordon, P.; et al. Transcription profiling of Candida albicans cells undergoing the yeast-to-hyphal transition. Mol. Biol. Cell 2002, 13, 3452–3465. [Google Scholar] [CrossRef]
- Chen, Y.V.; Rosli, R.; Fong, S.H.; Sidik, S.M.; Pei, C.P. Histopathological characteristics of experimental Candida tropicalis induced acute systemic candidiasis in BALB/c mice. Int. J. Zool. Res. 2012, 8, 12–22. [Google Scholar]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kadosh, D.; Mundodi, V. A Re-Evaluation of the Relationship between Morphology and Pathogenicity in Candida Species. J. Fungi 2020, 6, 13. https://doi.org/10.3390/jof6010013
Kadosh D, Mundodi V. A Re-Evaluation of the Relationship between Morphology and Pathogenicity in Candida Species. Journal of Fungi. 2020; 6(1):13. https://doi.org/10.3390/jof6010013
Chicago/Turabian StyleKadosh, David, and Vasanthakrishna Mundodi. 2020. "A Re-Evaluation of the Relationship between Morphology and Pathogenicity in Candida Species" Journal of Fungi 6, no. 1: 13. https://doi.org/10.3390/jof6010013
APA StyleKadosh, D., & Mundodi, V. (2020). A Re-Evaluation of the Relationship between Morphology and Pathogenicity in Candida Species. Journal of Fungi, 6(1), 13. https://doi.org/10.3390/jof6010013




