Candida glabrata’s Genome Plasticity Confers a Unique Pattern of Expressed Cell Wall Proteins
Abstract
1. Introduction
2. Role of Adherence during Infection
2.1. Adherence in Candida glabrata Is a Virulence Factor
2.2. Adherence Is Mediated by Cwps in C. glabrata
2.3. C. glabrata Shows High Variability in the Number and Sequence of CWP-Encoding Genes
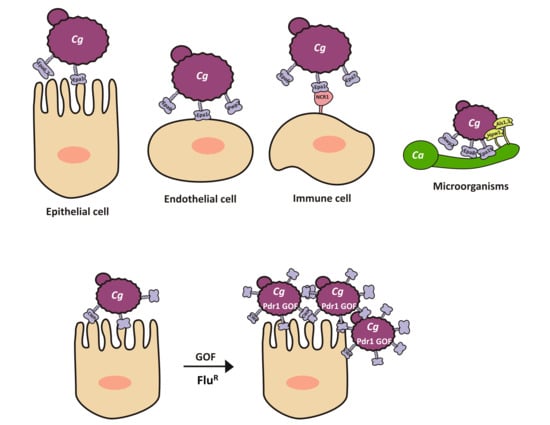
2.4. Adherence to a Variety of Surfaces Is Mediated by the Presence of a Large Family of (GPI)-Modified Proteins at the Cell Surface
2.5. EPA Family
3. Regulation of Expression of EPA Genes
3.1. Subtelomeric Silencing and Cis-Acting Elements Regulate the Expression of EPA Genes
3.2. EPA1 Is Tightly Regulated by Cis-Acting Elements
3.3. EPA1 Is Regulated by the Transcription Factor Pdr1
4. Large Variability in the Pattern of Proteins Present at the Cell Wall among C. glabrata Clinical Isolates
4.1. Different C. glabrata Clinical Isolates Display a Unique Pattern of Proteins at the Cell Surface
4.2. C. glabrata Shows Large Genomic Variability Involving CWP-Encoding Genes
5. Role of Epa1 in Host Cell Recognition of C. glabrata
Interactions with Cells from the Immune System: Neutrophils, Macrophages, Monocytes, Natural Killers (NK), and Dendritic Cells
6. Interactions with Other Microorganisms
7. Conclusions
Author Contributions
Acknowledgments
Conflicts of Interest
References
- Brown, G.D.; Denning, D.W.; Gow, N.A.; Levitz, S.M.; Netea, M.G.; White, T.C. Hidden killers: Human fungal infections. Sci. Transl. Med. 2012, 4, 165rv113. [Google Scholar] [CrossRef] [PubMed]
- Diekema, D.; Arbefeville, S.; Boyken, L.; Kroeger, J.; Pfaller, M. The changing epidemiology of healthcare-associated candidemia over three decades. Diagn. Microbiol. Infect. Dis. 2012, 73, 45–48. [Google Scholar] [CrossRef] [PubMed]
- Pfaller, M.A.; Andes, D.R.; Diekema, D.J.; Horn, D.L.; Reboli, A.C.; Rotstein, C.; Franks, B.; Azie, N.E. Epidemiology and outcomes of invasive candidiasis due to non-albicans species of Candida in 2,496 patients: Data from the Prospective Antifungal Therapy (PATH) registry 2004–2008. PLoS ONE 2014, 9, e101510. [Google Scholar] [CrossRef] [PubMed]
- Guinea, J. Global trends in the distribution of Candida species causing candidemia. Clin. Microbiol. Infect. 2014, 20, 5–10. [Google Scholar] [CrossRef] [PubMed]
- Khatib, R.; Johnson, L.B.; Fakih, M.G.; Riederer, K.; Briski, L. Current trends in candidemia and species distribution among adults: Candida glabrata surpasses C. albicans in diabetic patients and abdominal sources. Mycoses 2016, 59, 781–786. [Google Scholar] [CrossRef] [PubMed]
- Castanheira, M.; Messer, S.A.; Rhomberg, P.R.; Pfaller, M.A. Antifungal susceptibility patterns of a global collection of fungal isolates: Results of the SENTRY Antifungal Surveillance Program (2013). Diagn. Microbiol. Infect. Dis. 2016, 85, 200–204. [Google Scholar] [CrossRef] [PubMed]
- Carrete, L.; Ksiezopolska, E.; Pegueroles, C.; Gomez-Molero, E.; Saus, E.; Iraola-Guzman, S.; Loska, D.; Bader, O.; Fairhead, C.; Gabaldon, T. Patterns of Genomic Variation in the Opportunistic Pathogen Candida glabrata Suggest the Existence of Mating and a Secondary Association with Humans. Curr. Biol. 2018, 28, 15–27. [Google Scholar] [CrossRef] [PubMed]
- Gómez-Molero, E.; de Boer, A.D.; Dekker, H.L.; Moreno-Martínez, A.; Kraneveld, E.A.; Chauhan, N.; Weig, M.; de Soet, J.J.; de Koster, C.G.; Bader, O.; et al. Proteomic analysis of hyperadhesive Candida glabrata clinical isolates reveals a core wall proteome and differential incorporation of adhesins. FEMS Yeast Res. 2015, 15. [Google Scholar] [CrossRef] [PubMed]
- Ni, Q.; Wang, C.; Tian, Y.; Dong, D.; Jiang, C.; Mao, E.; Peng, Y. CgPDR1 gain-of-function mutations lead to azole-resistance and increased adhesion in clinical Candida glabrata strains. Mycoses 2018. [Google Scholar] [CrossRef] [PubMed]
- Vale-Silva, L.; Beaudoing, E.; Tran, V.D.T.; Sanglard, D. Comparative Genomics of Two Sequential Candida glabrata Clinical Isolates. G3 Genes Genomes Genet. 2017, 7, 2413–2426. [Google Scholar] [CrossRef] [PubMed]
- Cormack, B.P.; Ghori, N.; Falkow, S. An adhesin of the yeast pathogen Candida glabrata mediating adherence to human epithelial cells. Science 1999, 285, 578–582. [Google Scholar] [CrossRef] [PubMed]
- Martinez-Jimenez, V.; Ramirez-Zavaleta, C.Y.; Orta-Zavalza, E.; Diaz de Leon, G.; Gutierrez-Escobedo, G.; Ponce de Leon, A.; Sifuentes-Osornio, J.; Bobadilla Del Valle, M.; De Las Penas, A.; Castano, I. Sir3 Polymorphisms in Candida glabrata Clinical Isolates. Mycopathologia 2013, 175, 207–219. [Google Scholar] [CrossRef] [PubMed]
- Zupancic, M.L.; Frieman, M.; Smith, D.; Alvarez, R.A.; Cummings, R.D.; Cormack, B.P. Glycan microarray analysis of Candida glabrata adhesin ligand specificity. Mol. Microbiol. 2008, 68, 547–559. [Google Scholar] [CrossRef] [PubMed]
- Desai, C.; Mavrianos, J.; Chauhan, N. Candida glabrata Pwp7p and Aed1p are required for adherence to human endothelial cells. FEMS Yeast Res. 2011, 11, 595–601. [Google Scholar] [CrossRef] [PubMed]
- Torres-Tirado, D.; Knabb, M.; Castano, I.; Patron-Soberano, A.; De Las Penas, A.; Rubio, R. Candida glabrata binds to glycosylated and lectinic receptors on the coronary endothelial luminal membrane and inhibits flow sense and cardiac responses to agonists. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2016, 310, R24–R32. [Google Scholar] [CrossRef] [PubMed]
- Bourgeois, C.; Majer, O.; Frohner, I.E.; Lesiak-Markowicz, I.; Hildering, K.S.; Glaser, W.; Stockinger, S.; Decker, T.; Akira, S.; Muller, M.; et al. Conventional dendritic cells mount a type I IFN response against Candida spp. requiring novel phagosomal TLR7-mediated IFN-β signaling. J. Immunol. 2011, 186, 3104–3112. [Google Scholar] [CrossRef] [PubMed]
- Kuhn, D.M.; Vyas, V.K. The Candida glabrata adhesin Epa1p causes adhesion, phagocytosis, and cytokine secretion by innate immune cells. FEMS Yeast Res. 2012, 12, 398–414. [Google Scholar] [CrossRef] [PubMed]
- Vitenshtein, A.; Charpak-Amikam, Y.; Yamin, R.; Bauman, Y.; Isaacson, B.; Stein, N.; Berhani, O.; Dassa, L.; Gamliel, M.; Gur, C.; et al. NK Cell Recognition of Candida glabrata through Binding of NKp46 and NCR1 to Fungal Ligands Epa1, Epa6, and Epa7. Cell Host Microbe 2016, 20, 527–534. [Google Scholar] [CrossRef] [PubMed]
- Santoni, G.; Birarelli, P.; Hong, L.J.; Gamero, A.; Djeu, J.Y.; Piccoli, M. An alpha 5 beta 1-like integrin receptor mediates the binding of less pathogenic Candida species to fibronectin. J. Med. Microbiol. 1995, 43, 360–367. [Google Scholar] [CrossRef] [PubMed]
- Parnanen, P.; Kari, K.; Virtanen, I.; Sorsa, T.; Meurman, J.H. Human laminin-332 degradation by Candida proteinases. J. Oral Pathol. Med. 2008, 37, 329–335. [Google Scholar] [CrossRef] [PubMed]
- Parnanen, P.; Meurman, J.H.; Virtanen, I. Laminin-511 and fibronectin degradation with Candida yeast. J. Oral Pathol. Med. 2009, 38, 768–772. [Google Scholar] [CrossRef] [PubMed]
- Kojic, E.M.; Darouiche, R.O. Candida infections of medical devices. Clin. Microbiol. Rev. 2004, 17, 255–267. [Google Scholar] [CrossRef] [PubMed]
- Timmermans, B.; De Las Penas, A.; Castano, I.; Van Dijck, P. Adhesins in Candida glabrata. J. Fungi 2018, 4, 60. [Google Scholar] [CrossRef] [PubMed]
- Verstrepen, K.J.; Klis, F.M. Flocculation, adhesion and biofilm formation in yeasts. Mol. Microbiol. 2006, 60, 5–15. [Google Scholar] [CrossRef] [PubMed]
- De Groot, P.W.J.; Kraneveld, E.A.; Yin, Q.Y.; Dekker, H.L.; Gross, U.; Crielaard, W.; de Koster, C.G.; Bader, O.; Klis, F.M.; Weig, M. The Cell Wall of the Human Pathogen Candida glabrata: Differential Incorporation of Novel Adhesin-Like Wall Proteins. Eukaryot. Cell 2008, 7, 1951–1964. [Google Scholar] [CrossRef] [PubMed]
- Weig, M.; Jansch, L.; Gross, U.; De Koster, C.G.; Klis, F.M.; De Groot, P.W.J. Systematic identification in silico of covalently bound cell wall proteins and analysis of protein-polysaccharide linkages of the human pathogen Candida glabrata. Microbiology 2004, 150, 3129–3144. [Google Scholar] [CrossRef] [PubMed]
- Kapteyn, J.C.; Van Den Ende, H.; Klis, F.M. The contribution of cell wall proteins to the organization of the yeast cell wall. Biochim. Biophys. Acta Gen. Subj. 1999, 1426, 373–383. [Google Scholar] [CrossRef]
- De Groot, P.W.; Bader, O.; de Boer, A.D.; Weig, M.; Chauhan, N. Adhesins in human fungal pathogens: Glue with plenty of stick. Eukaryot. Cell 2013, 12, 470–481. [Google Scholar] [CrossRef] [PubMed]
- Maestre-Reyna, M.; Diderrich, R.; Veelders, M.S.; Eulenburg, G.; Kalugin, V.; Bruckner, S.; Keller, P.; Rupp, S.; Mosch, H.U.; Essen, L.O. Structural basis for promiscuity and specificity during Candida glabrata invasion of host epithelia. Proc. Natl. Acad. Sci. USA 2012, 109, 16864–16869. [Google Scholar] [CrossRef] [PubMed]
- Xie, X.; Lipke, P.N. On the evolution of fungal and yeast cell walls. Yeast 2010, 8, 479–488. [Google Scholar] [CrossRef] [PubMed]
- Dujon, B.; Sherman, D.; Fischer, G.; Durrens, P.; Casaregola, S.; Lafontaine, I.; De Montigny, J.; Marck, C.; Neuveglise, C.; Talla, E.; et al. Genome evolution in yeasts. Nature 2004, 430, 35–44. [Google Scholar] [CrossRef] [PubMed]
- Gabaldon, T.; Martin, T.; Marcet-Houben, M.; Durrens, P.; Bolotin-Fukuhara, M.; Lespinet, O.; Arnaise, S.; Boisnard, S.; Aguileta, G.; Atanasova, R.; et al. Comparative genomics of emerging pathogens in the Candida glabrata clade. BMC Genom. 2013, 14, 623. [Google Scholar] [CrossRef] [PubMed]
- De Las Penas, A.; Pan, S.J.; Castano, I.; Alder, J.; Cregg, R.; Cormack, B.P. Virulence-related surface glycoproteins in the yeast pathogen Candida glabrata are encoded in subtelomeric clusters and subject to RAP1- and SIR-dependent transcriptional silencing. Genes Dev. 2003, 17, 2245–2258. [Google Scholar] [CrossRef] [PubMed]
- Castano, I.; Pan, S.J.; Zupancic, M.; Hennequin, C.; Dujon, B.; Cormack, B.P. Telomere length control and transcriptional regulation of subtelomeric adhesins in Candida glabrata. Mol. Microbiol. 2005, 55, 1246–1258. [Google Scholar] [CrossRef] [PubMed]
- Gabaldon, T.; Carrete, L. The birth of a deadly yeast: Tracing the evolutionary emergence of virulence traits in Candida glabrata. FEMS Yeast Res. 2016, 16, fov110. [Google Scholar] [CrossRef] [PubMed]
- Vestrepen, K.; Fink, G.R. Genetic and epigenetic mechanisms underlying cell-surface variability in protozoa and fungi. Annu. Rev. Genet. 2009, 43, 1–24. [Google Scholar] [CrossRef] [PubMed]
- Kaur, R.; Domergue, R.; Zupancic, M.L.; Cormack, B.P. A yeast by any other name: Candida glabrata and its interaction with the host. Curr. Opin. Microbiol. 2005, 8, 378–384. [Google Scholar] [CrossRef] [PubMed]
- Kraneveld, E.A.; de Soet, J.J.; Deng, D.M.; Dekker, H.L.; de Koster, C.G.; Klis, F.M.; Crielaard, W.; de Groot, P.W.J. Identification and Differential Gene Expression of Adhesin-Like Wall Proteins in Candida glabrata Biofilms. Mycopathologia 2011, 172, 415–427. [Google Scholar] [CrossRef] [PubMed]
- Thierry, A.; Bouchier, C.; Dujon, B.; Richard, G.F. Megasatellites: A peculiar class of giant minisatellites in genes involved in cell adhesion and pathogenicity in Candida glabrata. Nucleic Acids Res. 2008, 36, 5970–5982. [Google Scholar] [CrossRef] [PubMed]
- Verstrepen, K.J.; Jansen, A.; Lewitter, F.; Fink, G.R. Intragenic tandem repeats generate functional variability. Nat. Genet. 2005, 37, 986–990. [Google Scholar] [CrossRef] [PubMed]
- Vale-Silva, L.; Ischer, F.; Leibundgut-Landmann, S.; Sanglard, D. Gain-of-function mutations in PDR1, a regulator of antifungal drug resistance in Candida glabrata, control adherence to host cells. Infect. Immun. 2013, 81, 1709–1720. [Google Scholar] [CrossRef] [PubMed]
- Vale-Silva, L.A.; Moeckli, B.; Torelli, R.; Posteraro, B.; Sanguinetti, M.; Sanglard, D. Upregulation of the Adhesin Gene EPA1 Mediated by PDR1 in Candida glabrata Leads to Enhanced Host Colonization. mSphere 2016, 1. [Google Scholar] [CrossRef] [PubMed]
- Alves, C.T.; Wei, X.Q.; Silva, S.; Azeredo, J.; Henriques, M.; Williams, D.W. Candida albicans promotes invasion and colonisation of Candida glabrata in a reconstituted human vaginal epithelium. J. Infect. 2014, 69, 396–407. [Google Scholar] [CrossRef] [PubMed]
- Domergue, R.; Castano, I.; De Las Penas, A.; Zupancic, M.; Lockatell, V.; Hebel, J.R.; Johnson, D.; Cormack, B.P. Nicotinic acid limitation regulates silencing of Candida adhesins during UTI. Science 2005, 308, 866–870. [Google Scholar] [CrossRef] [PubMed]
- De Las Penas, A.; Juarez-Cepeda, J.; Lopez-Fuentes, E.; Briones-Martin-Del-Campo, M.; Gutierrez-Escobedo, G.; Castano, I. Local and regional chromatin silencing in Candida glabrata: Consequences for adhesion and the response to stress. FEMS Yeast Res. 2015, 15, fov056. [Google Scholar] [CrossRef] [PubMed]
- Rosas-Hernandez, L.L.; Juarez-Reyes, A.; Arroyo-Helguera, O.E.; De Las Penas, A.; Pan, S.J.; Cormack, B.P.; Castano, I. yKu70/yKu80 and Rif1 Regulate Silencing Differentially at Telomeres in Candida glabrata. Eukaryot. Cell 2008, 7, 2168–2178. [Google Scholar] [CrossRef] [PubMed]
- Juarez-Cepeda, J.; Orta-Zavalza, E.; Canas-Villamar, I.; Arreola-Gomez, J.; Perez-Cornejo, G.P.; Hernandez-Carballo, C.Y.; Gutierrez-Escobedo, G.; Castano, I.; De Las Penas, A. The Epa2 adhesin encoding gene is responsive to oxidative stress in the opportunistic fungal pathogen Candida glabrata. Curr. Genet. 2015, 61, 529–544. [Google Scholar] [CrossRef] [PubMed]
- Roetzer, A.; Gregori, C.; Jennings, A.M.; Quintin, J.; Ferrandon, D.; Butler, G.; Kuchler, K.; Ammerer, G.; Schuller, C. Candida glabrata environmental stress response involves Saccharomyces cerevisiae Msn2/4 orthologous transcription factors. Mol. Microbiol. 2008, 69, 603–620. [Google Scholar] [CrossRef] [PubMed]
- Gallegos-Garcia, V.; Pan, S.J.; Juarez-Cepeda, J.; Ramirez-Zavaleta, C.Y.; Martin-del-Campo, M.B.; Martinez-Jimenez, V.; Castano, I.; Cormack, B.; De Las Penas, A. A novel downstream regulatory element cooperates with the silencing machinery to repress EPA1 expression in Candida glabrata. Genetics 2012, 190, 1285–1297. [Google Scholar] [CrossRef] [PubMed]
- Juarez-Reyes, A.; Ramirez-Zavaleta, C.Y.; Medina-Sanchez, L.; De Las Penas, A.; Castano, I. A Protosilencer of subtelomeric gene expression in Candida glabrata with unique properties. Genetics 2012, 190, 101–111. [Google Scholar] [CrossRef] [PubMed]
- Pfaller, M.A.; Messer, S.A.; Boyken, L.; Tendolkar, S.; Hollis, R.J.; Diekema, D.J. Variation in susceptibility of bloodstream isolates of Candida glabrata to fluconazole according to patient age and geographic location. J. Clin. Microbiol. 2003, 41, 2176–2179. [Google Scholar] [CrossRef] [PubMed]
- Pfaller, M.A.; Messer, S.A.; Hollis, R.J.; Boyken, L.; Tendolkar, S.; Kroeger, J.; Diekema, D.J. Variation in susceptibility of bloodstream isolates of Candida glabrata to fluconazole according to patient age and geographic location in the United States in 2001 to 2007. J. Clin. Microbiol. 2009, 47, 3185–3190. [Google Scholar] [CrossRef] [PubMed]
- Salazar, S.B.; Wang, C.; Munsterkotter, M.; Okamoto, M.; Takahashi-Nakaguchi, A.; Chibana, H.; Lopes, M.M.; Guldener, U.; Butler, G.; Mira, N.P. Comparative genomic and transcriptomic analyses unveil novel features of azole resistance and adaptation to the human host in Candida glabrata. FEMS Yeast Res. 2018, 18, fox079. [Google Scholar] [CrossRef] [PubMed]
- Vermitsky, J.P.; Earhart, K.D.; Smith, W.L.; Homayouni, R.; Edlind, T.D.; Rogers, P.D. Pdr1 regulates multidrug resistance in Candida glabrata: Gene disruption and genome-wide expression studies. Mol. Microbiol. 2006, 61, 704–722. [Google Scholar] [CrossRef] [PubMed]
- Vermitsky, J.P.; Edlind, T.D. Azole resistance in Candida glabrata: Coordinate upregulation of multidrug transporters and evidence for a Pdr1-like transcription factor. Antimicrob. Agents Chemother. 2004, 48, 3773–3781. [Google Scholar] [CrossRef] [PubMed]
- Thakur, J.K.; Arthanari, H.; Yang, F.J.; Pan, S.J.; Fan, X.C.; Breger, J.; Frueh, D.P.; Gulshan, K.; Li, D.K.; Mylonakis, E.; et al. A nuclear receptor-like pathway regulating multidrug resistance in fungi. Nature 2008, 452, 604. [Google Scholar] [CrossRef] [PubMed]
- Ferrari, S.; Ischer, F.; Calabrese, D.; Posteraro, B.; Sanguinetti, M.; Fadda, G.; Rohde, B.; Bauser, C.; Bader, O.; Sanglard, D. Gain of Function Mutations in CgPDR1 of Candida glabrata Not Only Mediate Antifungal Resistance but Also Enhance Virulence. PLoS Pathog. 2009, 5, e1000268. [Google Scholar] [CrossRef] [PubMed]
- Sanguinetti, M.; Posteraro, B.; Fiori, B.; Ranno, S.; Torelli, R.; Fadda, G. Mechanisms of azole resistance in clinical isolates of Candida glabrata collected during a hospital survey of antifungal resistance. Antimicrob. Agents Chemother. 2005, 49, 668–679. [Google Scholar] [CrossRef] [PubMed]
- Ottaviani, A.; Gilson, E.; Magdinier, F. Telomeric position effect: From the yeast paradigm to human pathologies? Biochimie 2008, 90, 93–107. [Google Scholar] [CrossRef] [PubMed]
- Polakova, S.; Blume, C.; Zarate, J.A.; Mentel, M.; Jorck-Ramberg, D.; Stenderup, J.; Piskur, J. Formation of new chromosomes as a virulence mechanism in yeast Candida glabrata. Proc. Natl. Acad. Sci. USA 2009, 106, 2688–2693. [Google Scholar] [CrossRef] [PubMed]
- Bader, O.; Schwarz, A.; Kraneveld, E.A.; Tangwattanachuleeporn, M.; Schmidt, P.; Jacobsen, M.D.; Gross, U.; De Groot, P.W.; Weig, M. Gross karyotypic and phenotypic alterations among different progenies of the Candida glabrata CBS138/ATCC2001 reference strain. PLoS ONE 2012, 7, e52218. [Google Scholar] [CrossRef] [PubMed]
- Xu, N.; Ye, C.; Chen, X.; Liu, J.; Liu, L.; Chen, J. Genome Sequencing of the Pyruvate-producing Strain Candida glabrata CCTCC M202019 and Genomic Comparison with Strain CBS138. Sci. Rep. 2016, 6, 34893. [Google Scholar] [CrossRef] [PubMed]
- Cuellar-Cruz, M.; Castano, I.; Arroyo-Helguera, O.; De Las Penas, A. Oxidative stress response to menadione and cumene hydroperoxide in the opportunistic fungal pathogen Candida glabrata. Mem. Inst. Oswaldo Cruz 2009, 104, 649–654. [Google Scholar] [CrossRef] [PubMed]
- Kaur, R.; Ma, B.; Cormack, B.P. A family of glycosylphosphatidylinositol-linked aspartyl proteases is required for virulence of Candida glabrata. Proc. Natl. Acad. Sci. USA 2007, 104, 7628–7633. [Google Scholar] [CrossRef] [PubMed]
- Seider, K.; Brunke, S.; Schild, L.; Jablonowski, N.; Wilson, D.; Majer, O.; Barz, D.; Haas, A.; Kuchler, K.; Schaller, M.; et al. The facultative intracellular pathogen Candida glabrata subverts macrophage cytokine production and phagolysosome maturation. J. Immunol. 2011, 187, 3072–3086. [Google Scholar] [CrossRef] [PubMed]
- Netea, M.G.; Joosten, L.A.; van der Meer, J.W.; Kullberg, B.J.; van de Veerdonk, F.L. Immune defence against Candida fungal infections. Nat. Rev. Immunol. 2015, 15, 630–642. [Google Scholar] [CrossRef] [PubMed]
- Lipke, P.; Ovalle, R. Cell wall architecture in yeast: New structure and new challenges. J. Bacteriol. 1998, 180, 3735–3740. [Google Scholar] [PubMed]
- Goodridge, H.S.; Reyes, C.N.; Becker, C.A.; Katsumoto, T.R.; Ma, J.; Wolf, A.J.; Bose, N.; Chan, A.S.; Magee, A.S.; Danielson, M.E.; et al. Activation of the innate immune receptor Dectin-1 upon formation of a ‘phagocytic synapse’. Nature 2011, 472, 471–475. [Google Scholar] [CrossRef] [PubMed]
- Branzk, N.; Lubojemska, A.; Hardison, S.E.; Wang, Q.; Gutierrez, M.G.; Brown, G.D.; Papayannopoulos, V. Neutrophils sense microbe size and selectively release neutrophil extracellular traps in response to large pathogens. Nat. Immunol. 2014, 15, 1017–1025. [Google Scholar] [CrossRef] [PubMed]
- Xu, S.; Huo, J.; Lee, K.G.; Kurosaki, T.; Lam, K.P. Phospholipase Cgamma2 is critical for Dectin-1-mediated Ca2+ flux and cytokine production in dendritic cells. J. Biol. Chem. 2009, 284, 7038–7046. [Google Scholar] [CrossRef] [PubMed]
- McGreal, E.P.; Rosas, M.; Brown, G.D.; Zamze, S.; Wong, S.Y.; Gordon, S.; Martinez-Pomares, L.; Taylor, P.R. The carbohydrate-recognition domain of Dectin-2 is a C-type lectin with specificity for high mannose. Glycobiology 2006, 16, 422–430. [Google Scholar] [CrossRef] [PubMed]
- Sato, K.; Yang, X.L.; Yudate, T.; Chung, J.S.; Wu, J.; Luby-Phelps, K.; Kimberly, R.P.; Underhill, D.; Cruz, P.D., Jr.; Ariizumi, K. Dectin-2 is a pattern recognition receptor for fungi that couples with the Fc receptor gamma chain to induce innate immune responses. J. Biol. Chem. 2006, 281, 38854–38866. [Google Scholar] [CrossRef] [PubMed]
- Ifrim, D.C.; Bain, J.M.; Reid, D.M.; Oosting, M.; Verschueren, I.; Gow, N.A.; van Krieken, J.H.; Brown, G.D.; Kullberg, B.J.; Joosten, L.A.; et al. Role of Dectin-2 for host defense against systemic infection with Candida glabrata. Infect. Immun. 2014, 82, 1064–1073. [Google Scholar] [CrossRef] [PubMed]
- Sem, X.; Le, G.T.; Tan, A.S.; Tso, G.; Yurieva, M.; Liao, W.W.; Lum, J.; Srinivasan, K.G.; Poidinger, M.; Zolezzi, F.; et al. Beta-glucan Exposure on the Fungal Cell Wall Tightly Correlates with Competitive Fitness of Candida Species in the Mouse Gastrointestinal Tract. Front. Cell. Infect. Microbiol. 2016, 6, 186. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.M.; Shen, H.; Zhang, T.; Huang, X.; Liu, X.Q.; Guo, S.Y.; Zhao, J.J.; Wang, C.F.; Yan, L.; Xu, G.T.; et al. Dectin-1 plays an important role in host defense against systemic Candida glabrata infection. Virulence 2017, 8, 1643–1656. [Google Scholar] [CrossRef] [PubMed]
- Swidergall, M.; Solis, N.V.; Lionakis, M.S.; Filler, S.G. EphA2 is an epithelial cell pattern recognition receptor for fungal beta-glucans. Nat. Microbiol. 2018, 3, 53–61. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Dongari-Bagtzoglou, A. Epithelial GM-CSF induction by Candida glabrata. J. Dent. Res. 2009, 88, 746–751. [Google Scholar] [CrossRef] [PubMed]
- Robert, R.; Nail, S.; Marot-Leblond, A.; Cottin, J.; Miegeville, M.; Quenouillere, S.; Mahaza, C.; Senet, J.M. Adherence of platelets to Candida species in vivo. Infect. Immun. 2000, 68, 570–576. [Google Scholar] [CrossRef] [PubMed]
- Tati, S.; Davidow, P.; McCall, A.; Hwang-Wong, E.; Rojas, I.G.; Cormack, B.; Edgerton, M. Candida glabrata Binding to Candida albicans Hyphae Enables Its Development in Oropharyngeal Candidiasis. PLoS Pathog. 2016, 12, e1005522. [Google Scholar] [CrossRef] [PubMed]
- Rossoni, R.D.; Barros, P.P.; Freire, F.; Santos, J.D.D.; Jorge, A.O.C.; Junqueira, J.C. Study of Microbial Interaction Formed by “Candida krusei” and “Candida glabrata”: “In Vitro” and “In Vivo” Studies. Braz. Dent. J. 2017, 28, 669–674. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.L.; Chu, W.L.; Lin, C.C.; Zhou, Z.L.; Chen, P.N.; Lo, H.J.; Hospitals, T. Mixed yeast infections in Taiwan. Med. Mycol. 2017. [Google Scholar] [CrossRef] [PubMed]
- Zajac, D.; Karkowska-Kuleta, J.; Bochenska, O.; Rapala-Kozik, M.; Kozik, A. Interaction of human fibronectin with Candida glabrata epithelial adhesin 6 (Epa6). Acta Biochim. Pol. 2016, 63, 417–426. [Google Scholar] [CrossRef] [PubMed]
- Munoz-Duarte, A.R.; Castrejon-Jimenez, N.S.; Baltierra-Uribe, S.L.; Perez-Rangel, S.J.; Carapia-Minero, N.; Castaneda-Sanchez, J.I.; Luna-Herrera, J.; Lopez-Santiago, R.; Rodriguez-Tovar, A.V.; Garcia-Perez, B.E. Candida glabrata survives and replicates in human osteoblasts. Pathog. Dis. 2016, 74, ftw030. [Google Scholar] [CrossRef] [PubMed]
- Bandara, H.M.; Yau, J.Y.; Watt, R.M.; Jin, L.J.; Samaranayake, L.P. Pseudomonas aeruginosa inhibits in-vitro Candida biofilm development. BMC Microbiol. 2010, 10, 125. [Google Scholar] [CrossRef] [PubMed]
- Baranska, M.; Kroll-Balcerzak, R.; Gil, L.; Rupa-Matysek, J.; Komarnicki, M. Successful treatment of pulmonary candidiasis and aspergillosis in patient with refractory Hodgkin lymphoma using micafungin —Case study and brief literature review. Cent. Eur. J. Immunol. 2017, 42, 111–115. [Google Scholar] [CrossRef] [PubMed]
- Skedros, J.G.; Keenan, K.E.; Updike, W.S.; Oliver, M.R. Failed Reverse Total Shoulder Arthroplasty Caused by Recurrent Candida glabrata Infection with Prior Serratia marcescens Coinfection. Case Rep. Infect. Dis. 2014, 2014, 142428. [Google Scholar] [PubMed]
- Klotz, S.A.; Chasin, B.S.; Powell, B.; Gaur, N.K.; Lipke, P.N. Polymicrobial bloodstream infections involving Candida species: Analysis of patients and review of the literature. Diagn. Microbiol. Infect. Dis. 2007, 59, 401–406. [Google Scholar] [CrossRef] [PubMed]
- Klotz, S.A.; Gaur, N.K.; De Armond, R.; Sheppard, D.; Khardori, N.; Edwards, J.E., Jr.; Lipke, P.N.; El-Azizi, M. Candida albicans Als proteins mediate aggregation with bacteria and yeasts. Med. Mycol. 2007, 45, 363–370. [Google Scholar] [CrossRef] [PubMed]
- Silva, S.; Henriques, M.; Hayes, A.; Oliveira, R.; Azeredo, J.; Williams, D.W. Candida glabrata and Candida albicans co-infection of an in vitro oral epithelium. J. Oral Pathol. Med. 2011, 40, 421–427. [Google Scholar] [CrossRef] [PubMed]


| Interaction with | Receptor in the Host or Microorganism b | Ligand on C. glabrata d | Reference | |
|---|---|---|---|---|
| Type of Cells | Specific Cell Line a | |||
| Epithelial cells | HEp2 (human), CHO-Lec2 (hamster) | ND c | Epa1 | [11] |
| Cardiac endothelium (guinea pig) | ND | ND | [15] | |
| Caco-2 (human) | ND | Epa1 | [29] | |
| Lec2 (hamster) | ND | Epa6, Epa7 | [33] | |
| OKF6/TERT-2 (human) | ND | β-glucan | [76] | |
| UVECS (human) | ND | Pwp7, Aed1 | [14] | |
| OKF6/TERT-2 (human) | CDw17 | ND | [77] | |
| Immune cells | Natural Killers (NK) | Nkp46 (human) NCR1 (murine) | Epa1, Epa6, Epa7 | [18] |
| Macrophages (human/murine) | Dectin1, Dectin2 | β-glucan | [73,74,75] | |
| ND | Epa1 | [17] | ||
| Dendritic Cells (murine) | ND | Epa1/ND | [16,17] | |
| Neutrophils (murine) | Dectin2 | β-glucan | [73] | |
| Platelets (murine) | ND | ND | [78] | |
| Candida spp. | C. albicans (hyphae) | Hwp1, Als3, Als1 | Epa8, Epa19, Awp2, Awp7, CAGL0F00181 | [79] |
| C. krusei | ND | ND | [80,81] | |
| Others | Fibronectin | Epa6 | [82] | |
| Osteoblast (human) | ND | ND | [83] | |
| S. marscesens, P. aeruginosa, A. fumigatus | NI e | NI | [84,85,86] | |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
López-Fuentes, E.; Gutiérrez-Escobedo, G.; Timmermans, B.; Van Dijck, P.; De Las Peñas, A.; Castaño, I. Candida glabrata’s Genome Plasticity Confers a Unique Pattern of Expressed Cell Wall Proteins. J. Fungi 2018, 4, 67. https://doi.org/10.3390/jof4020067
López-Fuentes E, Gutiérrez-Escobedo G, Timmermans B, Van Dijck P, De Las Peñas A, Castaño I. Candida glabrata’s Genome Plasticity Confers a Unique Pattern of Expressed Cell Wall Proteins. Journal of Fungi. 2018; 4(2):67. https://doi.org/10.3390/jof4020067
Chicago/Turabian StyleLópez-Fuentes, Eunice, Guadalupe Gutiérrez-Escobedo, Bea Timmermans, Patrick Van Dijck, Alejandro De Las Peñas, and Irene Castaño. 2018. "Candida glabrata’s Genome Plasticity Confers a Unique Pattern of Expressed Cell Wall Proteins" Journal of Fungi 4, no. 2: 67. https://doi.org/10.3390/jof4020067
APA StyleLópez-Fuentes, E., Gutiérrez-Escobedo, G., Timmermans, B., Van Dijck, P., De Las Peñas, A., & Castaño, I. (2018). Candida glabrata’s Genome Plasticity Confers a Unique Pattern of Expressed Cell Wall Proteins. Journal of Fungi, 4(2), 67. https://doi.org/10.3390/jof4020067