The Role of Non-Catalytic Region in Determining the Difference in Efficiency Between Two Cellobiohydrolases Revealed Through a Genetic Approach
Abstract
1. Introduction
2. Materials and Methods
2.1. Bioinformatics Analysis
2.2. Construction of Strains
2.3. Cultivation
2.4. Enzyme Assays and SDS-PAGE
2.5. Cellulose Saccharification
2.6. Measurement of Intracellular Proteins
2.7. Statistical Analysis
3. Results and Discussion
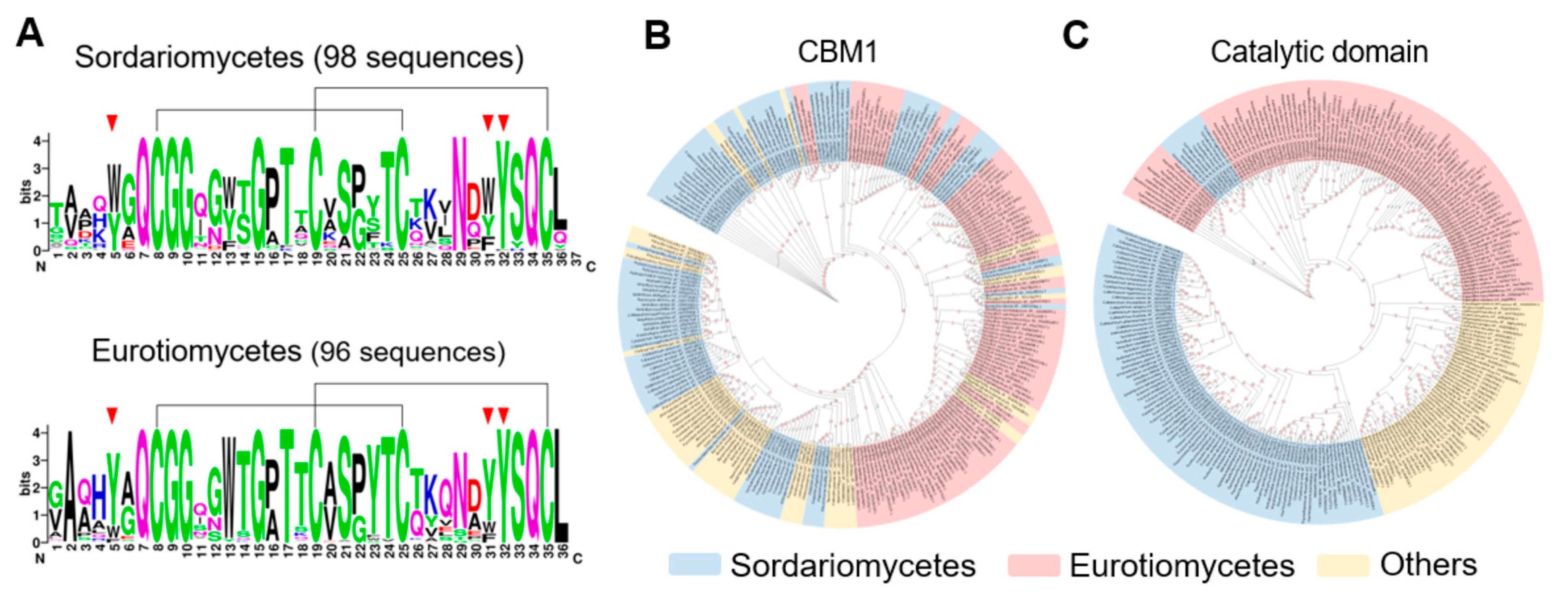
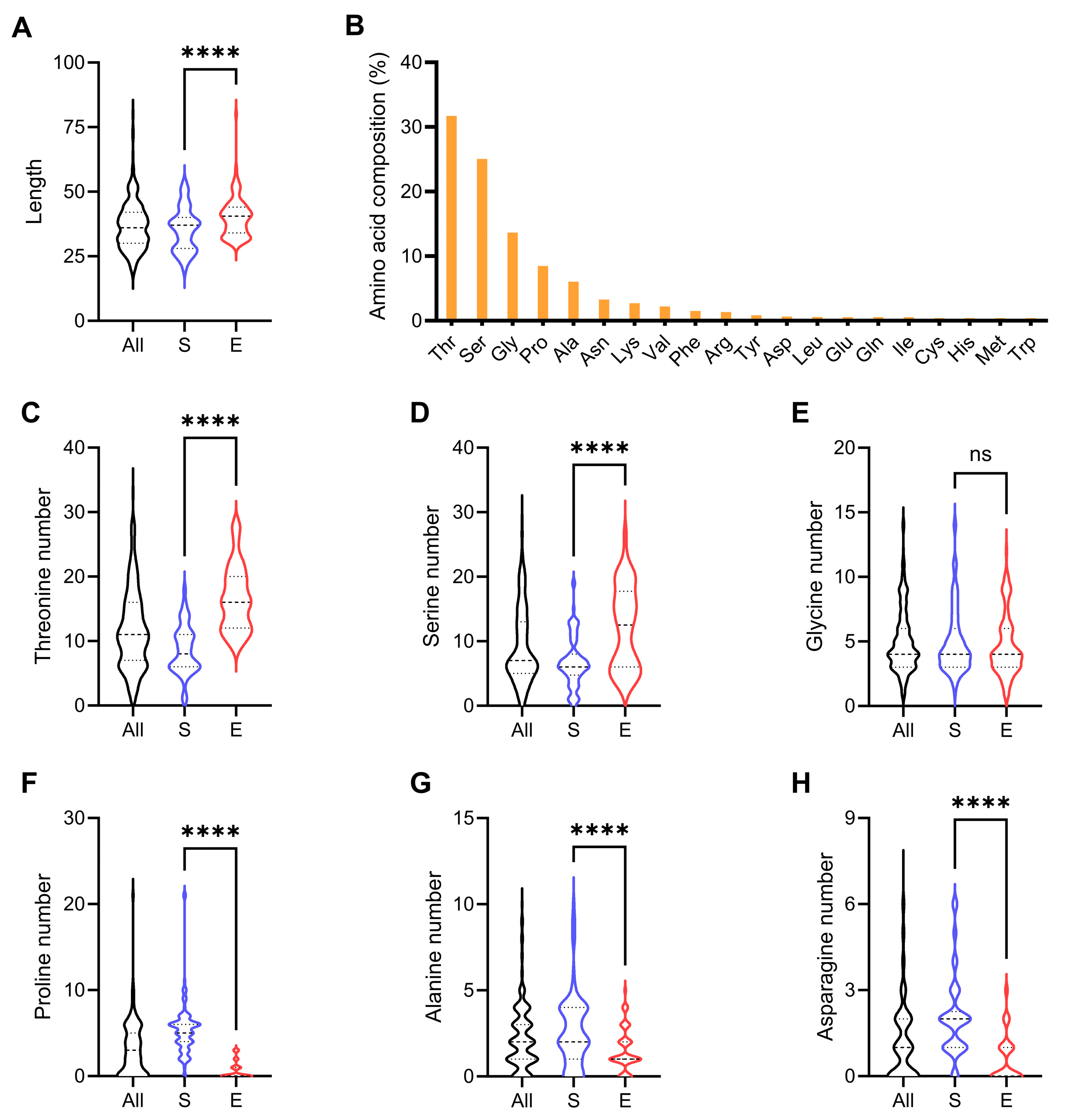
3.1. Sequence Characteristics of the Non-Catalytic Regions of GH7 CBHs
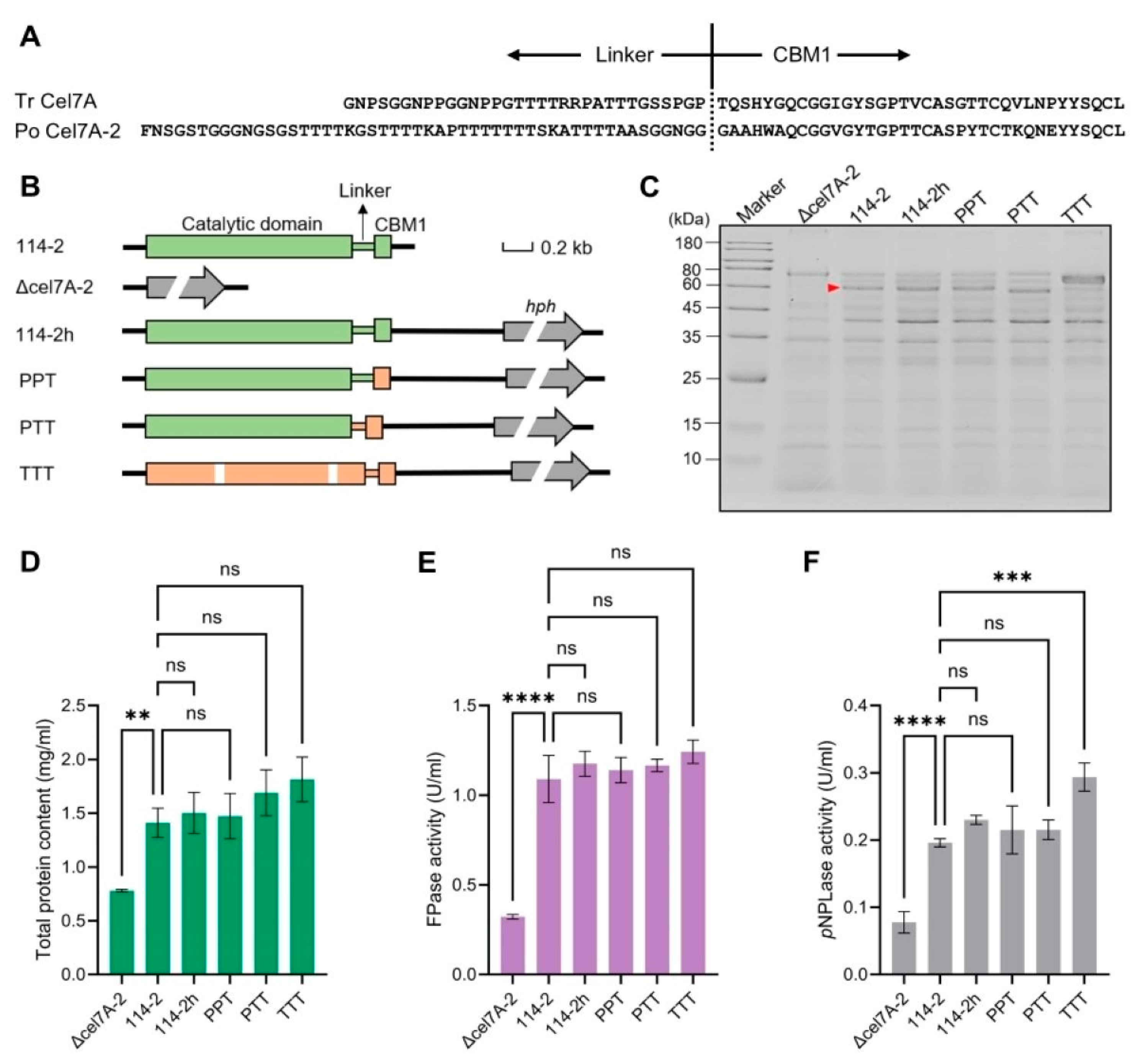
3.2. Effect of Cel7A-2 Engineering on Cellulase Production in P. oxalicum
3.3. Engineering CBM1 and Linker Region of Cel7A-2 Enhances the Degradation Efficiency of P. oxalicum Cellulase System
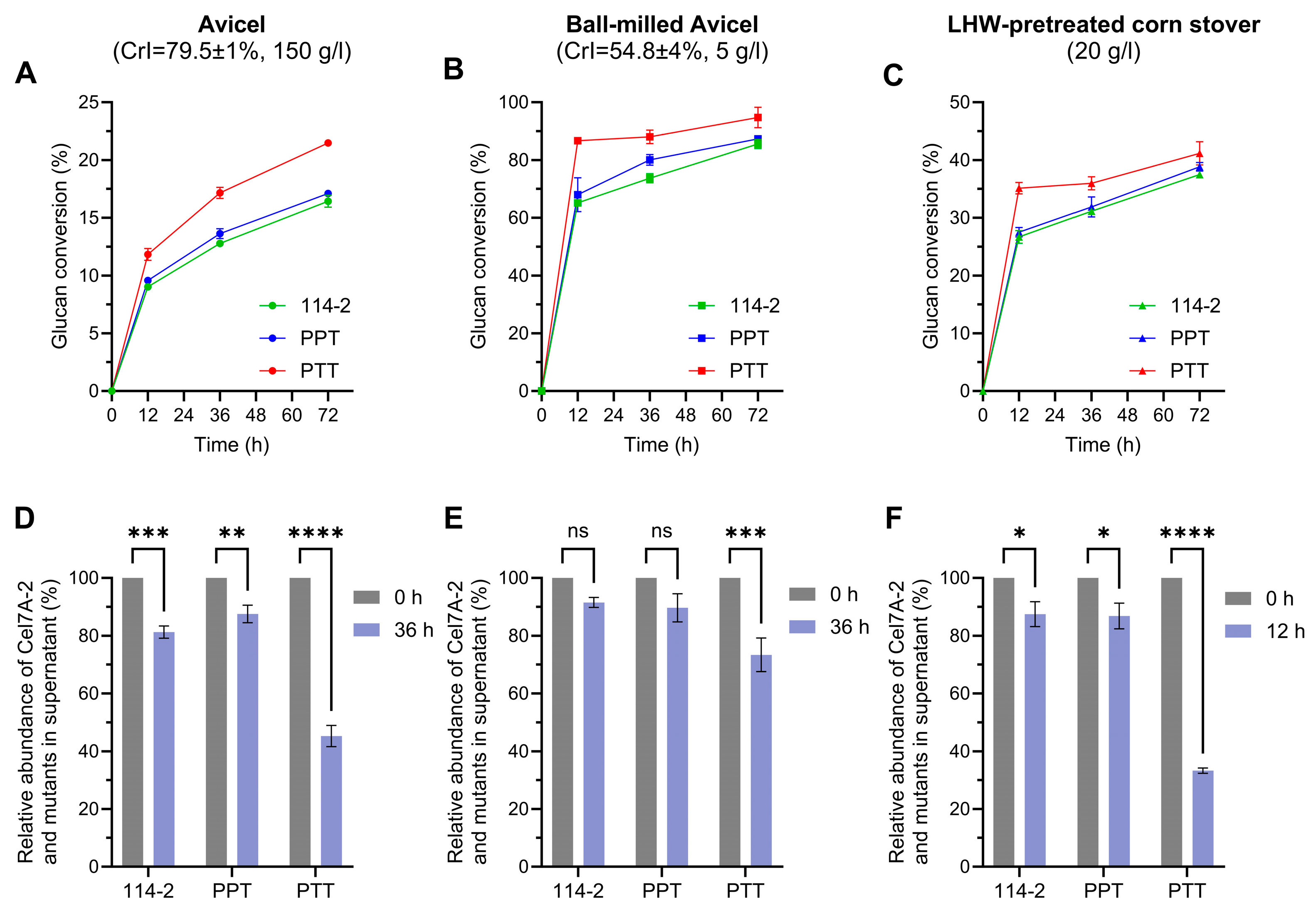
3.4. The Effect of Non-Catalytic Region Engineering of Cel7A-2 Is Dependent on the Concentration and Property of Substrate
3.5. Engineering of Non-Catalytic Region of Cel7A-2 Improves Cell Growth on Cellulose
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Cragg, S.M.; Beckham, G.T.; Bruce, N.C.; Bugg, T.D.H.; Distel, D.L.; Dupree, P.; Etxabe, A.G.; Goodell, B.S.; Jellison, J.; McGeehan, J.E.; et al. Lignocellulose degradation mechanisms across the tree of life. Curr. Opin. Chem. Biol. 2015, 29, 108–119. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.-C.; Dai, L.; Ma, L.; Guo, R.-T. Enzymatic degradation of plant biomass and synthetic polymers. Nat. Rev. Chem. 2020, 4, 114–126. [Google Scholar] [CrossRef] [PubMed]
- Chundawat, S.P.S.; Beckham, G.T.; Himmel, M.E.; Dale, B.E. Deconstruction of lignocellulosic biomass to fuels and chemicals. Annu. Rev. Chem. Biomol. Eng. 2011, 2, 121–145. [Google Scholar] [CrossRef] [PubMed]
- Su, C.; Cai, D.; Zhang, H.; Wu, Y.; Jiang, Y.; Liu, Y.; Zhang, C.; Li, C.; Qin, P.; Tan, T. Pilot-scale acetone-butanol-ethanol fermentation from corn stover. Green Carbon 2024, 2, 81–93. [Google Scholar] [CrossRef]
- Li, Z.; Waghmare, P.R.; Dijkhuizen, L.; Meng, X.; Liu, W. Research advances on the consolidated bioprocessing of lignocellulosic biomass. Eng. Microbiol. 2024, 4, 100139. [Google Scholar] [CrossRef] [PubMed]
- Teeri, T.T.; Koivula, A.; Linder, M.; Wohlfahrt, G.; Divne, C.; Jones, T.A. Trichoderma reesei cellobiohydrolases: Why so efficient on crystalline cellulose? Biochem. Soc. Trans. 1998, 26, 173–178. [Google Scholar] [CrossRef] [PubMed]
- Ren, M.; Wang, Y.; Liu, G.; Zuo, B.; Zhang, Y.; Wang, Y.; Liu, W.; Liu, X.; Zhong, Y. The effects of deletion of cellobiohydrolase genes on carbon source-dependent growth and enzymatic lignocellulose hydrolysis in Trichoderma reesei. J. Microbiol. 2020, 58, 687–695. [Google Scholar] [CrossRef] [PubMed]
- Lombard, V.; Golaconda Ramulu, H.; Drula, E.; Coutinho, P.M.; Henrissat, B. The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res. 2014, 42, D490–D495. [Google Scholar] [CrossRef] [PubMed]
- Igarashi, K.; Koivula, A.; Wada, M.; Kimura, S.; Penttilä, M.; Samejima, M. High speed atomic force microscopy visualizes processive movement of Trichoderma reesei cellobiohydrolase I on crystalline cellulose. J. Biol. Chem. 2009, 284, 36186–36190. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.-S.; Baker, J.O.; Zeng, Y.; Himmel, M.E.; Haas, T.; Ding, S.-Y. Cellobiohydrolase hydrolyzes crystalline cellulose on hydrophobic faces. J. Biol. Chem. 2011, 286, 11195–11201. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Liu, G. Harnessing cellulose-binding protein domains for the development of functionalized cellulose materials. Bioresour. Bioprocess. 2024, 11, 74. [Google Scholar] [CrossRef] [PubMed]
- Mello, B.; Polikarpov, I. Family 1 carbohydrate binding-modules enhance saccharification rates. AMB Express 2014, 4, 36. [Google Scholar] [CrossRef] [PubMed]
- Sammond, D.W.; Payne, C.M.; Brunecky, R.; Himmel, M.E.; Crowley, M.F.; Beckham, G.T. Cellulase linkers are optimized based on domain type and function: Insights from sequence analysis, biophysical measurements, and molecular simulation. PLoS ONE 2012, 7, e48615. [Google Scholar] [CrossRef] [PubMed]
- Reinikainen, T.; Ruohonen, L.; Nevanen, T.; Laaksonen, L.; Teeri, T.T. Investigation of the function of mutated cellulose-binding domains of Trichoderma reesei cellobiohydrolase I. Proteins 1992, 14, 475–482. [Google Scholar] [CrossRef] [PubMed]
- Srisodsuk, M.; Reinikainen, T.; Penttil, M.; Teeri, T.T. Role of the interdomain linker peptide of Trichoderma reesei cellobiohydrolase i in its interaction with crystalline cellulose. J. Biol. Chem. 1993, 268, 20756–20761. [Google Scholar] [CrossRef] [PubMed]
- Badino, S.F.; Bathke, J.K.; Sørensen, T.H.; Windahl, M.S.; Jensen, K.; Peters, G.H.J.; Borch, K.; Westh, P. The influence of different linker modifications on the catalytic activity and cellulose affinity of cellobiohydrolase cel7a from Hypocrea jecorina. Protein Eng. Des. Sel. 2017, 30, 495–501. [Google Scholar] [CrossRef] [PubMed]
- Voutilainen, S.P.; Puranen, T.; Siika-aho, M.; Lappalainen, A.; Alapuranen, M.; Kallio, J.; Hooman, S.; Viikari, L.; Vehmaanperä, J.; Koivula, A. Cloning, expression, and characterization of novel thermostable family 7 cellobiohydrolases. Biotechnol. Bioeng. 2008, 101, 515–528. [Google Scholar] [CrossRef] [PubMed]
- Taylor, L.E.; Knott, B.C.; Baker, J.O.; Alahuhta, P.M.; Hobdey, S.E.; Linger, J.G.; Lunin, V.V.; Amore, A.; Subramanian, V.; Podkaminer, K.; et al. Engineering enhanced cellobiohydrolase activity. Nat. Commun. 2018, 9, 1186. [Google Scholar] [CrossRef] [PubMed]
- Brunecky, R.; Knott, B.C.; Subramanian, V.; Linger, J.G.; Beckham, G.T.; Amore, A.; Taylor, L.E.; Vander Wall, T.A.; Lunin, V.V.; Zheng, F.; et al. Engineering of glycoside hydrolase family 7 cellobiohydrolases directed by natural diversity screening. J. Biol. Chem. 2024, 300, 105749. [Google Scholar] [CrossRef] [PubMed]
- Xue, J.; Jiang, X.; Li, A.; Li, J.; Su, X.; Huang, J.; Qin, L. Catalytic efficiency improvement in cellobiohydrolase I by cross-species domain exchange engineering. Int. J. Mol. Sci. 2025, 26, 4024. [Google Scholar] [CrossRef]
- Du, J.; Zhang, X.; Li, X.; Zhao, J.; Liu, G.; Gao, B.; Qu, Y. The cellulose binding region in Trichoderma reesei cellobiohydrolase I has a higher capacity in improving crystalline cellulose degradation than that of Penicillium oxalicum. Bioresour. Technol. 2018, 266, 19–25. [Google Scholar] [CrossRef] [PubMed]
- Madeira, F.; Pearce, M.; Tivey, A.R.N.; Basutkar, P.; Lee, J.; Edbali, O.; Madhusoodanan, N.; Kolesnikov, A.; Lopez, R. Search and sequence analysis tools services from EMBL-EBI in 2022. Nucleic Acids Res. 2022, 50, W276–W279. [Google Scholar] [CrossRef] [PubMed]
- Gado, J.E.; Harrison, B.E.; Sandgren, M.; Ståhlberg, J.; Beckham, G.T.; Payne, C.M. Machine learning reveals sequence-function relationships in family 7 glycoside hydrolases. J. Biol. Chem. 2021, 297, 100931. [Google Scholar] [CrossRef] [PubMed]
- Crooks, G.E.; Hon, G.; Chandonia, J.M.; Brenner, S.E. Weblogo: A sequence logo generator. Genome Res. 2004, 14, 1188–1190. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, L.-T.; Schmidt, H.A.; von Haeseler, A.; Minh, B.Q. IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive tree of life (iTOL) v5: An online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 2021, 49, W293–W296. [Google Scholar] [CrossRef] [PubMed]
- Liu, G.; Qin, Y.; Li, Z.; Qu, Y. Improving lignocellulolytic enzyme production with Penicillium: From strain screening to systems biology. Biofuels 2014, 4, 523–534. [Google Scholar] [CrossRef]
- Gao, L.; Li, Z.; Xia, C.; Qu, Y.; Liu, M.; Yang, P.; Yu, L.; Song, X. Combining manipulation of transcription factors and overexpression of the target genes to enhance lignocellulolytic enzyme production in Penicillium oxalicum. Biotechnol. Biofuels 2017, 10, 100. [Google Scholar] [CrossRef] [PubMed]
- Vogel, H.J.A. A convenient growth medium for neurospora (medium V). Microb. Genet. Bull. 1956, 13, 42–43. [Google Scholar]
- Schneider, C.A.; Rasband, W.S.; Eliceiri, K.W. NIH image to ImageJ: 25 years of image analysis. Nat. Methods 2012, 9, 671–675. [Google Scholar] [CrossRef] [PubMed]
- Rueden, C.T.; Schindelin, J.; Hiner, M.C.; DeZonia, B.E.; Walter, A.E.; Arena, E.T.; Eliceiri, K.W. ImageJ2: ImageJ for the next generation of scientific image data. BMC Bioinform. 2017, 18, 529. [Google Scholar] [CrossRef] [PubMed]
- Salem, K.S.; Kasera, N.K.; Rahman, M.A.; Jameel, H.; Habibi, Y.; Eichhorn, S.J.; French, A.D.; Pal, L.; Lucia, L.A. Comparison and assessment of methods for cellulose crystallinity determination. Chem. Soc. Rev. 2023, 52, 6417–6446. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Zaro, J.L.; Shen, W.-C. Fusion protein linkers: Property, design and functionality. Adv. Drug Deliv. Rev. 2013, 65, 1357–1369. [Google Scholar] [CrossRef] [PubMed]
- Liu, G.; Zhang, L.; Wei, X.; Zou, G.; Qin, Y.; Ma, L.; Li, J.; Zheng, H.; Wang, S.; Wang, C.; et al. Genomic and secretomic analyses reveal unique features of the lignocellulolytic enzyme system of Penicillium decumbens. PLoS ONE 2013, 8, e55185. [Google Scholar] [CrossRef] [PubMed]
- Payne, C.M.; Resch, M.G.; Chen, L.; Crowley, M.F.; Himmel, M.E.; Taylor, L.E.; Sandgren, M.; Ståhlberg, J.; Stals, I.; Tan, Z.; et al. Glycosylated linkers in multimodular lignocellulose-degrading enzymes dynamically bind to cellulose. Proc. Natl. Acad. Sci. USA 2013, 110, 14646–14651. [Google Scholar] [CrossRef] [PubMed]
- Chundawat, S.P.S.; Bellesia, G.; Uppugundla, N.; da Costa Sousa, L.; Gao, D.; Cheh, A.M.; Agarwal, U.P.; Bianchetti, C.M.; Phillips, G.N.; Langan, P.; et al. Restructuring the crystalline cellulose hydrogen bond network enhances its depolymerization rate. J. Am. Chem. Soc. 2011, 133, 11163–11174. [Google Scholar] [CrossRef] [PubMed]
- Shi, R.; Zhang, Z.; Zhang, J.; Chen, C.; Li, W.; Lin, Y.; Shi, X.; Zhao, P.; Zhang, T.; Yan, Q.; et al. A comparative study on enhanced enzymatic hydrolysis of diverse herbaceous and woody wastes by promising dilute acid and alkaline pretreatments. Bioresour. Bioprocess. 2025, 12, 36. [Google Scholar] [CrossRef] [PubMed]
- Le Costaouëc, T.; Pakarinen, A.; Várnai, A.; Puranen, T.; Viikari, L. The role of carbohydrate binding module (CBM) at high substrate consistency: Comparison of Trichoderma reesei and Thermoascus aurantiacus Cel7A (CBHI) and Cel5A (EGII). Bioresour. Technol. 2013, 143, 196–203. [Google Scholar] [CrossRef] [PubMed]
- Pakarinen, A.; Haven, M.; Djajadi, D.; Várnai, A.; Puranen, T.; Viikari, L. Cellulases without carbohydrate-binding modules in high consistency ethanol production process. Biotechnol. Biofuels 2014, 7, 27. [Google Scholar] [CrossRef] [PubMed]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yan, X.; Waghmare, P.R.; Meng, X.; Zhang, J.; Ding, S.; Lei, Y.; Yue, J.; Liu, G. The Role of Non-Catalytic Region in Determining the Difference in Efficiency Between Two Cellobiohydrolases Revealed Through a Genetic Approach. J. Fungi 2025, 11, 536. https://doi.org/10.3390/jof11070536
Yan X, Waghmare PR, Meng X, Zhang J, Ding S, Lei Y, Yue J, Liu G. The Role of Non-Catalytic Region in Determining the Difference in Efficiency Between Two Cellobiohydrolases Revealed Through a Genetic Approach. Journal of Fungi. 2025; 11(7):536. https://doi.org/10.3390/jof11070536
Chicago/Turabian StyleYan, Xinyuan, Pankajkumar Ramdas Waghmare, Xiaoli Meng, Jianhui Zhang, Shaoming Ding, Yu Lei, Jun Yue, and Guodong Liu. 2025. "The Role of Non-Catalytic Region in Determining the Difference in Efficiency Between Two Cellobiohydrolases Revealed Through a Genetic Approach" Journal of Fungi 11, no. 7: 536. https://doi.org/10.3390/jof11070536
APA StyleYan, X., Waghmare, P. R., Meng, X., Zhang, J., Ding, S., Lei, Y., Yue, J., & Liu, G. (2025). The Role of Non-Catalytic Region in Determining the Difference in Efficiency Between Two Cellobiohydrolases Revealed Through a Genetic Approach. Journal of Fungi, 11(7), 536. https://doi.org/10.3390/jof11070536





