Genomic and Functional Analysis of a Novel Yeast Cyberlindnera fabianii TBRC 4498 for High-Yield Xylitol Production
Abstract
1. Introduction
2. Materials and Methods
2.1. Strain, Chemicals, and Culture Media
2.2. Genome Sequence Analysis
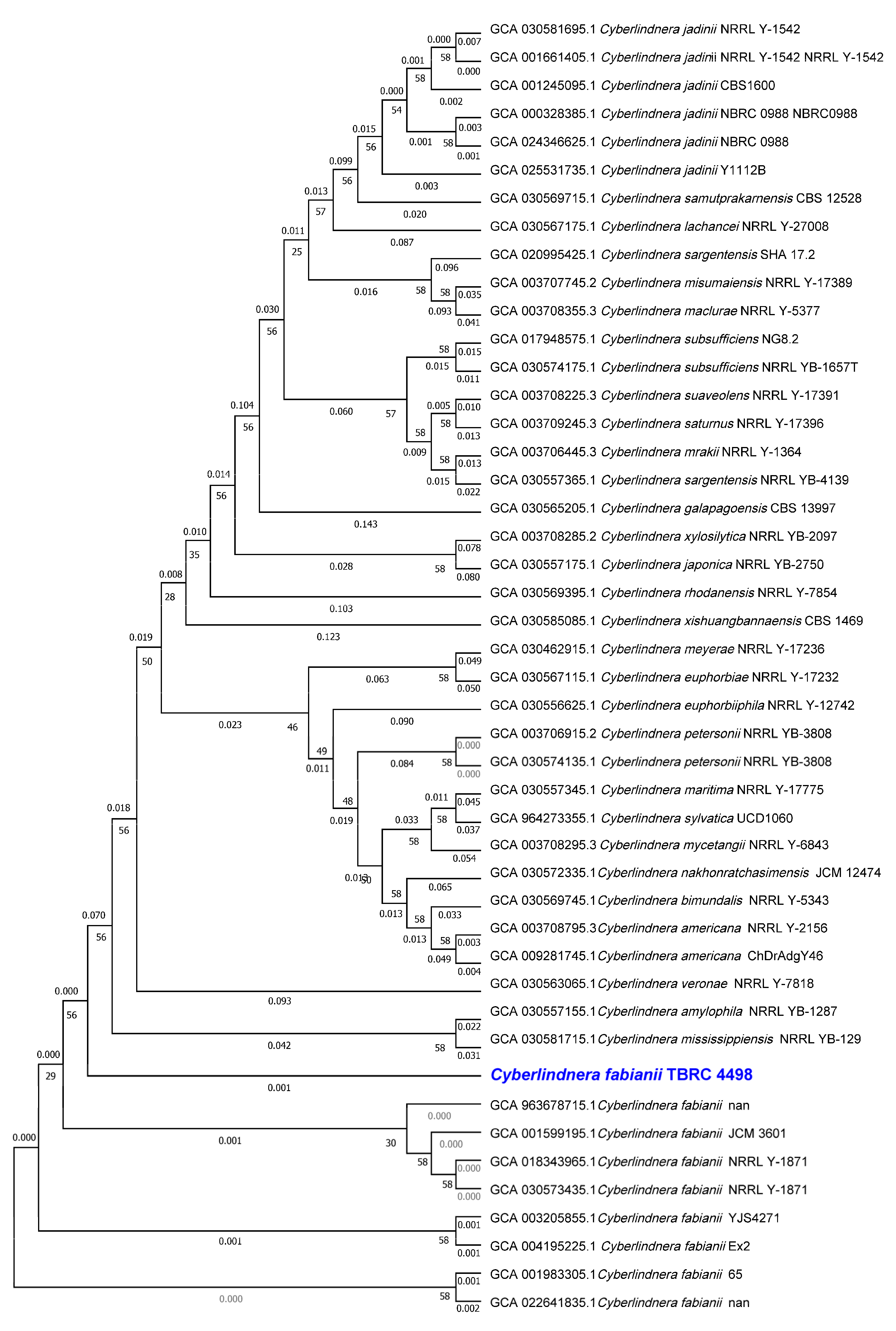
2.3. Taxonomic Classification
2.4. Xylitol Production
2.5. Product Profile Analysis Using HPLC
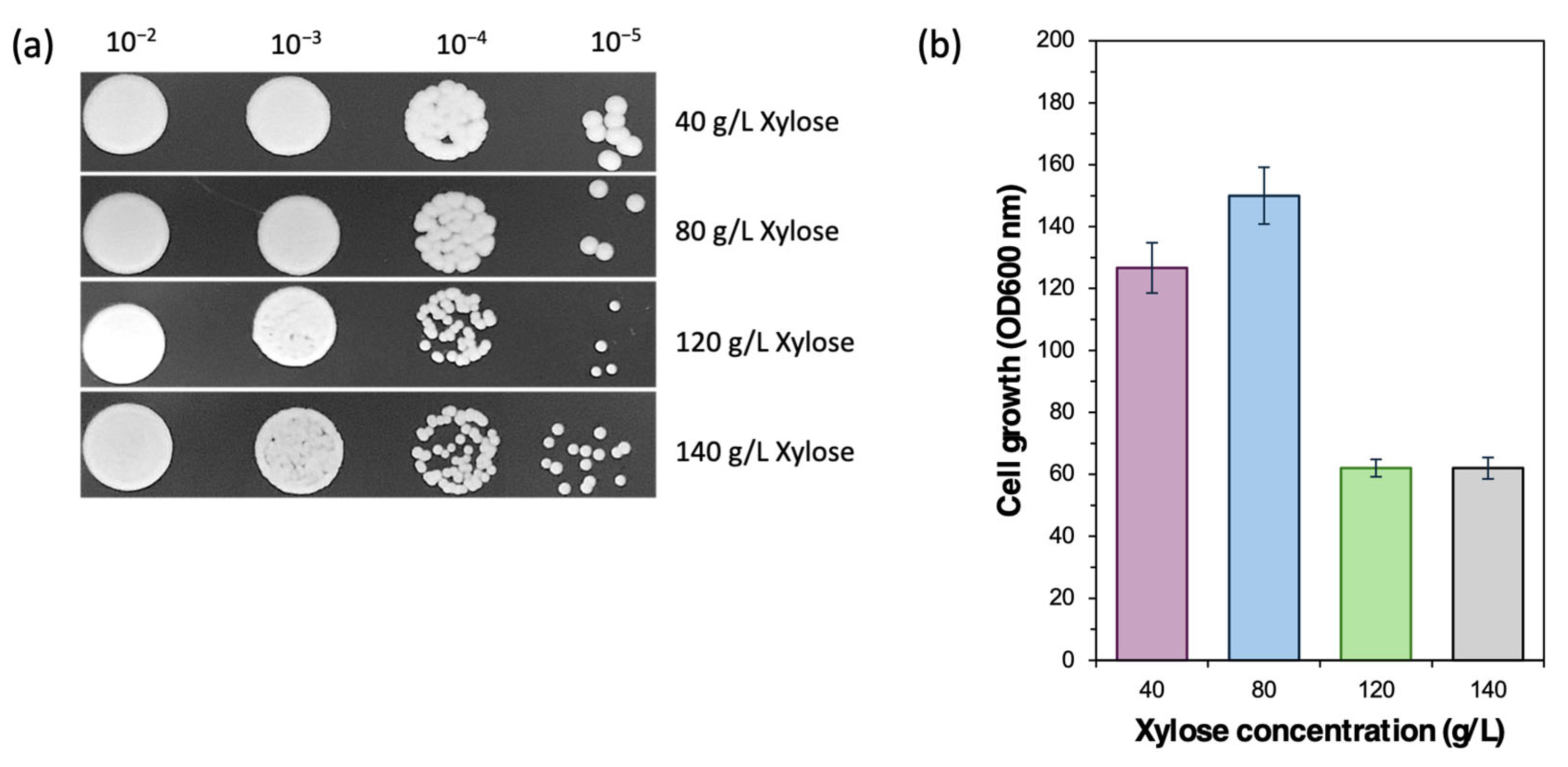
2.6. Cell Growth Comparison in Different Xylose Concentrations
3. Results and Discussion
3.1. Sugar-Utilizing Profile of Cy. fabianii TBRC 4498
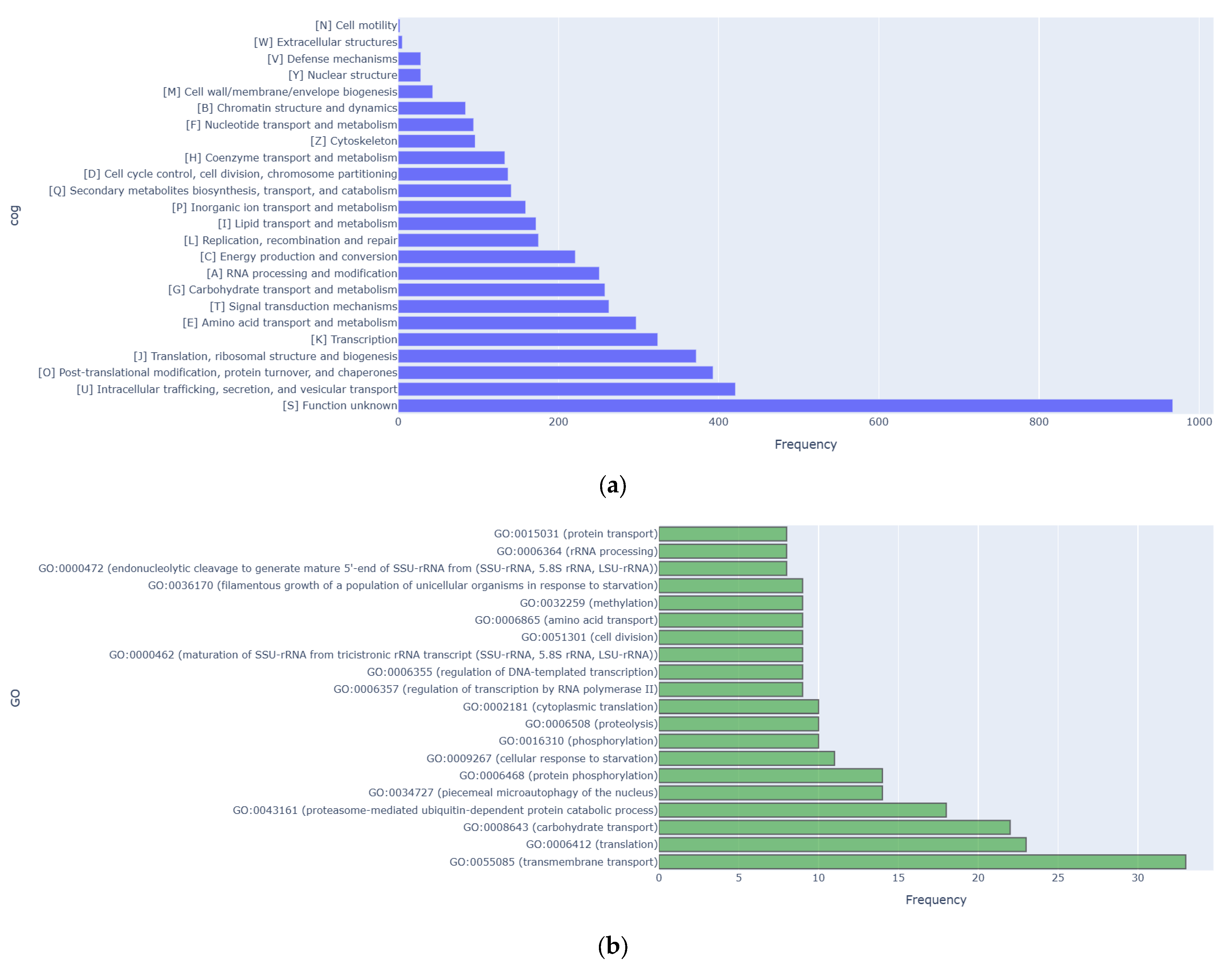
3.2. Genome Sequence and Functional Annotation
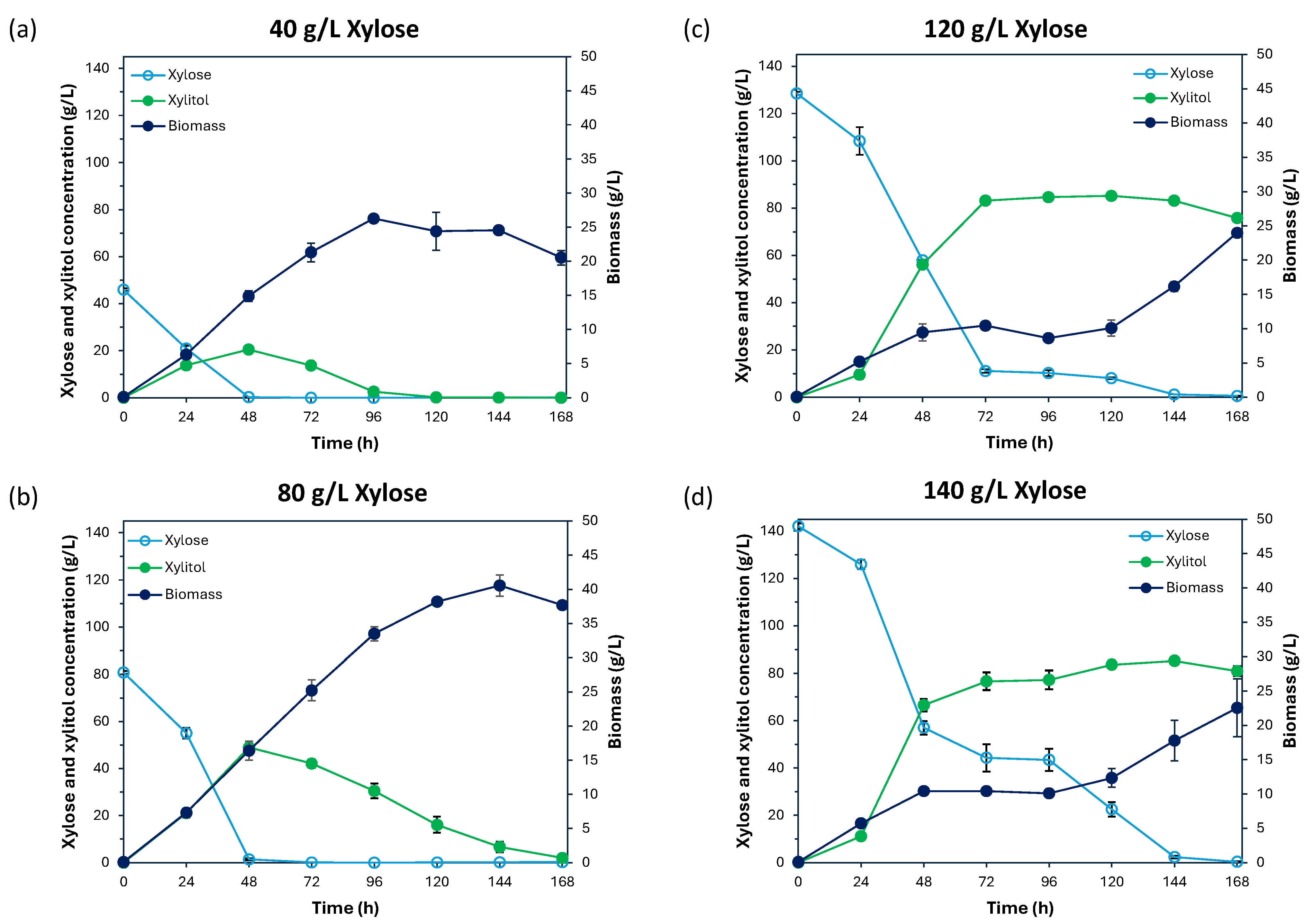
3.3. Xylitol Production Capability of Cy. fabianii TBRC 4498
| Yeast Strain | Xylose (g/L) | Xylitol (g/L) | Yield (g/g) (Based on Starting Xylose) | Yield (g/g) (Based on Consumed Xylose) | Productivity (g/L/h) | Conversion (%) (Based on Starting Xylose) | Time (h) | Risk Group | Reference |
|---|---|---|---|---|---|---|---|---|---|
| Cyberlindnera fabianii TBRC 4498 | 128.54 | 83.19 | 0.65 | 0.71 | 1.16 | 64.72 | 72 | 1 | This work |
| Cyberlindnera (Williopsis) saturnus | 150 | 38.63 | 0.26 | 0.54 | 0.27 | 25.75 | 144 | 1 | [50] |
| Cyberlindnera galapagoensis sp. nov. CLQCA-24SC-025 | 50 | 17.01 | 0.34 | 0.50 | 0.23 | 34.02 | 72 | n.r. | [7] |
| Cyberlindnera xylosilytica UFMG-CM-Y309 | 60.65 | 46.87 | 0.77 | 0.87 | 0.65 | 77.27 | 72 | n.r. | [9] |
| Meyerozyma guilliermondii B12 | 40 | 5.00 | 0.13 | 0.33 | 0.11 | 12.50 | 44 | 2 | [8] |
| Meyerozyma guilliermondii IM/UFRJ | 109 | 40.8 | 0.37 | 0.71 | 0.85 | 37.43 | 48 | 2 | [51] |
| Wickerhamomyces anomalus 740 | 40 | 24.75 | 0.62 | 0.75 | 0.56 | 61.88 | 44 | 1 | [8] |
| Wickerhamomyces rabaulensis UFMG-CM-Y3716 | 60.65 | 39.66 | 0.65 | 0.69 | 0.55 | 65.39 | 72 | n.r. | [9] |
| Candida tropicalis CLQCA-24F-125 | 50 | 25.63 | 0.51 | 0.67 | 0.36 | 51.26 | 72 | 2 | [7] |
| Candida xylopsoci SL6 | 50 | 5.00 | 0.10 | n.r. | 0.07 | 10.00 | 72 | n.r. | [10] |
| Candida tropicalis isolate Balki1 | 20 | 4.25 | 0.21 | n.r. | 0.06 | 21.25 | 72 | 2 | [52] |
| Candida guilliermondii FTI20037 | 57.7 | 38.4 | 0.67 | 0.73 | 1.92 | 66.55 | 20 | 2 | [46] |
| Candida tropicalis As 2.1776 | 80 | 58.3 | 0.72 | 0.74 | 0.61 | 72.87 | 96 | 2 | [53] |
| Candida tropicalis JH030 | 45.8 | 31.1 | 0.68 | 0.71 | 0.44 | 67.90 | 70 | 2 | [45] |
| Pichia kudriavzevii R5 | 50 | 5.50 | 0.11 | n.r. | 0.08 | 11.00 | 72 | 2 | [10] |
| Pichia stiptis CBS 5773 | 52 | 31.68 | 0.61 | 0.96 | 0.44 | 60.92 | 72 | 1 | [54] |
| Kluyveromyces marxianus CCA510 | 36.44 | 12.27 | 0.35 | 0.38 | 0.106 | 34.93 | 120 | 1 | [55] |
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Chen, X.; Jiang, Z.H.; Chen, S.; Qin, W. Microbial and bioconversion production of D-xylitol and its detection and application. Int. J. Biol. Sci. 2010, 6, 834–844. [Google Scholar] [CrossRef] [PubMed]
- Tiefenbacher, K.F. Technology of Main Ingredients—Sweeteners and Lipids. Wafer Waffle 2017, 123–225. [Google Scholar] [CrossRef]
- Ur-Rehman, S.; Mushtaq, Z.; Zahoor, T.; Jamil, A.; Murtaza, M.A. Xylitol: A review on bioproduction, application, health benefits, and related safety issues. Crit. Rev. Food Sci. Nutr. 2015, 55, 1514–1528. [Google Scholar] [CrossRef]
- Mussatto, S.I. Application of xylitol in food formulations and benefits for health. In D-xylitol: Fermentative production, Application and Commercialization; da Silva, S.S., Chandel, A.K., Eds.; Springer: Berlin/Heidelberg, Germany, 2012; pp. 309–323. [Google Scholar]
- Pappu, S.M.J.; Gummadi, S.N. Effect of cosubstrate on xylitol production by Debaryomyces nepalensis NCYC 3413: A cybernetic modelling approach. Process Biochem. 2018, 69, 12–21. [Google Scholar] [CrossRef]
- Morais, W.G., Jr.; Pacheco, T.F.; Trichez, D.; Almeida, J.R.M.; Gonçalves, S.B. Xylitol production on sugarcane biomass hydrolysate by newly identified JA2 strain. Yeast 2019, 36, 349–361. [Google Scholar] [CrossRef]
- Guamán-Burneo, M.C.; Dussán, K.J.; Cadete, R.M.; Cheab, M.A.M.; Portero, P.; Carvajal-Barriga, E.J.; da Silva, S.S.; Rosa, C.A. Xylitol production by yeasts isolated from rotting wood in the Galápagos Islands, Ecuador, and description of Cyberlindnera galapagoensis f.a., sp. nov. Anton. Leeuw. Int. J. G. 2015, 108, 919–931. [Google Scholar] [CrossRef]
- Carneiro, C.V.G.C.; Silva, F.C.d.P.e.; Almeida, J.R.M. Xylitol production: Identification and comparison of new producing yeasts. Microorganisms 2019, 7, 484. [Google Scholar] [CrossRef]
- Palladino, F.; Rodrigues, R.C.L.B.; Cadete, R.M.; Barros, K.O.; Rosa, C.A. Novel potential yeast strains for the biotechnological production of xylitol from sugarcane bagasse. Biofuel Bioprod. Biorefining 2021, 15, 690–702. [Google Scholar] [CrossRef]
- Kusumawati, N.; Sumarlan, S.H.; Zubaidah, E.; Wardani, A.K. Isolation of xylose-utilizing yeasts from oil palm waste for xylitol and ethanol production. Bioresour. Bioprocess. 2023, 10, 71. [Google Scholar] [CrossRef]
- van Rijswijck, I.M.H.; Dijksterhuis, J.; Wolkers-Rooijackers, J.C.M.; Abee, T.; Smid, E.J. Nutrient limitation leads to penetrative growth into agar and affects aroma formation in Pichia fabianii, P. kudriavzevii and Saccharomyces cerevisiae. Yeast 2015, 32, 89–101. [Google Scholar]
- van Rijswijck, I.M.H.; Derks, M.F.L.; Abee, T.; de Ridder, D.; Smid, E.J. Genome sequences of Cyberlindnera fabianii 65, Pichia kudriavzevii 129, and Saccharomyces cerevisiae 131 isolated from fermented masau fruits in Zimbabwe. Genome Announc. 2017, 5, 10–1128. [Google Scholar] [CrossRef] [PubMed]
- Liang, J.; Yuan, H.; Fei, Y.; Wang, H.; Qu, C.; Bai, W.; Liu, G. Effects of Saccharomyces cerevisiae and Cyberlindnera fabianii inoculation on rice-flavor baijiu fermentation. Foods 2024, 13, 3175. [Google Scholar] [CrossRef] [PubMed]
- van Rijswijck, I.M.H.; van Mastrigt, O.; Pijffers, G.; Wolkers—Rooijackers, J.C.M.; Abee, T.; Zwietering, M.H.; Smid, E.J. Dynamic modelling of brewers’ yeast and Cyberlindnera fabianii co-culture behaviour for steering fermentation performance. Food Microbiol. 2019, 83, 113–121. [Google Scholar] [CrossRef] [PubMed]
- Danouche, M.; Ferioun, M.; Bahafid, W.; El Ghachtouli, N. Mycoremediation of azo dyes using Cyberlindnera fabianii yeast strain: Application of designs of experiments for decolorization optimization. Water Environ. Res. 2021, 93, 1402–1416. [Google Scholar] [CrossRef]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef]
- Prjibelski, A.; Antipov, D.; Meleshko, D.; Lapidus, A.; Korobeynikov, A. Using SPAdes De Novo Assembler. Curr. Protoc. Bioinform. 2020, 70, e102. [Google Scholar] [CrossRef]
- Alonge, M.; Lebeigle, L.; Kirsche, M.; Jenike, K.; Ou, S.; Aganezov, S.; Wang, X.; Lippman, Z.B.; Schatz, M.C.; Soyk, S. Automated assembly scaffolding using RagTag elevates a new tomato system for high-throughput genome editing. Genome Biol. 2022, 23, 258. [Google Scholar] [CrossRef]
- Gurevich, A.; Saveliev, V.; Vyahhi, N.; Tesler, G. QUAST: Quality assessment tool for genome assemblies. Bioinformatics 2013, 29, 1072–1075. [Google Scholar] [CrossRef]
- Manni, M.; Berkeley, M.R.; Seppey, M.; Zdobnov, E.M. BUSCO: Assessing genomic data quality and beyond. Curr. Protoc. 2021, 1, e323. [Google Scholar] [CrossRef]
- Stanke, M.; Morgenstern, B. AUGUSTUS: A web server for gene prediction in eukaryotes that allows user-defined constraints. Nucleic Acids Res. 2005, 33, W465–W467. [Google Scholar] [CrossRef]
- Haas, B.J.; Salzberg, S.L.; Zhu, W.; Pertea, M.; Allen, J.E.; Orvis, J.; White, O.; Buell, C.R.; Wortman, J.R. Automated eukaryotic gene structure annotation using EVidenceModeler and the program to assemble spliced alignments. Genome Biol. 2008, 9, R7. [Google Scholar] [CrossRef] [PubMed]
- Consortium, T.U. UniProt: The Universal Protein Knowledgebase in 2023. Nucleic Acids Res. 2022, 51, D523–D531. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Yohe, T.; Huang, L.; Entwistle, S.; Wu, P.; Yang, Z.; Busk, P.K.; Xu, Y.; Yin, Y. dbCAN2: A meta server for automated carbohydrate-active enzyme annotation. Nucleic Acids Res. 2018, 46, W95–W101. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Sato, Y.; Morishima, K. BlastKOALA and GhostKOALA: KEGG tools for functional characterization of genome and metagenome sequences. J. Mol. Biol. 2016, 428, 726–731. [Google Scholar] [CrossRef]
- Törönen, P.; Medlar, A.; Holm, L. PANNZER2: A rapid functional annotation web server. Nucleic Acids Res. 2018, 46, W84–W88. [Google Scholar] [CrossRef]
- Huerta-Cepas, J.; Szklarczyk, D.; Heller, D.; Hernández-Plaza, A.; Forslund, S.K.; Cook, H.; Mende, D.R.; Letunic, I.; Rattei, T.; Jensen, L.J.; et al. eggNOG 5.0: A hierarchical, functionally and phylogenetically annotated orthology resource based on 5090 organisms and 2502 viruses. Nucleic Acids Res. 2018, 47, D309–D314. [Google Scholar] [CrossRef]
- Marchler-Bauer, A.; Derbyshire, M.K.; Gonzales, N.R.; Lu, S.; Chitsaz, F.; Geer, L.Y.; Geer, R.C.; He, J.; Gwadz, M.; Hurwitz, D.I.; et al. CDD: NCBI’s conserved domain database. Nucleic Acids Res. 2014, 43, D222–D226. [Google Scholar] [CrossRef]
- Teufel, F.; Almagro Armenteros, J.J.; Johansen, A.R.; Gíslason, M.H.; Pihl, S.I.; Tsirigos, K.D.; Winther, O.; Brunak, S.; von Heijne, G.; Nielsen, H. SignalP 6.0 predicts all five types of signal peptides using protein language models. Nat. Biotechnol. 2022, 40, 1023–1025. [Google Scholar] [CrossRef]
- Bengtsson-Palme, J.; Ryberg, M.; Hartmann, M.; Branco, S.; Wang, Z.; Godhe, A.; De Wit, P.; Sánchez-García, M.; Ebersberger, I.; de Sousa, F.; et al. Improved software detection and extraction of ITS1 and ITS2 from ribosomal ITS sequences of fungi and other eukaryotes for analysis of environmental sequencing data. Methods Ecol. Evol. 2013, 4, 914–919. [Google Scholar] [CrossRef]
- Jain, C.; Rodriguez-R, L.M.; Phillippy, A.M.; Konstantinidis, K.T.; Aluru, S. High throughput ANI analysis of 90K prokaryotic genomes reveals clear species boundaries. Nat. Commun. 2018, 9, 5114. [Google Scholar] [CrossRef]
- Kim, D.; Gilchrist, C.L.M.; Chun, J.; Steinegger, M. UFCG: Database of universal fungal core genes and pipeline for genome-wide phylogenetic analysis of fungi. Nucleic Acids Res. 2022, 51, D777–D784. [Google Scholar] [CrossRef] [PubMed]
- Minter, D.W. Cyberlindnera, a replacement name for Lindnera Kurtzman et al., nom. illegit. Mycotaxon 2009, 110, 473–476. [Google Scholar] [CrossRef]
- Kurtzman, C.P.; Robnett, C.J.; Basehoar-Powers, E. Phylogenetic relationships among species of Pichia, Issatchenkia and Williopsis determined from multigene sequence analysis, and the proposal of Barnettozyma gen. nov., Lindnera gen. nov. and Wickerhamomyces gen. nov. FEMS Yeast Res. 2008, 8, 939–954. [Google Scholar] [CrossRef]
- Bellut, K.; Michel, M.; Zarnkow, M.; Hutzler, M.; Jacob, F.; Atzler, J.J.; Hoehnel, A.; Lynch, K.M.; Arendt, E.K. Screening and application of Cyberlindnera yeasts to produce a fruity, non-alcoholic beer. Fermentation 2019, 5, 103. [Google Scholar] [CrossRef]
- Zhai, X.-T.; Wang, H.-Q.; Han, L.; Wang, F.-Y.; Zhang, Y.-H.; Li, J.; He, C.-A.; Tan, B.; Wang, M. Impact of solid-state fermentation by Aspergillus niger 4Q and Cyberlindnera fabianii J2 on antioxidant and flavor properties of whole grain barley. Cereal Chem. 2023, 100, 914–926. [Google Scholar] [CrossRef]
- Sousa-Silva, M.; Vieira, D.; Soares, P.; Casal, M.; Soares-Silva, I. Expanding the knowledge on the skillful yeast Cyberlindnera jadinii. J. Fungi 2021, 7, 36. [Google Scholar] [CrossRef]
- Freel, K.C.; Sarilar, V.; Neuveglise, C.; Devillers, H.; Friedrich, A.; Schacherer, J. Genome sequence of the yeast Cyberlindnera fabianii (Hansenula fabianii). Genome Announc. 2014, 2, e00638-14. [Google Scholar] [CrossRef]
- Gárdonyi, M.; Jeppsson, M.; Lidén, G.; Gorwa-Grauslund, M.F.; Hahn-Hägerdal, B. Control of xylose consumption by xylose transport in recombinant Saccharomyces cerevisiae. Biotechnol. Bioeng. 2003, 82, 818–824. [Google Scholar] [CrossRef]
- Leandro, M.J.; Fonseca, C.; Gonçalves, P. Hexose and pentose transport in ascomycetous yeasts: An overview. FEMS Yeast Res. 2009, 9, 511–525. [Google Scholar] [CrossRef]
- Bueno, J.G.R.; Borelli, G.; Corrêa, T.L.R.; Fiamenghi, M.B.; José, J.; de Carvalho, M.; de Oliveira, L.C.; Pereira, G.A.G.; dos Santos, L.V. Novel xylose transporter Cs4130 expands the sugar uptake repertoire in recombinant Saccharomyces cerevisiae strains at high xylose concentrations. Biotechnol. Biofuels 2020, 13, 145. [Google Scholar] [CrossRef]
- Krallish, I.; Jeppsson, H.; Rapoport, A.; Hahn-Hägerdal, B. Effect of xylitol and trehalose on dry resistance of yeasts. Appl. Microbiol. Biotechnol. 1997, 47, 447–451. [Google Scholar] [CrossRef] [PubMed]
- He, Y.; Li, H.; Chen, L.; Zheng, L.; Ye, C.; Hou, J.; Bao, X.; Liu, W.; Shen, Y. Production of xylitol by Saccharomyces cerevisiae using waste xylose mother liquor and corncob residues. Microb. Biotechnol. 2021, 14, 2059–2071. [Google Scholar] [CrossRef] [PubMed]
- Umai, D.; Kayalvizhi, R.; Kumar, V.; Jacob, S. Xylitol: Bioproduction and applications—A review. Front. Sustain. 2022, 3, 826190. [Google Scholar] [CrossRef]
- Huang, C.-F.; Jiang, Y.-F.; Guo, G.-L.; Hwang, W.-S. Development of a yeast strain for xylitol production without hydrolysate detoxification as part of the integration of co-product generation within the lignocellulosic ethanol process. Bioresour. Technol. 2011, 102, 3322–3329. [Google Scholar] [CrossRef]
- Cortez, D.; Roberto, I. Optimization of D-xylose to xylitol biotransformation by Candida guilliermondii cells permeabilized with Triton X-100. Biocatal. Biotransform. 2013, 32, 34–38. [Google Scholar] [CrossRef]
- Nidetzky, B.; Brüggler, K.; Kratzer, R.; Mayr, P. Multiple forms of xylose reductase in Candida intermedia: Comparison of their functional properties using quantitative structure-activity relationships, steady-state kinetic analysis, and pH studies. J. Agric. Food Chem. 2003, 51, 7930–7935. [Google Scholar] [CrossRef]
- Zha, J.; Yuwen, M.; Qian, W.; Wu, X. Yeast-based biosynthesis of natural products from xylose. Front. Bioeng. Biotechnol. 2021, 9, 634919. [Google Scholar] [CrossRef]
- Jeffries, T.W.; Grigoriev, I.V.; Grimwood, J.; Laplaza, J.M.; Aerts, A.; Salamov, A.; Schmutz, J.; Lindquist, E.; Dehal, P.; Shapiro, H.; et al. Genome sequence of the lignocellulose-bioconverting and xylose-fermenting yeast Pichia stipitis. Nat. Biotechnol. 2007, 25, 319–326. [Google Scholar] [CrossRef]
- Kamat, S.; Gaikwad, S.; Ravi Kumar, A.; Gade, W.N. Xylitol production by Cyberlindnera (Williopsis) saturnus, a tropical mangrove yeast from xylose and corn cob hydrolysate. J. Appl. Microbiol. 2013, 115, 1357–1367. [Google Scholar] [CrossRef]
- Faria, L.F.F.; Gimenes, M.A.P.; Nobrega, R.; Pereira, N. Influence of oxygen availability on cell growth and xylitol production by Candida guilliermondii. Appl. Biochem. Biotechnol. 2002, 98, 449–458. [Google Scholar] [CrossRef]
- Rengaraju, B.; Ponnuswamy, R.; Malayandi, S.; Lakshminarayanan, V. Molecular identification of novel yeast isolate associated with xylitol production and its physicochemical parameters optimization. J. Pure Appl. Microbiol. 2014, 8, 3037–3042. [Google Scholar]
- Li, M.; Meng, X.; Diao, E.; Du, F. Xylitol production by Candida tropicalis from corn cob hemicellulose hydrolysate in a two-stage fed-batch fermentation process. J. Chem. Technol. Biotechnol. 2012, 87, 387–392. [Google Scholar] [CrossRef]
- Neeru, C.; Chandrajit, B.; Vidyasagar, J. Biological production of xylitol from corn husk and switchgrass by Pichia stiptis. Res. J. Chem. Sci. 2013, 2231, 606X. [Google Scholar]
- de Albuquerque, T.L.; Gomes, S.D.L.; Marques, J.E., Jr.; da Silva, I.J., Jr.; Rocha, M.V.P. Xylitol production from cashew apple bagasse by Kluyveromyces marxianus CCA510. Catal. Today 2015, 255, 33–40. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bonthong, P.; Bunterngsook, B.; Mhuantong, W.; Aiewviriyasakul, K.; Sritusnee, W.; Champreda, V.; Lekakarn, H. Genomic and Functional Analysis of a Novel Yeast Cyberlindnera fabianii TBRC 4498 for High-Yield Xylitol Production. J. Fungi 2025, 11, 453. https://doi.org/10.3390/jof11060453
Bonthong P, Bunterngsook B, Mhuantong W, Aiewviriyasakul K, Sritusnee W, Champreda V, Lekakarn H. Genomic and Functional Analysis of a Novel Yeast Cyberlindnera fabianii TBRC 4498 for High-Yield Xylitol Production. Journal of Fungi. 2025; 11(6):453. https://doi.org/10.3390/jof11060453
Chicago/Turabian StyleBonthong, Pawarin, Benjarat Bunterngsook, Wuttichai Mhuantong, Katesuda Aiewviriyasakul, Wipawee Sritusnee, Verawat Champreda, and Hataikarn Lekakarn. 2025. "Genomic and Functional Analysis of a Novel Yeast Cyberlindnera fabianii TBRC 4498 for High-Yield Xylitol Production" Journal of Fungi 11, no. 6: 453. https://doi.org/10.3390/jof11060453
APA StyleBonthong, P., Bunterngsook, B., Mhuantong, W., Aiewviriyasakul, K., Sritusnee, W., Champreda, V., & Lekakarn, H. (2025). Genomic and Functional Analysis of a Novel Yeast Cyberlindnera fabianii TBRC 4498 for High-Yield Xylitol Production. Journal of Fungi, 11(6), 453. https://doi.org/10.3390/jof11060453










