Characterization of Homeodomain Proteins at the Aβ Sublocus in Schizophyllum commune and Their Role in Sexual Compatibility and Development
Abstract
1. Introduction
2. Materials and Methods
2.1. Strains, Plasmids, and Culture Conditions
2.2. De Novo Genome Sequencing and Assembly
2.3. Phylogenetic Analysis and Annotation of Mating-Type Genes
2.4. Protein–Protein Interaction (PPI) Prediction Analysis
2.5. Polymerase Chain Reaction (PCR) Amplification of Mating-Type Genes
2.6. Construction of BiFC Plasmids
2.7. Construction of Protein Expression Plasmids
2.8. Protoplast Preparation and Transformation of S. commune
2.9. Mating Experiments and In Vivo Functional Validation
2.10. Confocal Microscopy and Quantitative Image Analysis
2.11. His Pull-Down Assays and Western Blot Analyses
2.12. Statistical Analysis
3. Results
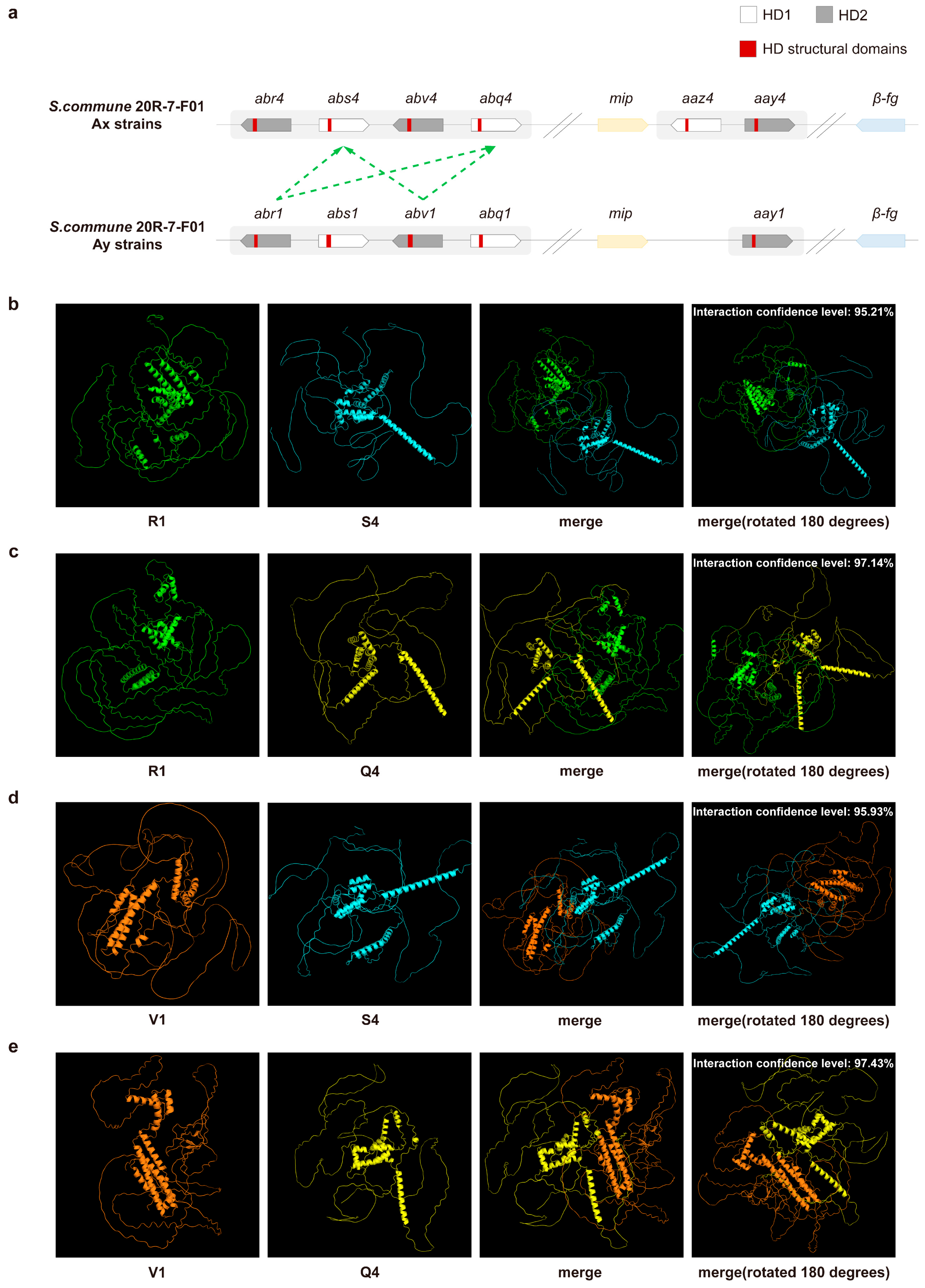
3.1. Identification of the A Mating-Type Locus in S. commune 20R-7-F01 Strains
3.2. PPI Prediction of Aβ Mating-Type HD Proteins
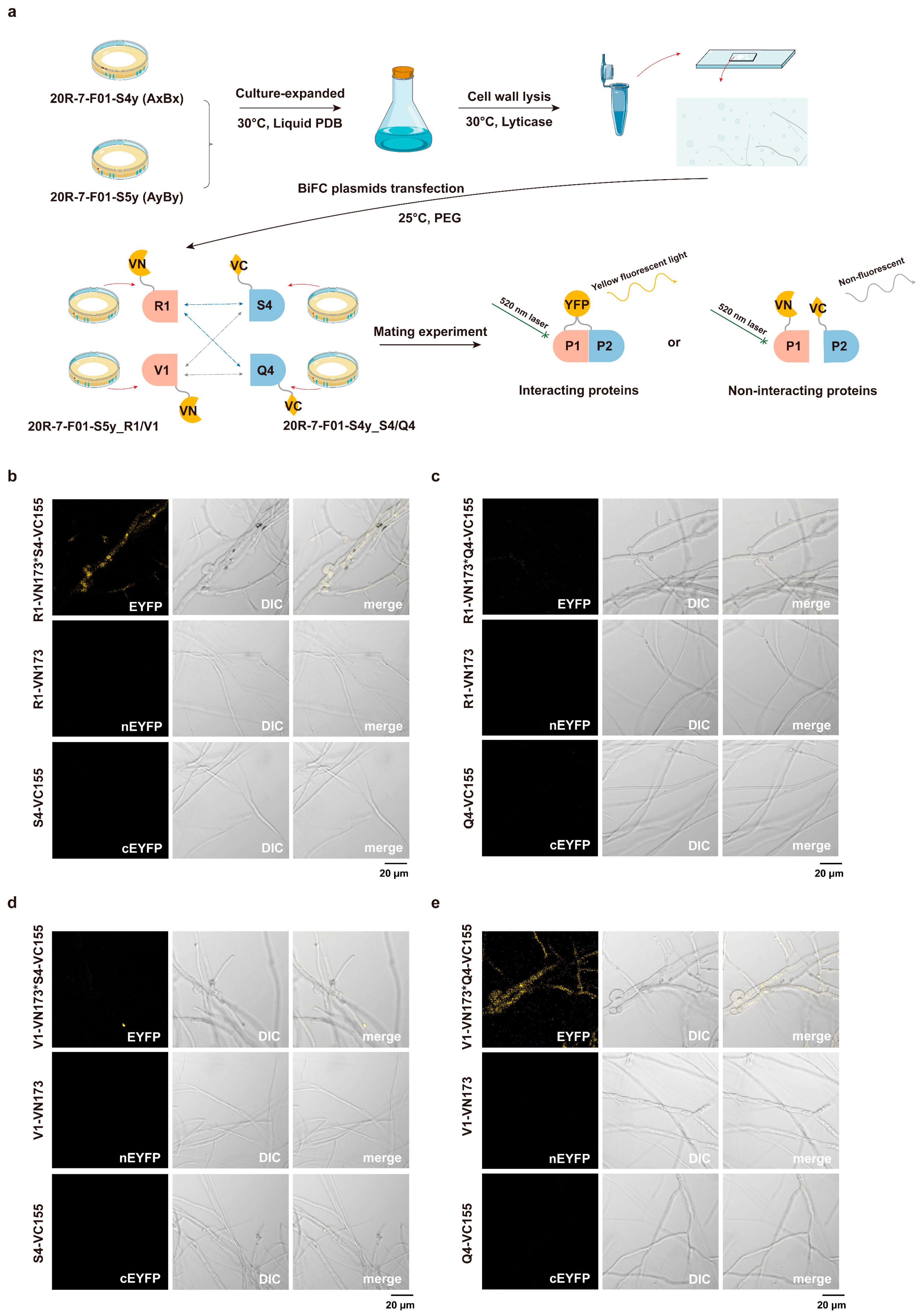
3.3. BiFC Interaction of Aβ Mating-Type HD Proteins
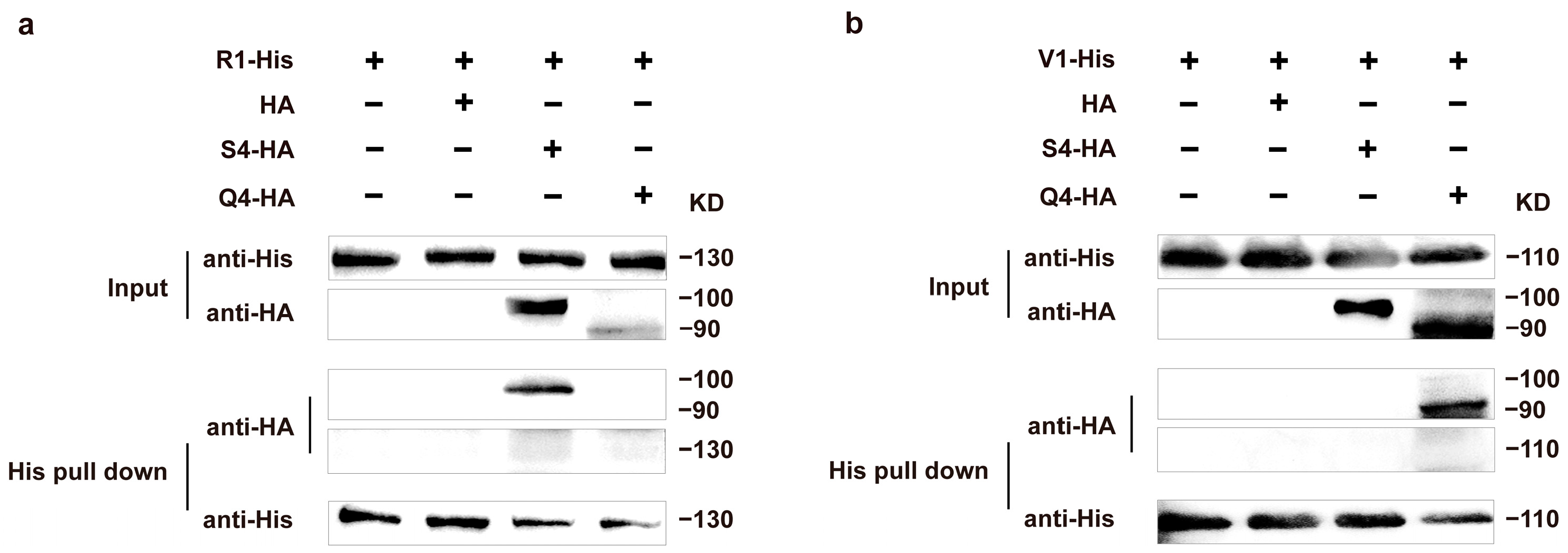
3.4. His Pull-Down Assays and Western Blot Analysis of Aβ Mating-Type HD Proteins
3.5. Validation of In Vivo Interactions for Fertility
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| HD | Homeodomain |
| Mip | Mitochondrial intermediate peptidase |
| β-fg | Beta-flanking |
| BiFC | Bimolecular Fluorescence Complementation |
| IODP | International Ocean Discovery Program |
| MM | Minimal medium |
| LB | Luria–Bertani |
| IPTG | Isopropyl β-D-1-thiogalactopyranoside |
| NGS | Next-generation sequencing |
| Nr | Non-redundant protein database |
| KOG | Eukaryotic Clusters of Orthologous Groups |
| GO | Gene Ontology |
| KEGG | Kyoto Encyclopedia of Genes and Genomes |
| TIGRFAM | TIGR defined protein families |
| PFAM | Protein families |
| PPI(s) | Protein–protein interaction(s) |
| PCR | Polymerase Chain Reaction |
| MCS | Multiple cloning sites |
| PEG | Polyethylene glycol |
| SDS-PAGE | Sodium dodecyl sulphate–polyacrylamide gel electrophoresis |
| PVDF | Polyvinylidene fluoride |
| TBST | Tris-buffered saline containing 0.1% Tween-20 |
| HRP | Horseradish peroxidase |
| ANOVA | Analysis of variance |
| LSD | Least significant difference |
References
- Hyde, K.; Noorabadi, M.; Thiyagaraja, V.; Mq, H.; Johnston, P.; Wijesinghe, S.; Armand, A.; Biketova, A.; Kwt, C.; Erdoğdu, M. The 2024 Outline of Fungi and fungus-like taxa. Mycosphere 2024, 15, 5146–6239. [Google Scholar] [CrossRef]
- Ohm, R.A.; De Jong, J.F.; Lugones, L.G.; Aerts, A.; Kothe, E.; Stajich, J.E.; De Vries, R.P.; Record, E.; Levasseur, A.; Baker, S.E. Genome sequence of the model mushroom Schizophyllum commune. Nat. Biotechnol. 2010, 28, 957–963. [Google Scholar] [CrossRef] [PubMed]
- Khardziani, T.; Metreveli, E.; Didebulidze, K.; Elisashvili, V.I. Screening of Georgian medicinal mushrooms for their antibacterial activity and optimization of cultivation conditions for the split gill medicinal mushroom, Schizophyllum commune BCC64 (Agaricomycetes). Int. J. Med. Mushrooms 2020, 22, 659–669. [Google Scholar] [CrossRef]
- Hamzah, H.N.; Mohammad, A. A study of wild mushrooms utilised for food and medicine by the orang asli communities of Sungai Berua, Terengganu. In Resource Use and Sustainability of Orang Asli: Indigenous Communities in Peninsular Malaysia, 1st ed.; Abdullah, M.T., Bartholomew, C.V., Mohammad, A., Eds.; CRC Press: Boca Raton, FL, USA; Springer: Cham, Switzerland, 2021; Volume 1, pp. 75–84. [Google Scholar]
- Chen, T.; Liu, Y.; Ma, B.; Sun, B.; Pan, Y.; Ou, Y.; Yu, H.; She, Z.; Long, Y. Anti-Inflammatory Sesquiterpenes from Fruiting Bodies of Schizophyllum commune. J. Agric. Food Chem. 2024, 72, 5416–5427. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Chu, C.; Zhao, M.; Hou, S.; Ji, R.; Liu, C. Nitric Oxide-Mediated Regulation of Chitinase Activity and Cadmium Sequestration in the Response of Schizophyllum commune to Cadmium Stress. Microorganisms 2025, 13, 470. [Google Scholar] [CrossRef] [PubMed]
- Mišković, J.; Karaman, M.; Rašeta, M.; Krsmanović, N.; Berežni, S.; Jakovljević, D.; Piattoni, F.; Zambonelli, A.; Gargano, M.L.; Venturella, G. Comparison of two Schizophyllum commune strains in production of acetylcholinesterase inhibitors and antioxidants from submerged cultivation. J. Fungi 2021, 7, 115. [Google Scholar] [CrossRef]
- Bora, P.K.; Kemprai, P.; Hussain, S.; Baishya, R.; Jadhav, D.; Haldar, S. Mushroom-based biocatalysts for the synthesis of aroma and flavours from exogenous organic molecules: A review of two decades. Int. J. Food Sci. Technol. 2024, 59, 84–94. [Google Scholar] [CrossRef]
- Takemoto, S.; Nakamura, H.; IMAMURA, Y.; SHIMANE, T. Schizophyllum commune as a ubiquitous plant parasite. Jpn. Agric. Res. Q. JARQ 2010, 44, 357–364. [Google Scholar] [CrossRef]
- Ueyama, M.; Mizuno, K.; Hirose, D.; Kamei, K.; Ohta, K. A case of allergic fungal rhinosinusitis caused by Schizophyllum commune identified in both patient’s nasal sputum and veranda’s soil samples. J. Infect. Chemother. 2021, 27, 759–765. [Google Scholar] [CrossRef]
- Raper, J.R.; San Antonio, J.P. Heterokaryotic mutagenesis in Hymenomycetes. I. Heterokaryosis in Schizophyllum commune. Am. J. Bot. 1954, 41, 69–86. [Google Scholar] [CrossRef]
- Kothe, E. Mating types and pheromone recognition in the homobasidiomycete Schizophyllum commune. Fungal Genet. Biol. 1999, 27, 146–152. [Google Scholar] [CrossRef] [PubMed]
- Raper, C.A.; Raper, J.R. Mutations affecting heterokaryosis in Schizophyllum commune. Am. J. Bot. 1964, 51, 503–512. [Google Scholar] [CrossRef]
- Fowler, T.J.; Mitton, M.F.; Rees, E.I.; Raper, C.A. Crossing the boundary between the Bα and Bβ mating-type loci in Schizophyllum commune. Fungal Genet. Biol. 2004, 41, 89–101. [Google Scholar] [CrossRef] [PubMed]
- Raper, J.R.; Baxter, M.G.; Ellingboe, A.H. The genetic structure of the incompatibility factors of Schizophyllum commune: The A-factor. Proc. Natl. Acad. Sci. USA 1960, 46, 833–842. [Google Scholar] [CrossRef]
- Specht, C.A.; Stankis, M.M.; Giasson, L.; Novotny, C.P.; Ullrich, R.C. Functional analysis of the homeodomain-related proteins of the A alpha locus of Schizophyllum commune. Proc. Natl. Acad. Sci. USA 1992, 89, 7174–7178. [Google Scholar] [CrossRef]
- Kües, U.; Casselton, L.A. Homeodomains and regulation of sexual development in basidiomycetes. Trends Genet. 1992, 8, 154–155. [Google Scholar] [CrossRef]
- Stankis, M.M.; Specht, C.A.; Yang, H.; Giasson, L.; Ullrich, R.C.; Novotny, C.P. The A alpha mating locus of Schizophyllum commune encodes two dissimilar multiallelic homeodomain proteins. Proc. Natl. Acad. Sci. USA 1992, 89, 7169–7173. [Google Scholar] [CrossRef]
- James, T.Y.; Kües, U.; Rehner, S.A.; Vilgalys, R. Evolution of the gene encoding mitochondrial intermediate peptidase and its cosegregation with the A mating-type locus of mushroom fungi. Fungal Genet. Biol. 2004, 41, 381–390. [Google Scholar] [CrossRef] [PubMed]
- Bao, D.-P. Research progress on the mating-typing locus structures of basidiomycete mushrooms. Mycosystema 2020, 38, 2061–2077. [Google Scholar] [CrossRef]
- Au, C.H.; Wong, M.C.; Bao, D.; Zhang, M.; Song, C.; Song, W.; Law, P.T.W.; Kües, U.; Kwan, H.S. The genetic structure of the A mating-type locus of Lentinula edodes. Gene 2014, 535, 184–190. [Google Scholar] [CrossRef]
- Luo, Y.; Ullrich, R.C.; Novotny, C.P. Only one of the paired Schizophyllum commune Aα mating-type, putative homeobox genes encodes a homeodomain essential for Aα-regulated development. Mol. Gen. Genet. MGG 1994, 244, 318–324. [Google Scholar] [CrossRef] [PubMed]
- Magae, Y.; Novotny, C.; Ullrich, R. Interaction of the Aα Y and Z mating-type homeodomain proteins of Schizophyllum commune detected by the two-hybrid system. Biochem. Biophys. Res. Commun. 1995, 211, 1071–1076. [Google Scholar] [CrossRef] [PubMed]
- Robertson, C.I.; Bartholomew, K.A.; Novotny, C.P.; Ullrich, R.C. Deletion of the Schizophyllum commune Aα locus: The roles of Aα Y and Z mating-type genes. Genetics 1996, 144, 1437–1444. [Google Scholar] [CrossRef]
- Shen, G.-P.; Park, D.-C.; Ullrich, R.C.; Novotny, C.P. Cloning and characterization of a Schizophyllum gene with Aβ6 mating-type activity. Curr. Genet. 1996, 29, 136–142. [Google Scholar] [CrossRef]
- Chu, C.; Li, D.; Gu, L.; Yang, S.; Liu, C. Evidence for the Existence of Mating Subtypes Within the Schizophyllum commune: Mating Behavior and Genetic Divergence. J. Fungi 2025, 11, 277. [Google Scholar] [CrossRef]
- Liu, C.H.; Huang, X.; Xie, T.N.; Duan, N.; Xue, Y.R.; Zhao, T.X.; Lever, M.A.; Hinrichs, K.U.; Inagaki, F. Exploration of cultivable fungal communities in deep coal-bearing sediments from ∼1.3 to 2.5 km below the ocean floor. Environ. Microbiol. 2017, 19, 803–818. [Google Scholar] [CrossRef] [PubMed]
- Jerpseth, B. XL1-Blue MRF’E. coli cells: McrA-, mcrCB-, mcrF-, mrr-, hsdR-derivative of XL1-Blue cells. Strategies 1992, 5, 81–83. [Google Scholar]
- Kim, S.; Jeong, H.; Kim, E.-Y.; Kim, J.F.; Lee, S.Y.; Yoon, S.H. Genomic and transcriptomic landscape of Escherichia coli BL21 (DE3). Nucleic Acids Res. 2017, 45, 5285–5293. [Google Scholar] [CrossRef]
- Yamamoto, K.; Toya, S.; Sabidi, S.; Hoshiko, Y.; Maeda, T. Diluted Luria-Bertani medium vs. sewage sludge as growth media: Comparison of community structure and diversity in the culturable bacteria. Appl. Microbiol. Biotechnol. 2021, 105, 3787–3798. [Google Scholar] [CrossRef]
- Pöggeler, S.; Masloff, S.; Hoff, B.; Mayrhofer, S.; Kück, U. Versatile EGFP reporter plasmids for cellular localization of recombinant gene products in filamentous fungi. Curr. Genet. 2003, 43, 54–61. [Google Scholar] [CrossRef]
- Li, L.; Li, H.; Tian, Q.; Ge, B.; Xu, X.; Chi, Y.; Zhao, H.; Liu, Y.; Jia, N.; Zhou, T. Expression and purification of soluble recombinant β-lactamases using Escherichia coli as expression host and pET-28a as cloning vector. Microb. Cell Factories 2022, 21, 244. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef]
- Waterhouse, R.M.; Seppey, M.; Simão, F.A.; Manni, M.; Ioannidis, P.; Klioutchnikov, G.; Kriventseva, E.V.; Zdobnov, E.M. BUSCO applications from quality assessments to gene prediction and phylogenomics. Mol. Biol. Evol. 2018, 35, 543–548. [Google Scholar] [CrossRef]
- Gurevich, A.; Saveliev, V.; Vyahhi, N.; Tesler, G. QUAST: Quality assessment tool for genome assemblies. Bioinformatics 2013, 29, 1072–1075. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Xu, S.; Zhang, J.; Li, M. Assembly and annotation of whole-genome sequence of Fusarium equiseti. Genomics 2021, 113, 2870–2876. [Google Scholar] [CrossRef]
- Altschul, S.F.; Madden, T.L.; Schäffer, A.A.; Zhang, J.; Zhang, Z.; Miller, W.; Lipman, D.J. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997, 25, 3389–3402. [Google Scholar] [CrossRef]
- Thompson, J.D.; Gibson, T.J.; Plewniak, F.; Jeanmougin, F.; Higgins, D.G. The CLUSTAL_X windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997, 25, 4876–4882. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Nei, M.; Dudley, J.; Tamura, K. MEGA: A biologist-centric software for evolutionary analysis of DNA and protein sequences. Brief. Bioinform. 2008, 9, 299–306. [Google Scholar] [CrossRef]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Žídek, A.; Potapenko, A. Highly accurate protein structure prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef]
- Xie, S.; Xie, X.; Zhao, X.; Liu, F.; Wang, Y.; Ping, J.; Ji, Z. HNSPPI: A hybrid computational model combing network and sequence information for predicting protein–protein interaction. Brief. Bioinform. 2023, 24, bbad261. [Google Scholar] [CrossRef] [PubMed]
- Krissinel, E.; Henrick, K. Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 2007, 372, 774–797. [Google Scholar] [CrossRef] [PubMed]
- Subotić, A.; Swinnen, E.; Demuyser, L.; De Keersmaecker, H.; Mizuno, H.; Tournu, H.; Van Dijck, P. A bimolecular fluorescence complementation tool for identification of protein-protein interactions in Candida albicans. G3 Genes Genomes Genet. 2017, 7, 3509–3520. [Google Scholar] [CrossRef]
- Li, P.; Wang, Y.; Zhang, L.; Ning, Y.; Zan, L. The expression pattern of PLIN2 in differentiated adipocytes from Qinchuan cattle analysis of its protein structure and interaction with CGI-58. Int. J. Mol. Sci. 2018, 19, 1336. [Google Scholar] [CrossRef]
- Okabe, T.; Katayama, T.; Mo, T.; Mori, N.; Jin, F.J.; Fujii, I.; Iwashita, K.; Kitamoto, K.; Maruyama, J.-I. BiFC-based visualisation system reveals cell fusion morphology and heterokaryon incompatibility in the filamentous fungus Aspergillus oryzae. Sci. Rep. 2018, 8, 2922. [Google Scholar] [CrossRef] [PubMed]
- Van Peer, A.F.; De Bekker, C.; Vinck, A.; Wösten, H.A.; Lugones, L.G. Phleomycin increases transformation efficiency and promotes single integrations in Schizophyllum commune. Appl. Environ. Microbiol. 2009, 75, 1243–1247. [Google Scholar] [CrossRef]
- Inciuraite, R.; Gedgaudas, R.; Lukosevicius, R.; Tilinde, D.; Ramonaite, R.; Link, A.; Kasetiene, N.; Malakauskas, M.; Kiudelis, G.; Jonaitis, L.V. Constituents of stable commensal microbiota imply diverse colonic epithelial cell reactivity in patients with ulcerative colitis. Gut Pathog. 2024, 16, 16. [Google Scholar] [CrossRef]
- Xiong, Y.; Zhong, Y.; Yim, H.; Yang, X.; Park, K.-S.; Xie, L.; Poulikakos, P.I.; Han, X.; Xiong, Y.; Chen, X. Bridged proteolysis targeting chimera (PROTAC) enables degradation of undruggable targets. J. Am. Chem. Soc. 2022, 144, 22622–22632. [Google Scholar] [CrossRef]
- Yue, C.; Osier, M.; Novotny, C.P.; Ullrich, R.C. The specificity determinant of the Y mating-type proteins of Schizophyllum commune is also essential for YZ protein binding. Genetics 1997, 145, 253–260. [Google Scholar] [CrossRef]
- Pomerantz, J.L.; Sharp, P.A. Homeodomain determinants of major groove recognition. Biochemistry 1994, 33, 10851–10858. [Google Scholar] [CrossRef]
- Frenz, C.M.; Lefebvre, P.P. Presence of pKa Perturbations Among Homeodomain Residues Facilitates DNA Binding. arXiv 2009, arXiv:0907.4819. [Google Scholar] [CrossRef]
- Fujioka, M.; Gebelein, B.; Cofer, Z.C.; Mann, R.S.; Jaynes, J.B. Engrailed cooperates directly with Extradenticle and Homothorax on a distinct class of homeodomain binding sites to repress sloppy paired. Dev. Biol. 2012, 366, 382–392. [Google Scholar] [CrossRef] [PubMed]
- Stamberg, J.; Koltin, Y. The organisation of the incompatibility factors in higher fungi: The effect of structure and symmetry on breeding. Heredity 1973, 30, 15–26. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chu, C.; Li, D.; Liu, C. Characterization of Homeodomain Proteins at the Aβ Sublocus in Schizophyllum commune and Their Role in Sexual Compatibility and Development. J. Fungi 2025, 11, 451. https://doi.org/10.3390/jof11060451
Chu C, Li D, Liu C. Characterization of Homeodomain Proteins at the Aβ Sublocus in Schizophyllum commune and Their Role in Sexual Compatibility and Development. Journal of Fungi. 2025; 11(6):451. https://doi.org/10.3390/jof11060451
Chicago/Turabian StyleChu, Chen, Dongxu Li, and Changhong Liu. 2025. "Characterization of Homeodomain Proteins at the Aβ Sublocus in Schizophyllum commune and Their Role in Sexual Compatibility and Development" Journal of Fungi 11, no. 6: 451. https://doi.org/10.3390/jof11060451
APA StyleChu, C., Li, D., & Liu, C. (2025). Characterization of Homeodomain Proteins at the Aβ Sublocus in Schizophyllum commune and Their Role in Sexual Compatibility and Development. Journal of Fungi, 11(6), 451. https://doi.org/10.3390/jof11060451





