Abstract
Tannase, a highly adaptive biocatalyst, plays a pivotal role in diverse bioconversion reactions in nature. This enzyme exhibits numerous applications across various industrial sectors, including food, pharmaceuticals, chemicals, and beverages. This study aimed to screen and characterize fungal endophytes isolated from mangrove plants for their enzyme tannase-producing ability. Eighty-five filamentous endophytic fungi were isolated from different mangrove samples and subsequently identified. These fungal strains were initially screened using the tannic acid agar plate method. Out of the screened strains, 13 fungal isolates demonstrated tannase production ability. The quantitative estimation of extracellular tannase was performed using the submerged fermentation technique. Among the studied endophytes, eight isolates, namely LV_084 (21.21 IU/mL), LV_074 (15.41 IU/mL), LV_078 (6.98 IU/mL), LV_038 (6.97 IU/mL), LV_077 (6.32 IU/mL), LV_016 and LV_066 (6.37 IU/mL), and LV_060 (6.18 IU/mL) exhibited excellent tannase activity. Among these isolates, LV_084 Phyllosticta capitalensis and LV_074 Aspergillus chevalieri showed the highest enzyme-producing ability. These isolates were authenticated using ITS rDNA sequencing, followed by BLAST search and phylogenetic analysis. Furthermore, the physical and chemical conditions for the maximum enzyme production were optimized. This is the first report of enzyme tannase production by Phyllosticta capitalensis and Aspergillus chevalieri.
1. Introduction
Mangrove ecosystems, at the intersection of terrestrial and marine environments, are well-known for their unique biodiversity and resilience. The microbial diversity, including bacteria, fungi, and actinomycetes, is rich in mangrove ecosystems and plants. Fungal communities in the mangrove habitat are diverse and are essential in nutrient recycling. Endophytic fungi are recognized for their ability to produce a range of bioactive compounds and enzymes that significantly contribute to the host plant’s defense mechanisms and ecological balance [1,2,3]. The harsh environment of mangrove habitats induces the fungal physiology, producing several biochemicals, including enzymes, by endophytes. Research on screening endophytic fungi for their potential to produce enzymes is gaining attention in this biotechnological era. The enzymes cellulases, proteases, amylases, lipases, laccases, and chitinases are industrially important enzymes obtained from fungal sources [4,5,6,7]. Tannase is less explored among the reports of important hydrolytic enzymes from mangrove endophytic isolates. However, some soil fungi, such as Aspergillus, Penicillium, and Paecilomyces, are known to produce extracellular tannase. Tannase is particularly noteworthy because it breaks down tannins into gallic acid, a compound with numerous applications in pharmaceuticals, food processing, and environmental cleanup.
Tannins are naturally occurring polyphenolic compounds present in various plant species [8]. They possess strong antibacterial properties and can precipitate proteins by forming complexes with enzymes and other proteins [9]. The enzymatic degradation of tannins by tannase offers an environment-friendly alternative to chemical catalysts, highlighting the potential of microbial tannase in sustainable industrial applications. Several fungi, such as Penicillium, Aspergillus, Fusarium, Rhizopus, Trichoderma, Candida, and Saccharomyces, are commonly explored for tannase enzyme production [10,11]. Tannase-producing fungi, such as Aspergillus and Penicillium, have been extensively studied for their biotechnological potential [12,13,14]. However, exploring mangrove endophytes as sources of tannase-producing fungi presents a promising opportunity to discover novel strains with enhanced enzymatic activities.
Mangrove endophytes, which are adapted to the challenging conditions of mangrove environments, may exhibit unique metabolic capabilities that enable them to thrive in these ecosystems. Isolating and characterizing tannase-producing fungi from mangrove endophytes could offer valuable insights into their enzymatic diversity and potential applications. This study aimed to investigate the diversity of tannase-producing fungi within mangrove endophytes, evaluate their enzymatic properties, and characterize the enzyme tannase.
2. Materials and Methods
2.1. Study Sites
The conserved mangrove region of the Godrej Mangrove Forest, Vikhroli, India (21.62° N, 108.23° E) was selected for this study (Figure 1). This site has a mean annual temperature of 27.2 °C, an average precipitation of 242.2 cm (95.35 inches), and a soil salinity of 0.5–1.2%. The dominant mangrove species observed at the site are Avicennia marina Vierh., Acanthus ilicifolius L., Suaeda maritima (L.) Dumort., Derris indica (Lam.), Salvodora persica L., Acanthus ilicifolius L., Rhizophora mucronata Lam., and Derris trifoliata Lour.
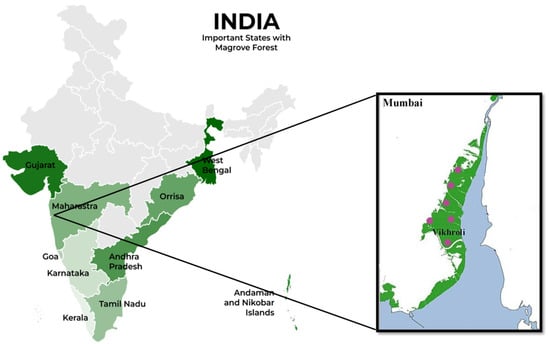
Figure 1.
The map represents the mangrove ecosystem in different states of India . The green zone in the box represents Mumbai coastal region. Pink coloured spots represent sample collection sites in the Godrej Mangrove Forest at Vikhroli.
2.2. Sample Collection
The samples such as fresh and healthy leaves, twigs, and pneumatophores of all the abovementioned plants were collected from the Godrej Mangrove Forest, Vikhroli, India, from 2021 to 2023. A total of 40 samples were collected in three different seasons from selected plants and brought to the P.G. Research Laboratory, Department of Botany of the Smt. C.H.M. College, Ulhasnagar, Maharashtra, India. The samples were stored in a refrigerator at 4 °C until further isolation of endophytic fungi.
2.3. Isolation of Endophytic Fungi
The samples were washed with tap water to remove the dust and dirt from the surface and surface-sterilized using 1% sodium hypochlorite (NaOCl) and 75% ethanol solution for one minute, followed by washing with sterilized distilled water to remove traces of the sterilizing agent [15]. The surface-sterilized leaves, stems, and pneumatophores were cut into small segments (0.5 cm). Five segments of each plant part were aseptically placed onto the potato dextrose agar (PDA), potato carrot agar (PCA), Czapek–Dox agar (CzDA), and malt extract agar (MEA) media. Chloramphenicol (50 mg/L, HiMedia, Mumbai, India) was added to the media to suppress bacterial growth. All the plates were incubated at 30 °C for 10 days, and periodic observations were undertaken for the emergence of hyphae from the seeded segments. A small piece of growing hyphal tip from the segments was placed on fresh PDA plates for further growth and purification.
2.4. Morphological Characterization and Identification of Isolates
The pure isolates were transferred to PDA plates for colony characterization and growth study. A small portion of 5-day-old mycelia was aseptically placed on slides mounted with lactophenol cotton blue stain and sterile distilled water, then observed under a light microscope (Magnus MX21i-LED, Magnus Opto Systems India Pvt. Ltd., Noida, India). Images of fungal structures were captured using a MagcamHDL5MP camera, Magnus Opto Systems India Pvt. Ltd., Noida, India. The isolates were identified morpho-taxonomically using standard books and monographs [16,17,18,19,20,21,22,23,24,25,26]. All the identified fungal strains were deposited at the National Fungal Culture Collection of India, Agharkar Research Institute, Pune, Maharashtra, India.
2.5. Screening of the Isolates for Tannase Production
2.5.1. Qualitative Screening (Plate Method)
The primary screening of tannase-producing isolates was performed on tannin agar media as per Pinto et al. [27]. The freshly grown 5-day-old fungal mycelia were centrally inoculated on tannin acid agar (TAA) media: peptone (5 g/L), sodium chloride (5.0 g/L), beef extract (1.5 g/L), yeast extract (1.5 g/L), tannic acid (5 g/L), and agar (20 g/L). The plates were incubated at 30 °C for 5–7 days. The isolates capable of utilizing tannic acid exhibited growth on TAA media. A dark zone around the colony indicated hydrolysis of tannins by isolates, which is considered a positive result. The zone diameters around the fungal colonies were measured at 72 h and 120 h, and the enzymatic index (EI) was calculated using the following formula:
2.5.2. Quantitative Screening (Liquid Broth)
The positive isolates were further screened for tannase production in a liquid broth. The isolates were inoculated in separate flasks containing 100 mL of liquid broth media containing peptone (5.0 g/L), sodium chloride (5.0 g/L), beef extract (1.5 g/L), yeast extract (1.5 g/L), and 1% tannic acid as the sole carbon source (pH 5.5). Five-day-old pure cultures of selected isolates were inoculated (5 mycelial plugs of 0.5 cm) in separate flasks containing sterilized production media. The inoculated flasks were incubated at 35 °C for 96 h at 150 rpm on a rotary shaker. After the incubation period, cultures were harvested, and the mycelial biomass was filtered using Whatman No. 1 filter paper. The free cell filtrate was used as a source of extracellular tannase and taken to determine tannase activity.
2.6. Enzyme Tannase Assay
The enzyme assay was based on the formation of the chromogen due to the reaction between gallic acid and rhodamine as described by Sharma et al. [28]. Enzyme tannase from the crude filtrate was quantified spectrophotometrically at 520 nm using rhodamine. A gallic acid standard curve was prepared by varying concentrations ranging from 0.125 mM to 4 mM. Methyl gallate was used as a substrate. One unit (U) of enzyme activity was determined as the amount of enzyme required to release one µmol of gallic acid per minute under standard assay conditions. The specific activity of the enzyme was calculated by dividing the enzyme activity by the protein concentration.
2.7. Protein Assay
The protein concentration of the samples (crude extract) was measured using Bradford’s method, with bovine serum albumin (BSA) as the standard [29]. The specific activity of the enzyme tannase was expressed as enzyme activity (IU) per milligram of protein (mg) in each sample.
2.8. Optimization of the Production of Extracellular Tannase Under SmF
Based on quantitative screening, hyper-tannase-producing isolates were cultured using submerged fermentation (SmF). The parameters such as incubation period, temperature, and pH were optimized for higher yields of a stable enzyme using the univariate parameter optimization method [30]. All the isolates were inoculated separately in 100 mL of production media (tannic acid broth with 1% w/v tannic acid) and incubated on a rotary shaker at 120 rpm.
2.8.1. Incubation Period
A loopful culture of selected isolates was separately inoculated in flasks containing 100 mL of production media with a pH of 5.5. The inoculated flasks were incubated at 35 °C. The tannase production was estimated by periodically withdrawing 5 mL of the fermentation broth at 48 h, 72 h, 96 h, and 120 h. This experiment was performed in triplicate.
2.8.2. Temperature
To determine the optimal temperature for enzyme production, sets of inoculated flasks with 100 mL of production media (pH 5.5) were incubated at different temperatures, 30 °C, 35 °C, 37 °C, and 40 °C, for the optimum incubation period, i.e., 96 h.
2.8.3. pH
To determine the optimal pH, all the selected isolates were inoculated in production media with different pH levels (3.5, 4.5, 5.5, 6.5, and 7.5) and incubated at the optimum temperature (i.e., 35 °C) and incubation period (96 h).
2.9. Determination of Fungal Biomass
The fresh (wet) weight of fungal biomass was measured after 120 h of incubation at 35 °C in production media. At the end of the incubation time, mycelia were filtered through a circular Whatman No. 1 filter paper and weighed using a digital balance.
2.10. Molecular Identification of LV_074 and LV_084
The hyper-enzyme-producing isolates were authenticated using molecular methods. Five-day-old cultures grown on PDA media at 30 °C were used for DNA extraction. DNA was extracted using the CTAB method [31]. The targeted DNA regions were amplified using the polymerase chain reaction (PCR) by outsourcing the samples to Barcode Bioscience Pvt. Ltd., Bangalore, India. The ITS region was amplified using the primers ITS4/ITS5 [32,33]. The amplified products were purified using a Qiagen (Cat. No. 28104) PCR kit, QIAGEN Pvt. Ltd., Germantown, MD, USA and sent for sequencing to Barcode Bioscience Pvt. Ltd., Bangalore, India. The sequences of the isolates were compared with the NCBI GenBank database using BLASTn (https://blast.ncbi.nlm.nih.gov/Blast.cgi?PROGRAM=blastn&PAGE_TYPE=BlastSearch&LINK_LOC=blasthome, accessed on 20 March 2025) to identify the closest matching sequences.
2.11. Sequence Alignment and Phylogenetic Analysis
ITS sequences of all the closely matched sequences were downloaded and assembled using sequences of all the available species described in the database. The datasets were aligned using MAFFT v.7 (https://mafft.cbrc.jp/alignment/server; accessed on 25 March 2025) [34]. The sequences were manually edited using MEGA version 7 [35]. Maximum likelihood (ML) analysis was performed with IQTree v2.4.0 [36]. The phylogenetic trees were viewed and arranged using Interactive Tree of Life (iTOL) v4 (https://itol.embl.de/; accessed on 25 March 2025) [37]. The newly obtained sequences were deposited in the GenBank database.
2.12. Statistical Analysis
Assays were performed in triplicates, and the values were expressed as the means ± standard deviations using MS Excel 2019.
3. Results
3.1. Isolation of Tannase-Producing Fungi
In this study, 85 endophytic fungi were isolated from mangrove plant species from the Godrej Mangrove Forest, Vikhroli, India. Most of the endophytic fungal strains were recovered from A. marina and A. officinale which covered 31.6% and 14.1% of the total isolates. These two plants are true mangrove plants and more dominant in the study area (Figure 2). This region has a lower abundance of Rhizophora mucronata, Salvadora persica, Derris indica, and Suaeda maritima. However, a good number of endophytic fungi were also obtained from different parts of the selected plants. Based on morphological and microscopic observations, 25 genera and 56 species from the different classes of fungi were identified. The predominant genera were Chaetomium, Aspergillus, Scopulariopsis, Microascus, Alternaria, and Fusarium. Some isolates were unable to produce reproductive structures even after a long period of incubation on different culture media. To identify such isolates morphologically, clamp connections were seen in the mycelium. Therefore, they were considered as basidiomycete isolates. All 85 isolates were screened for their tannase-producing ability. Among them, 13 active isolates showed a positive result (Table 1).
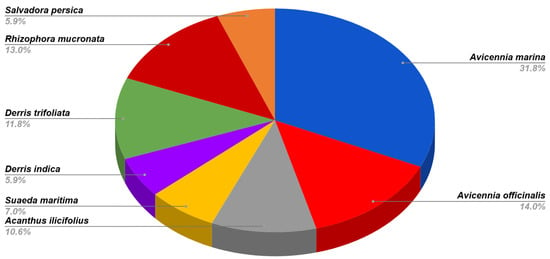
Figure 2.
Diversity of fungal endophytes in A. marina, A. officinalis, Acanthus ilicifolius, S. maritima, D. indica, D. trifoliata, R. mucronate, and Salvadora persica.

Table 1.
Tannase-producing isolates and their tannin-hydrolyzing ability (enzymatic index) in plate assay.
The morphological study on all tannase-producing fungi was carried out on PDA media. The selected isolates were morphologically distinct from each other. Based on morphological characters, the isolates were identified, and the authenticity of identification was confirmed by the Fungal Identification Service, Agharkar Research Institute, Pune. GenBank accession numbers are listed in Table 2. In this study, eight different genera were isolated from different mangrove plants, including dematiaceous fungi; most isolates belonged to ascomycetes and anamorphic ascomycetes. Morphological studies enabled the identification of dematiaceous fungi Curvularia brachyspora, Curvularia lunata, Alternaria tenuissima, Corynespora cassiicola, and Cladosporium sp. based on colony color, conidiophores, and conidial morphology. In contrast, the ascomycetous genus Chaetomium was determined based on ascomata, asci, and ascospores. The genera such as Aspergillus, Penicillium, and Phyllosticta are very complex, with more species; hence, they were initially determined at the genus level based on colony morphology and conidial characters. Furthermore, the fungi that showed hyperactivity of the enzyme tannase were authenticated using ITS rDNA sequence analysis. The authenticated fungi were deposited for accession number in the National Fungal Culture Collection of India, ARI, Pune.

Table 2.
List of tannase producing-isolates.
3.2. Primary Screening of Tannase-Producing Fungi
Tannic acid agar (TAA) plates were used for primary screenings of isolates for their tannase-producing ability as described by Bradoo et al. [38]. Approximately 13 isolates could grow on TAA medium containing 1% tannic acid as the carbon source. After 72 and 120 h of incubation, the tannase hydrolysis activity in a dark zone around the colony was observed. The diameter of the hydrolysis zone was measured, and the enzymatic index was calculated (Table 1). The photographs showing the dark zone around the colony produced by isolates are shown in Figure 3. With the increase in time, the isolates’ colony diameter and zone diameter increased, confirming the utilization of tannic acid as the carbon source. The enzymatic activity of the isolates differed significantly in terms of the enzymatic index (Table 1). The enzymatic index after 72 h of incubation ranged from 0.17 to 2.28; however, after 120 h, it was not possible to calculate the enzymatic index for isolates LV_002, LV_022, LV_038, LV_060, and LV_074 due to mycelial growth covering the entire plate.

Figure 3.
Fungi showing tannic acid degradation leading to the formation of a hydrolysis zone after 72 h: Penicillium sp. (LV_001); Chaetomium sp. (LV_002); Chaetomium sp. (LV_004); Aspergillus sp. (LV_010); Aspergillus sp. (LV_016); Curvularia brachyspora (LV_019); Alternaria tenuissima (LV_022); Chaetomium sp. (LV_038); Corynespora cassiicola (LV_042); Cladosporium limoniforme (LV_047); Curvularia lunata (LV_053); and control.
The enzymatic index of some isolates could only be measured after 120 h of incubation due to a slow growth rate. After 120 h of incubation, four isolates exhibited an enzyme index greater than 1.0. Among them, isolates LV_047 and LV_084 showed the highest indices of 1.54 and 1.45, while isolates LV_002 and LV_074 demonstrated the maximum indices of 2.28 and 2.1 after 72 h of incubation and were characterized by very rapid growth.
The plate assay is considered an easy and quick screening method that indicates the microbial capacity to use tannic acid as the carbon source [27]. Although 13 fungal strains could thrive on the TAA medium containing 1% tannic acid, the possibility of producing maximum enzymes under submerged fermentation for extracellular tannase was to be examined. All the positive isolates were further analyzed for extracellular enzyme tannase production in a liquid broth.
3.3. Quantitative Estimation of Tannase Under SmF
After inoculating all 13 isolates individually in a liquid broth and incubating on a rotary shaker for 96 h, extracellular tannase assays were carried out. A total of 4 isolates exhibited excellent tannase activity in the liquid (Figure 4); among them, isolates LV_074 and LV_084 were hyper tannase producers. The enzyme tannase activity results showed that fungal isolates LV_084 and LV_074 isolated from Avicennia marina showed the highest tannase activity with 21.21 IU/mL and 15.41 IU/mL at 96 h, followed by other fungal isolates LV_038 (6.97 IU/mL) and LV_016 (6.37 IU/mL).
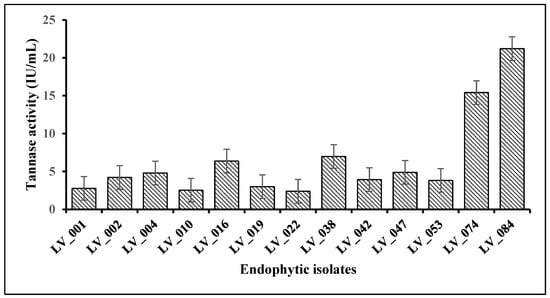
Figure 4.
Quantitative estimation of the tannase enzyme activity of fungal isolates under submerged fermentation.
The highest tannase activity for all the fungal strains was observed after 96 h. The low tannase activity recorded after 48 h of incubation was because of less mycelial growth in the early stage in the production media; thus, less extracellular tannase was synthesized to break down the tannic acid present in the media. Due to their highest tannase activity, the LV_084 and LV_074 isolates were chosen as the best tannase-producing fungi. Furthermore, physicochemical parameters of tannase production by isolates LV_084 and LV_074 were performed in control conditions.
3.4. Optimization of Conditions for Tannase Activity of LV_074 and LV_084
3.4.1. Optimization of the Incubation Period
The optimization of the incubation period for the maximum tannase activity was carried out at 48, 72, 96, and 120 h of incubation (Figure 5). The results revealed that from 48 h to 96 h, both isolates showed a significant increase in tannase activity at 96 h (from 1.5 IU/mL to 15.48 IU/mL in LV_074 and from 2.5 IU/mL to 21.21 IU/mL in LV_084). Later, the tannase activity decreased significantly at 120 h (11.5 IU/mL in LV_074 and 15.96 IU/mL in LV_084). The optimum incubation period for the maximum tannase activity for both isolates was 96 h.
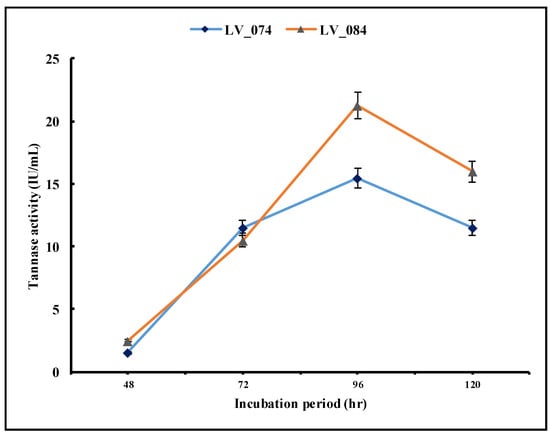
Figure 5.
Optimization of the incubation period (48 h, 72 h, 96 h, and 120 h) for the maximum tannase activity of LV_074 and LV_084.
3.4.2. Optimization of the Incubation Temperature
Temperature is another important factor in the growth and metabolic activities of fungi, consequently in extracellular enzyme production. This work examined four different temperatures: 30, 35, 40, and 45 °C (Figure 6). The results showed that at 30 °C, the enzyme activity was lowest, while the highest activity was observed at 35 °C. Generally, the concentration of the enzymes gradually decreases at 40 °C and 45 °C.
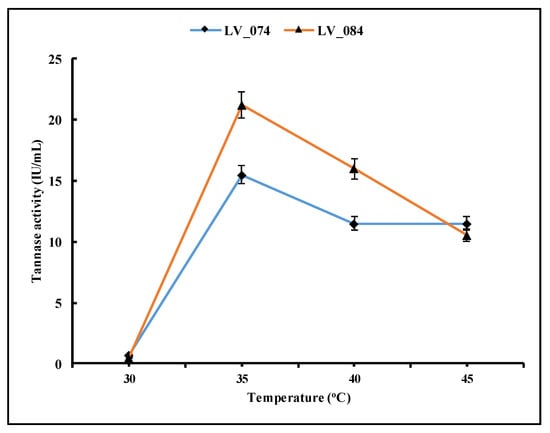
Figure 6.
Optimization of the optimum temperature for the maximum tannase activity of LV_074 and LV_084.
3.4.3. Optimization of the pH Level
The pH level of microbial growth media is one of the most important factors for microbial growth and enzyme production. However, the optimum pH for microbial growth is not necessarily the same as the optimum pH for enzyme activity. The pH level of the growth medium plays a vital role in enzyme secretion by microorganisms. This study applied five different pH levels: 3.5, 4.5, 5.5, 6.5, and 7.5. The tannase activity was low at 3.5 pH after 96 h of incubation (Figure 7) and increased in both isolates at pH 5.5 (15.48 IU/mL in LV_074 and 21.21 IU/mL in LV_084). However, after pH 5.5, it decreased. Thus, pH 5.5 was the optimum condition for the maximum tannase activity.
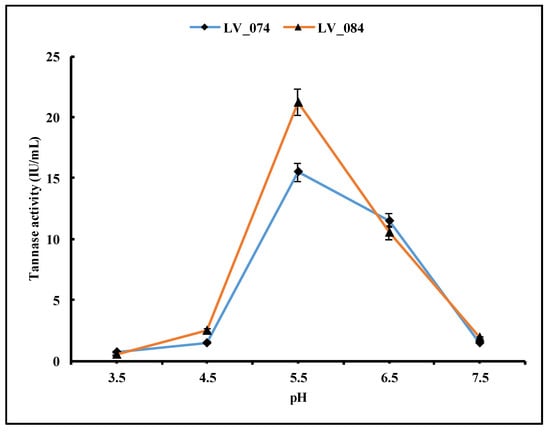
Figure 7.
Optimization of the pH level of the growth medium for the maximum tannase activity in LV_074 and LV_084.
3.5. Biomass Production
The biomass dynamics of isolates during the extracellular secretion of tannase were illustrated in Figure 8. As time progressed, the biomass of the isolates increased, confirming the utilization of tannic acid as the carbon source. The biomass of both isolates LV_074 and LV_084 increased with incubation period, and the maximum biomass was measured to be 4.292 mg and 3.492 mg, respectively, at 96 h. However, thereafter, there was no significant change in biomass.
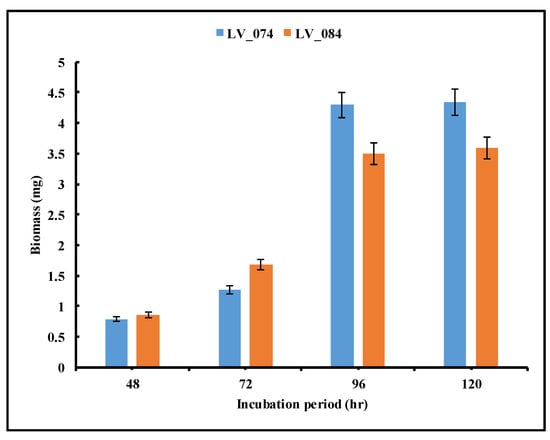
Figure 8.
Optimization of the incubation period for the maximum biomass production of isolates LV_074 and LV_084.
3.6. Molecular Identification of the Maximum Tannase Producer: LV_074 and LV_084
The isolated strains LV_074 and LV_084 were identified as Aspergillus chevalieri and Phyllosticta capitalensis using sequencing of the ITS-4 and ITS-5 of ribosomal DNA and then BLASTn. Furthermore, the phylogeny based on the ITS rDNA gene sequence confirmed the identity of isolates LV_074 and LV_084.
- LV_074. Aspergillus chevalieri var. chevalieri (L. Mangin) Thom & Church
The BLAST (https://blast.ncbi.nlm.nih.gov/Blast.cgi?PROGRAM=blastn&PAGE_TYPE=BlastSearch&LINK_LOC=blasthome, accessed on 20 March 2025) search results on the ITS sequence of LV_074 showed 100% sequence similarity with Aspergillus chevalieri accession No. LN482478.1 and Aspergillus montevidansis accession No. OW987704.1. The phylogenetic relationship within the restriction section of the genus Aspergillus revealed that LV_074 forms a topology with Aspergillus chevalieri, so isolate LV_074 is assigned as Aspergillus chevalieri and taxonomically described (Figure 9), GenBank accession No. PV054919. The phylogram is given in Figure 10.
Description. Colonies: on PDA, sulfur yellow-to-orange-colored, velvety to slightly sulcate, slow-growing (15–25 mm in diam.), in 15 days, at 35 °C. Mycelium: wide, septate, granulate, shiny structure on the surface of the colony due to formation of ascomata, irregular margins, sporulation sparse to moderately dense, with green conidial mass. Conidiophores: short (70–120 × 4–6 μm), hyaline, and smooth-walled. Vesicles: pyriform, 20–48 μm in diameter. Phialides: ampulliform, 5.5–7.5(–10) × 3–5 μm, uniseriate, conidial head radiating with conidial mass. Conidia: 3–4(–6) × 2.5–3.5(–5) μm, globose to subglobose, hyaline, rough-walled. Ascomata: 100–250 μm in diameter, cleistothecial, superficial, yellow, globose to subglobose. Ascospores: globose to subglobose, smooth to slightly verruculose, 3.5–5.5 × 3–4 μm, lenticular furrow present, crests of 0.5–1 μm, smooth-walled to roughened.
Habitat. Isolated from mangrove plant Avicennia marina, Thane-Mumbra creek, Mumbai, Maharashtra India
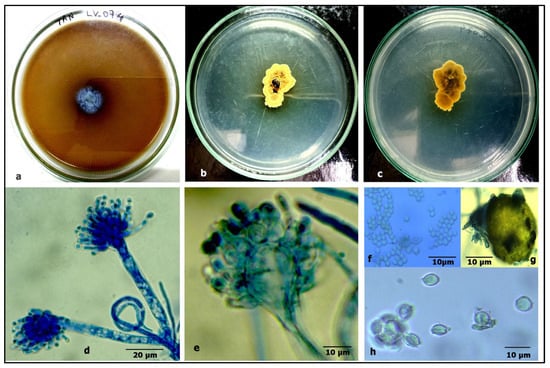
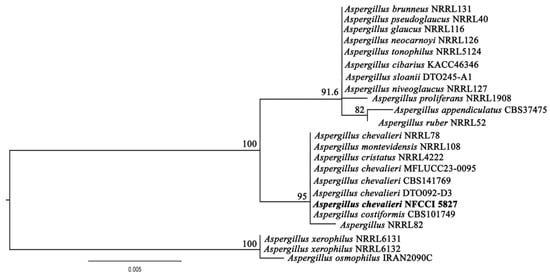

Figure 9.
Aspergillus chevalieri: (a) growth on TAA showing a dark zone around the colony; (b) colony on PDA verse; (c) PDA reverse; (d) a conidiophore with a vesicle; (e) a vesicle with sterigmata; (f) conidia; (g) ascomata; and (h) ascospores.

Figure 10.
Phylogram of Aspergillus resulting from the maximum likelihood (RAxML) tree using ITS sequences. Confidence values for ML ≥50% (UFboot2/RAxML) are included near the nodes. The specimens described in this study are highlighted in bold. Aspergillus xerophilus NRRL6131, Aspergillus xerophilus NRRL6132, and Aspergillus osmophilus IRAN2090C were used as an outgroup.
- LV_084. Phyllosticta capitalensis Henn.
Based on the BLAST search and phylogenetic analysis of the ITS sequence, the isolate LV_084 is described as Phyllosticta capitalensis (Figure 11), GenBank accession No. PV054914. The phylogram is given in Figure 12.
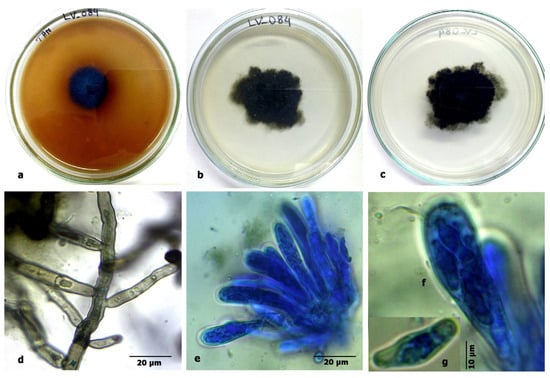
Figure 11.
Phyllosticta capitalensis Henn.: (a) growth on tannic acid agar showing a dark zone around the colony; (b) colony on PDA verse; (c) PDA reverse; (d) mycelium; (e) an ascus; (f,g) asci and ascospores.
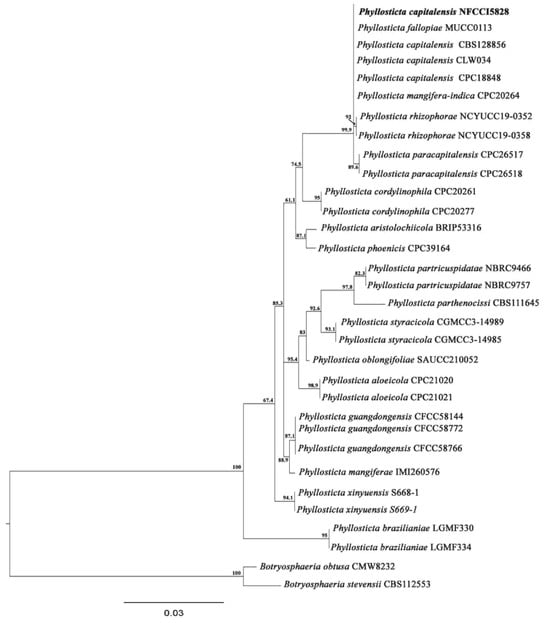
Figure 12.
Phylogram of Phyllosticta resulting from the maximum likelihood (RAxML) tree using ITS sequences. Confidence values for ML ≥50% (UFboot2/RAxML) are included near the nodes. The specimens described in this study are highlighted in bold. B. obtusa (CMW 8232) and B. stevensii (CBS112553) were used as an outgroup.
Description. Colonies: on PDA, greenish-black to light grey, slow-growing (30–40 mm), in 15 days, at 35 °C. Reverse black, irregular in shape, with sparse mycelium. Mycelium: wide, branched, septate, light dematiaceous and globulate. Conidiomata: pycnidial type, black-to-grey-colored, round to elongated (up to 35 μm), pycnidial walls multilayered, dark brown with textura angularis. Ascomata: similar to conidiomata. Asci: clavate to broadly ellipsoid, bitunicate, hyaline, 45–85 × 9–13 μm. Ascospores: hyaline, aseptate, bi- to multi-seriate, unicellular, broader in the middle, with obtuse colorless ends with a terminal mucoid appendage, smooth-walled, 15–18 × 6–7 μm.
Habitat. Isolated from mangrove plant Avicennia marina, Kalher creek, Mumbai, Maharashtra, India.
4. Discussion
Fungal endophytes represent a significant component of fungal communities. Interest in endophytic fungi is predicated on several factors; they constitute the primary reservoir of biodiversity within the fungal kingdom [39], encompassing a diverse array of taxa from eumycota, predominantly ascomycetes and their coelomycetous anamorphs, along with the occasional occurrence of basidiomycetes. In the present study, it was found that ascomycetous fungi predominantly colonized mangrove plants. The ascomycete fungi were isolated from all the selected mangrove plants frequently. In anamorphic ascomycete fungi, dematiaceous fungi were dominant along with coelomycetous anamorphs. These results support the findings of Du et al. [40], Mishra et al. [41], and Pecoraro et al. [42]. They reported that ascomycetes were the most abundant members of endophytic fungal communities isolated using traditional separation techniques. However, it is essential to note that diversity analyses of endophytes based on culture-dependent methodologies underestimate actual diversity. Many isolates may still be identified using culture-independent methods [43].
The cultural characteristics of endophytic fungi were significantly different from the strains isolated from soil or any other habitat. The mycelial growth and reproductive structure formation by endophytic fungi were slow; some isolates exhibited reduced sporulation. Therefore, morphological identifications are somewhat tricky. Table 2 revealed that the tannase-producing isolates were from eight different genera; the predominant genera were Chaetomium, Aspergillus, and Curvularia. The fungal genus Phyllosticta, a plant pathogen, isolated from the healthy leaves of A. mariana, causes leaf spot disease.
Studying fungi and their role in biotechnological applications has proven significant in industry and sustainable agriculture. Several noteworthy strains of fungi are known for their beneficial secondary metabolites and ability to produce enzymes. Exploring potent and industrially important fungi from various habitats has been an ongoing global research. Mangrove endophytic fungi have attracted considerable attention in biotechnology due to their capacity to produce bioactive substances with diverse applications. Researchers have explored mangrove-associated fungi for enzymes such as amylases, cellulases, pectinases, proteases, and chitinases [4,5,6,7]. Enzymes extracted from these organisms have many commercial and industrial uses [44]. Due to its wide applications, tannase stands out among the array of enzymes as a potential game-changer in various industrial sectors. However, the mangrove ecosystem is the least exploited among the habitats explored for tannase-producing fungi.
In the present study, 13 isolates were obtained as potent tannase enzyme producers (Table 1 and Table 2). The most tannase-producing genera were identified as Chaetomium, Aspergillus, Curvularia, Alternaria, Corynespora, and Phyllosticta. The records of tannase producers from mangrove habitats are meagre and restricted to Aspergillus niger, A. japonicas, A.aculeatus, A. fumigatous, and Penicillium species [45,46]. To the best of our knowledge, this is the first report demonstrating the ability of the mangrove endophytic Alternaria tenuissima, Curvularia brachyspora, Curvularia lunata, Corynespora cassiicola, and Phyllosticta capitalensis to produce tannase, thereby expanding the microbial sources for this industrially significant enzyme from a unique habitat. The specific tannase enzyme production ability under the specified conditions by the isolates Alternaria tenuissima (2.38 IU/mL), Curvularia brachyspora (3.0 IU/mL), Curvularia lunata (3.81 IU/mL), Corynespora cassiicola (3.93 IU/mL), and Phyllosticta capitalensis (21.22 IU/mL), respectively, was very comparable to known tannase-producing isolates. The mangrove ecosystem is rich with tannin-containing plants. The soil contains more organic matter due to this specific feature of the ecosystem, which supports microbial growth. Microbes have the potential to degrade tannins as an energy source [45].
Several tannase-producing fungi have been reported; among them, Aspergillus sp. and Penicillium sp. are the most common filamentous fungi involved in tannin bioconversion, individually or in co-cultures [12,47]. Furthermore, various other fungi from the genera Trichoderma, Fusarium, Chaetomium, Rhizoctonia, Candida, and Saccharomyces have also been reported for their ability to degrade tannins, particularly hydrolyzable tannins [10,11,48,49]. However, the genus Aspergillus has been the most potent and extensively studied tannase producer among the existing fungal sources [13,14,50]. Batra and Saxena [12] screened 35 Aspergillus and 25 Penicillium isolates for their tannase-producing ability, and it was found that 25 Aspergillus and 20 Penicillium isolates exhibited tannase activity in solid as well as in a liquid broth. Furthermore, endophytic Penicillium rolfsii CCMB 714 isolated from cocoa (Theobroma cacao) leaves also showed high tannase production when using tannic acid as a substrate (9.4 U/mL) [51]. In this study, Aspergillus was the predominant genus with three different isolates: LV_010 Aspergillus flavus (2.522 IU/mL), LV_016 Aspergillus sp. (6.37 IU/mL), and Aspergillus chevalieri (15.41 IU/mL) exhibited significant tannase activity in a liquid broth. This finding validates the previous records reported by researchers. The genus Aspergillus is robust, able to tolerate extreme conditions, and known for several industrially applicable metabolites, enzymes, and organic acids. This may be due to its adaptability to thrive, genetic potential, and evolutionary pressure to produce diverse metabolites to survive. The genus Chaetomium, commonly considered for its vigorous cellulolytic activity, also predominantly colonized mangrove plants and exhibited good tannase activity in this study [4]. The isolate LV_038 Chaetomium globosum showed high tannase production (6.97 IU/mL).
The most potent tannase producers LV_074 and LV_084 were identified as Aspergillus chevalieri and Phyllosticta capitalensis, with GenBank accession numbers PV054919 and PV054914. The tannase activity of the above isolates was confirmed qualitatively and quantitatively. Further optimization of tannase production by LV_074 and LV_084, such as of the pH level, temperature, and duration, was studied in a liquid medium. The optimization conditions revealed that tannase synthesized by both isolates exhibited the highest gallic acid production at 35 °C and a pH of 5.5 (15.41 IU/mL and 21.21 IU/mL) after 96 h of incubation; they are best-suited for tannase production. Both isolates are mesophilic, thriving at temperatures between 25 °C and 38 °C; at 35 °C, the maximum growth was recorded. On the other hand, slightly acidic media (pH 5.5) supported optimal growth; hence, the above physicochemical conditions were very suitable for enzyme secretion. In agreement with this finding, the optimal activity of the purified tannase from endophytic Aspergillus niger, Penicillium samsonii, and P. minioluteum was obtained at 35 °C and pH 5.5 [52,53,54]. The incubation at 40 °C and pH 6.0 led to a reduction in the activity. These findings also align with endophytic Aspergillus sydowii, which showed an optimal pH range of 5.0 to 6.0. [55].
5. Conclusions
This study explored several tannase-producing endophytic fungal strains isolated from mangrove plants for the identification of novel sources of the tannase enzyme. These strains demonstrated significant tannase production when cultivated in submerged fermentation using 1% tannic acid as a substrate. The potential isolates Aspergillus chevaleris and Phyllosticta capitalensis exhibited the highest stable tannase-producing ability across a wide range of pH and temperature conditions, highlighting their potential for biotechnological applications. In addition, other isolates such as Alternaria tenuissima, Curvularia brachyspora, Curvularia lunata, and Corynespora cassiicola were reported for the first time as tannase producers. However, the optimization of physicochemical parameters for the maximum production of a stable tannase enzyme from these isolates must be standardized. The exploration of tannase-producing organisms is crucial due to the growing demand for cost-effective and stable enzymes derived from untapped biological sources. This study provides a pioneering report on tannase-producing endophytic isolates A. chevaleris and P. capitalensis from the mangrove plant Avicennia marina, contributing valuable insights into the potential of these fungi for future applications in enzyme production. More research studies are required on optimizing large-scale, cost-effective production of tannase and exploring marker genes involved in the biosynthesis of tannase.
Author Contributions
Conceptualization, L.S.Y.; methodology, L.S.Y. and V.K.; software, V.K. and L.S.Y.; validation, L.S.Y. and G.C.N.; formal analysis, J.R.P. and L.S.Y.; investigation, L.S.Y.; writing—original draft preparation, V.K.; writing—review and editing, J.R.P., L.S.Y. and G.C.N.; supervision, L.S.Y.; project administration, L.S.Y. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The original contributions presented in this study are included in the article. Further inquiries can be directed to the corresponding authors.
Acknowledgments
L.S.Y., V.K. and J.R.P. are thankful to the Principal of Smt. C.H.M. College, Ulhasnagar, for providing a necessary laboratory facility.
Conflicts of Interest
The authors declare no conflicts of interest.
Abbreviations
The following abbreviations are used in this manuscript:
| TAA | Tannic acid agar |
| SmF | Submerged fermentation |
| ITS | Internal transcribed spacer |
| BLAST | Basic local alignment search tool |
| CTAB | Cetrimonium bromide |
References
- Hawar, S.N. Extracellular enzyme of endophytic fungi isolated from Ziziphus spina leaves as medicinal plant. Int. J. Biomater. 2022, 2022, 2135927. [Google Scholar] [CrossRef] [PubMed]
- Shankar Naik, B. Functional roles of fungal endophytes in host fitness during stress conditions. Symbiosis 2019, 79, 99–115. [Google Scholar] [CrossRef]
- Sonawane, H.; Borde, M.; Nikalje, G.; Terkar, A.; Math, S. HR-LC-MS based metabolic profiling of Fusarium solani a fungal endophyte associated with Avicennia officinalis. Curr. Res. Environ. Appl. Mycol. 2020, 10, 262–273. [Google Scholar] [CrossRef]
- Kushwaha, V.; Yadav, L.S. Endophytic fungi from mangrove plant Avicennia marina and their enzyme cellulase activity. Int. J. Sci. Math. Technol. Learn. 2023, 30, 117–130. [Google Scholar]
- Paliga, L.R.; Bonatto, C.; Camargo, A.F.; Cadamuro, R.D.; da Silveira Bastos, I.M.A.; de Freitas, A.C.O.; da Silva Rosa, M.; Silva, I.T.; Robl, D.; Stoco, P.H.; et al. Extraction of enzymes produced by endophytic fungi isolated from mangroves. J. Chem. Technol. Biotechnol. 2024, 99, 695–703. [Google Scholar] [CrossRef]
- Sopalun, K.; Iamtham, S. Isolation and screening of extracellular enzymatic activity of endophytic fungi isolated from Thai orchids. S. Afr. J. Bot. 2020, 134, 273–279. [Google Scholar] [CrossRef]
- De Paula, N.M.; da Silva, K.; Brugnari, T.; Haminiuk, C.W.I.; Maciel, G.M. Biotechnological potential of fungi from a mangrove ecosystem: Enzymes, salt tolerance and decolorization of a real textile effluent. Microbiol. Res. 2022, 254, 126899. [Google Scholar] [CrossRef]
- Bhat, T.K.; Singh, B.; Sharma, O.P. Microbial degradation of tannins—A current perspective. Biodegradation 1998, 9, 343–357. [Google Scholar] [CrossRef]
- Putra, I.P.Y.A.; Utami, K.S.; Hardini, J.; Wirasuta, I.M.A.G.; Ujam, N.T.; Ariantari, N.P. Fermentation, bioactivity and molecular identification of endophytic fungi isolated from mangrove Ceriops tagal. Biodiversitas 2023, 24, 3091–3098. [Google Scholar] [CrossRef]
- Prigione, V.; Trocini, B.; Spina, F.; Poli, A.; Romanisio, D.; Giovando, S.; Varese, G.C. Fungi from industrial tannins: Potential application in biotransformation and bioremediation of tannery wastewaters. Appl. Microbiol. Biotechnol. 2018, 102, 4203–4216. [Google Scholar] [CrossRef]
- Ahmed, A.I.; Abou-Taleb, K. Implementation of different fermentation techniques for induction of tannase and gallic acid using agro-residues substrates. Egypt. J. Microbiol. 2019, 54, 39–54. [Google Scholar] [CrossRef][Green Version]
- Batra, A.; Saxena, R.K. Potential tannase producers from the genera Aspergillus and Penicillium. Process Biochem. 2005, 40, 1553–1557. [Google Scholar] [CrossRef]
- Farag, A.M.; Hassan, S.W.; El-Says, A.M.; Ghanem, K.M. Purification, Characterization and Application of Tannase Enzyme Isolated from Marine Aspergillus nomius GWA5. J. Pure Appl. Microbiol. 2018, 12, 1939–1949. [Google Scholar] [CrossRef]
- Saad, M.M.; Saad, A.M.; Hassan, H.M.; Ibrahim, E.I.; Abdelraof, M.; Ali, B.A. Optimization of tannase production by Aspergillus glaucus in solid-state fermentation of black tea waste. Bioresour. Bioprocess. 2023, 10, 73. [Google Scholar] [CrossRef]
- Guo, L.D.; Hyde, K.D.; Liew, E.C.Y. Identification of endophytic fungi from Livistona chinensis based on morphology and rDNA sequences. New Phytol. 2000, 147, 617–630. [Google Scholar] [CrossRef]
- Raper, K.B.; Fennell, D.I. The Genus Aspergillus; The Williams and Wilkins Company: Baltimore, MD, USA, 1965; p. 612. [Google Scholar]
- Ellis, M.B. Dematiaceous Hyphomycetes; Commonwealth Mycological Institute: Kew, UK, 1971. [Google Scholar]
- Barnett, H.L.; Hunter, B.B. Illustrated Genera of Imperfect Fungi; Minneapolis, Burgess Publishing Company: Minneapolis, MN, USA, 1972. [Google Scholar]
- Von Arx, J.A. On Thielavia and some similar genera of Ascomycetes. Stud. Mycol. 1975, 8, 1–29. [Google Scholar]
- Ellis, M.B. More Dematiaceous Hyphomycetes; Commonwealth Mycological Institute: Kew, UK, 1976. [Google Scholar]
- Barron, G.L. The Nematode-Destroying Fungi; Canadian Biological Publications Ltd.: Guelph, ON, Canada, 1977; p. 140. [Google Scholar]
- Pitt, J.I. The Genus Penicillium and Its Teleomorphic States Eupenicillium and Talaromyces; Academic Press Inc. Ltd.: London, UK, 1979; p. 634. [Google Scholar]
- Carmichael, J.W.; Kendrick, B.; Conners, I.L.; Sigler, L. Genera of Hyphomycetes; University of Alberta Press: Edmonton, AB, Canada, 1980. [Google Scholar]
- Sutton, B.C. The Coelomycetes, Fungi Imperfecti with Pycnidia, Acervuli and Stromata; Commonwealth Mycological Institute: Kew, UK, 1980. [Google Scholar]
- Domsch, K.H.; Gams, W.; Anderson, T.H. Compendium of Soil Fungi; Academic Press: London, UK, 1980; Volume 1–2. [Google Scholar]
- Kirk, P.M.; Cannon, P.F.; Minter, D.W.; Stalpers, J.A. Dictionary of the Fungi, 10th ed.; CAB International: Wallingford, UK, 2008. [Google Scholar]
- Pinto, G.A.; Leite, S.G.; Terzi, S.C.; Couri, S. Selection of tannase-producing Aspergillus niger strains. Braz. J. Microbiol. 2001, 32, 24–26. [Google Scholar] [CrossRef]
- Sharma, S.; Bhat, T.K.; Dawra, R.K. A spectrophotometric method for assay of tannase using rhodamine. Anal. Biochem. 2000, 279, 85–89. [Google Scholar] [CrossRef]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Manjit; Yadav, A.; Aggarwal, N.K.; Kumar, K.; Kumar, A. Tannase production by Aspergillus fumigatus MA under solid-state fermentation. World J. Microbiol. Biotechnol. 2008, 24, 3023–3030. [Google Scholar] [CrossRef]
- Doyle, J.J.; Doyle, J.L. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem. Bull. 1987, 19, 11–15. [Google Scholar]
- White, T.J.; Bruns, T.; Lee, S.J.W.T.; Taylor, J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In PCR Protocols: A Guide to Methods and Applications; Innis, M.A., Gelfand, D.H., Sninsky, J.J., White, T.J., Eds.; Academic Press: Cambridge, MA, USA, 1990; Volume 18, pp. 315–322. [Google Scholar]
- Vilgalys, R.; Hester, M. Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. J. Bacteriol. 1990, 172, 4238–4246. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef]
- Ronquist, F.; Teslenko, M.; Van Der Mark, P.; Ayres, D.L.; Darling, A.; Höhna, S.; Larget, B.; Liu, L.; Suchard, M.A.; Huelsenbeck, J.P. MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive Tree of Life (iTOL) v4: Recent updates and new developments. Nucleic Acids Res. 2019, 47, W256–W259. [Google Scholar] [CrossRef]
- Bradoo, S.; Gupta, R.; Saxena, R.K. Screening of extracellular tannase producing fungi: Development of a rapid and simple plate assay. J. Gen. Appl. Microbiol. 1996, 42, 325–329. [Google Scholar] [CrossRef]
- Dreyfuss, M.M.; Chapela, I.H. Potential of fungi in the discovery of novel, low-molecular weight pharmaceuticals. In Discovery of Novel Natural Products with Therapeutic Potential; Gullo, V.P., Ed.; Newnes: Bathurst, Australia, 1994; pp. 49–80. [Google Scholar]
- Du, W.; Yao, Z.; Li, J.; Sun, C.; Xia, J.; Wang, B.; Shi, D.; Ren, L. Diversity and antimicrobial activity of endophytic fungi isolated from Securinega suffruticosa in the Yellow River Delta. PLoS ONE 2020, 15, e0229589. [Google Scholar] [CrossRef]
- Mishra, V.K.; Singh, G.; Passari, A.K.; Yadav, M.K.; Gupta, V.K.; Singh, B.P. Distribution and antimicrobial potential of endophytic fungi associated with ethnomedicinal plant Melastoma malabathricum L. J. Environ. Biol. 2016, 37, 229. [Google Scholar]
- Pecoraro, L.; Caruso, T.; Cai, L.; Gupta, V.K.; Liu, Z.J. Fungal networks and orchid distribution: New insights from above-and below-ground analyses of fungal communities. IMA Fungus 2018, 9, 1–11. [Google Scholar] [CrossRef]
- Dos Reis, J.B.A.; Lorenzi, A.S.; do Vale, H.M.M. Methods used for the study of endophytic fungi: A review on methodologies and challenges, and associated tips. Arch. Microbiol. 2022, 204, 675. [Google Scholar] [CrossRef] [PubMed]
- Kumaresan, V.; Suryanarayanan, T.S.; Johnson, J.A. Ecology of mangrove endophytes. In Fungi in Marine Environments; Fungal Diversity Research Series; Fungal Diversity Press: Hong Kong, 2002; pp. 145–166. [Google Scholar]
- George, D.S.; Ong, C.B. Improvement of tannase production under submerged fermentation by Aspergillus niger FBT1 isolated from a mangrove forest. BioTechnologia. J. Biotechnol. Comput. Biol. Bionanotechnol. 2013, 94, 451–456. [Google Scholar] [CrossRef]
- Neethu, R.S.; Pradeep, S. Isolation and characterization of potential tannase producing fungi from mangroves and tanneries. Indian J. Appl. Microbiol. 2018, 21, 1–13. [Google Scholar] [CrossRef]
- Ahmed, A.I.; Abou-Taleb, K.A.; Abd-Elhalim, B.T. Characterization and application of tannase and gallic acid produced by co-fungi of Aspergillus niger and Trichoderma viride utilizing agro-residues substrates. Sci. Rep. 2023, 13, 16755. [Google Scholar] [CrossRef] [PubMed]
- Malgireddy, N.R.; Nimma, L.N.R. Optimal conditions for production of tannase from newly isolated Aspergillus terrus under solidstate fermentation. Eur. J. Biotechnol. Biosci. 2015, 3, 56–64. [Google Scholar]
- Cruz, R.; de Lima, J.S.; Fonseca, J.C.; Gomes, J.E.G.; Silva, J.I.D.S., Jr.; Moreira, K.A.; de Souza-Motta, C.M. Promising substrates to increase the production of tannase under solid state fermentation (SSF) by Penicillium spp. Afr. J. Biotechnol. 2017, 16, 2121–2126. [Google Scholar]
- Dhiman, S.; Mukherjee, G.; Singh, A.K. Recent trends and advancements in microbial tannase-catalyzed biotransformation of tannins: A review. Int. Microbiol. 2018, 21, 175–195. [Google Scholar] [CrossRef]
- Andrade, P.M.; Baptista, L.; Bezerra, C.O.; Peralta, R.M.; Góes-Neto, A.; Uetanabaro, A.P.; Costa, A.M. Immobilization and characterization of tannase from Penicillium rolfsii CCMB 714 and its efficiency in apple juice clarification. J. Food Meas. Charact. 2021, 15, 1005–1013. [Google Scholar] [CrossRef]
- D’Souza, P.S.; Mansy, T.K.; Preethi, T.C.; Gunashree, B.S. Tamarindus indica seed induced tannase production from Aspergillus niger. Biomedicine 2022, 42, 752–756. [Google Scholar]
- Gayen, S.; Ghosh, U. Purification and characterization of tannin acyl hydrolase produced by mixed solid-state fermentation of wheat bran and marigold flower by Penicillium notatum NCIM 923. BioMed Res. Int. 2013, 2013, 596380. [Google Scholar] [CrossRef]
- Sivashanmugam, K.; Jayaraman, G. Production and partial purification of extracellular tannase by Klebsiella pneumoniae MTCC 7162 isolated from tannery effluent. Afr. J. Biotechnol. 2011, 10, 1364–1374. [Google Scholar]
- Albuquerque, K.K.; Albuquerque, W.W.; Costa, R.M.; Batista, J.M.S.; Marques, D.A.; Bezerra, R.P.; Herculano, P.N.; Porto, A.L. Biotechnological potential of a novel tannase-acyl hydrolase from Aspergillus sydowii using waste coir residue: Aqueous two-phase system and chromatographic techniques. Biocatal. Agric. Biotechnol. 2020, 23, 101453. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
