Morphology and Phylogeny Reveal New Species and Records of Diplodia, Dothiorella, and Phaeobotryon Associated with Tree Cankers in Xizang, China
Abstract
1. Introduction
2. Materials and Methods
2.1. Specimens and Strains
2.2. Morphological Observations
2.3. DNA Extraction and Amplification
2.4. Molecular Phylogeny
3. Results
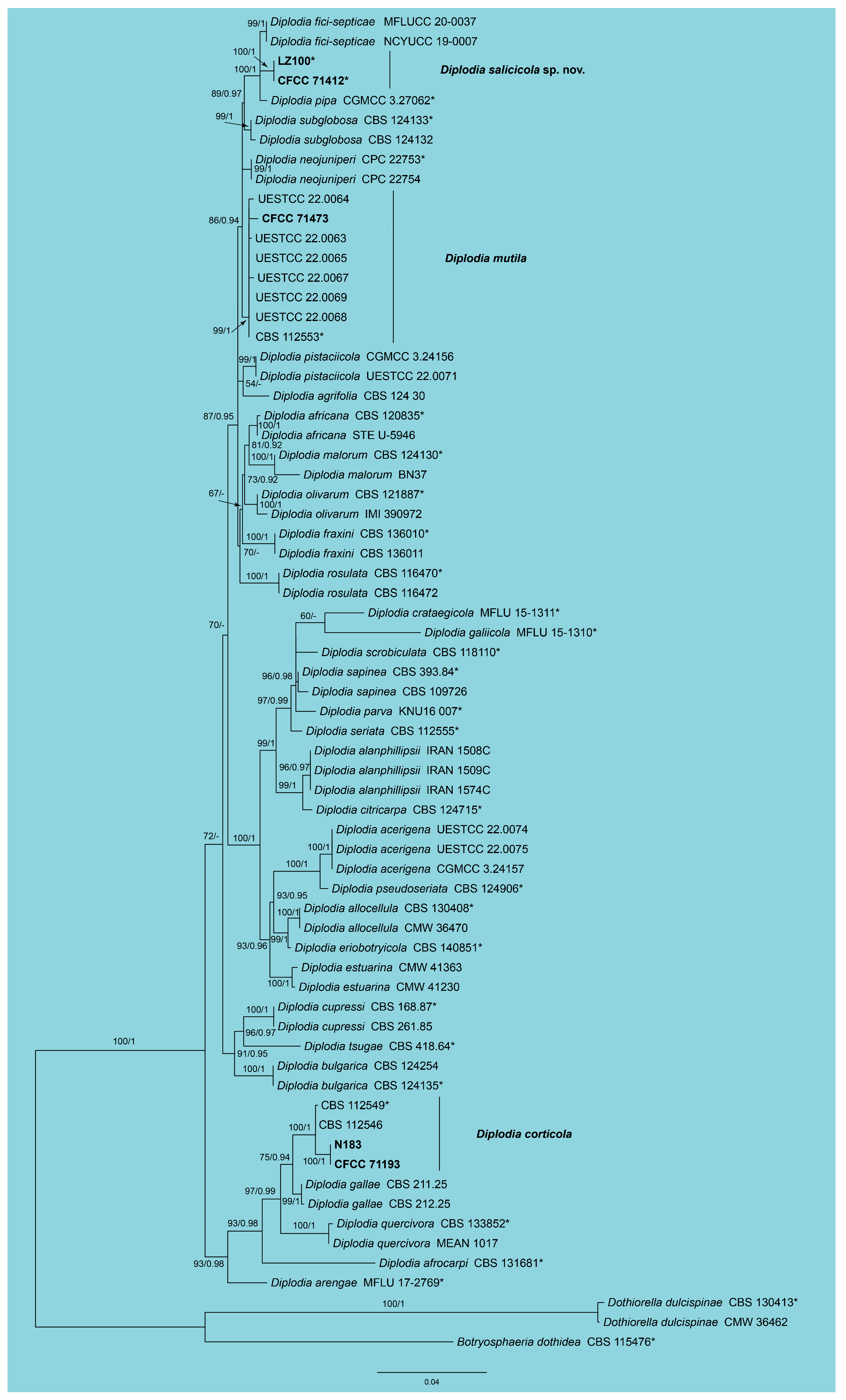
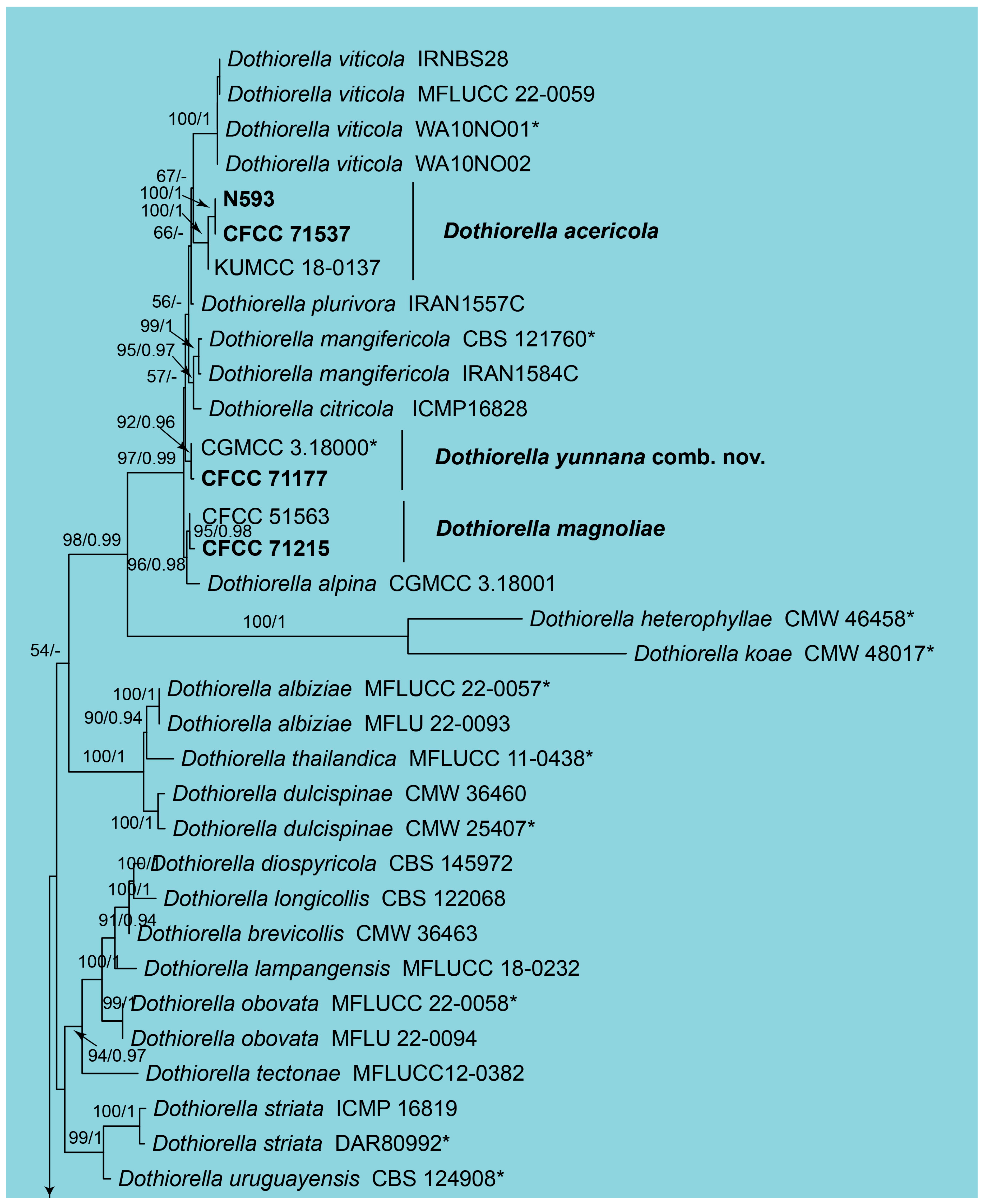
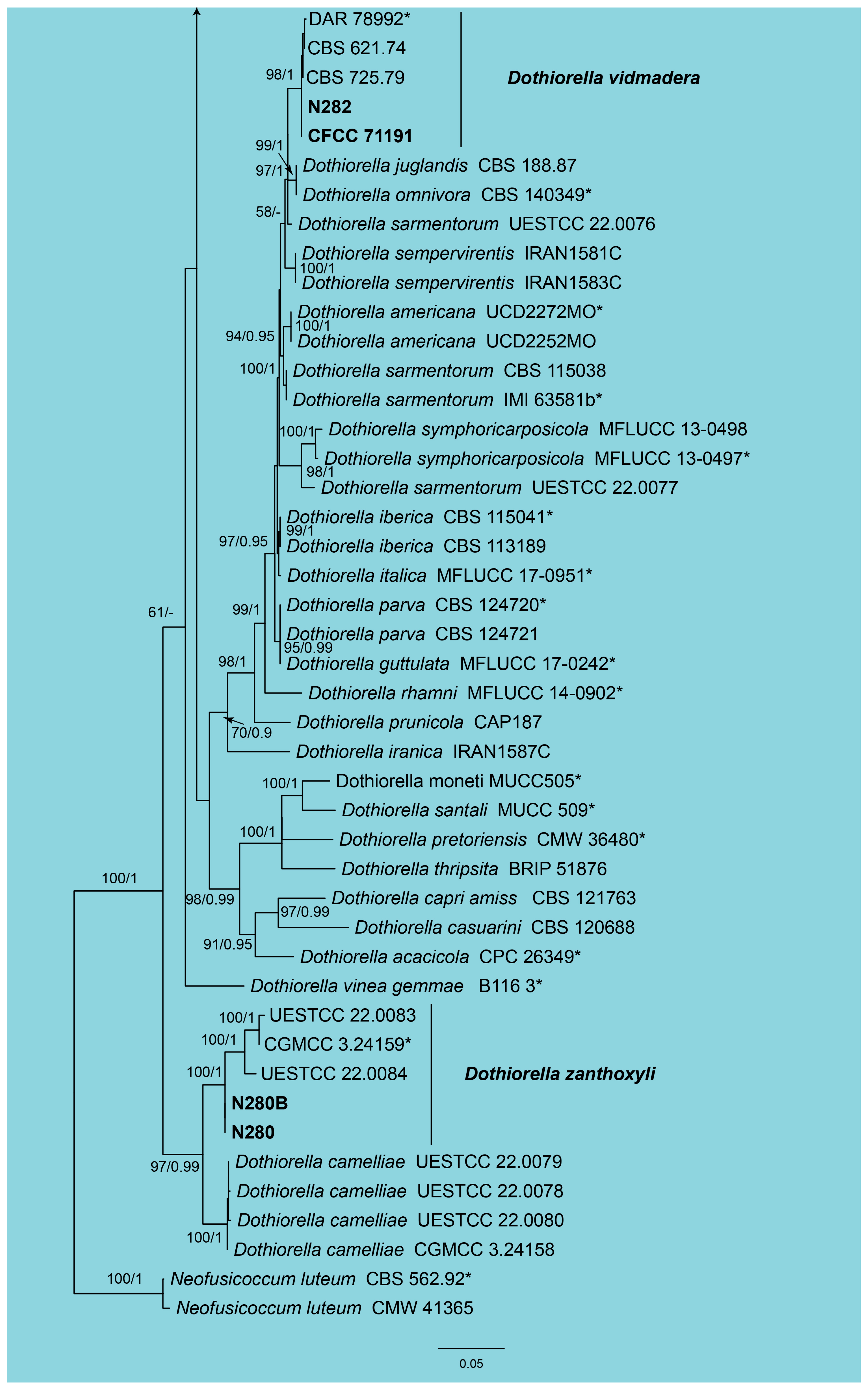
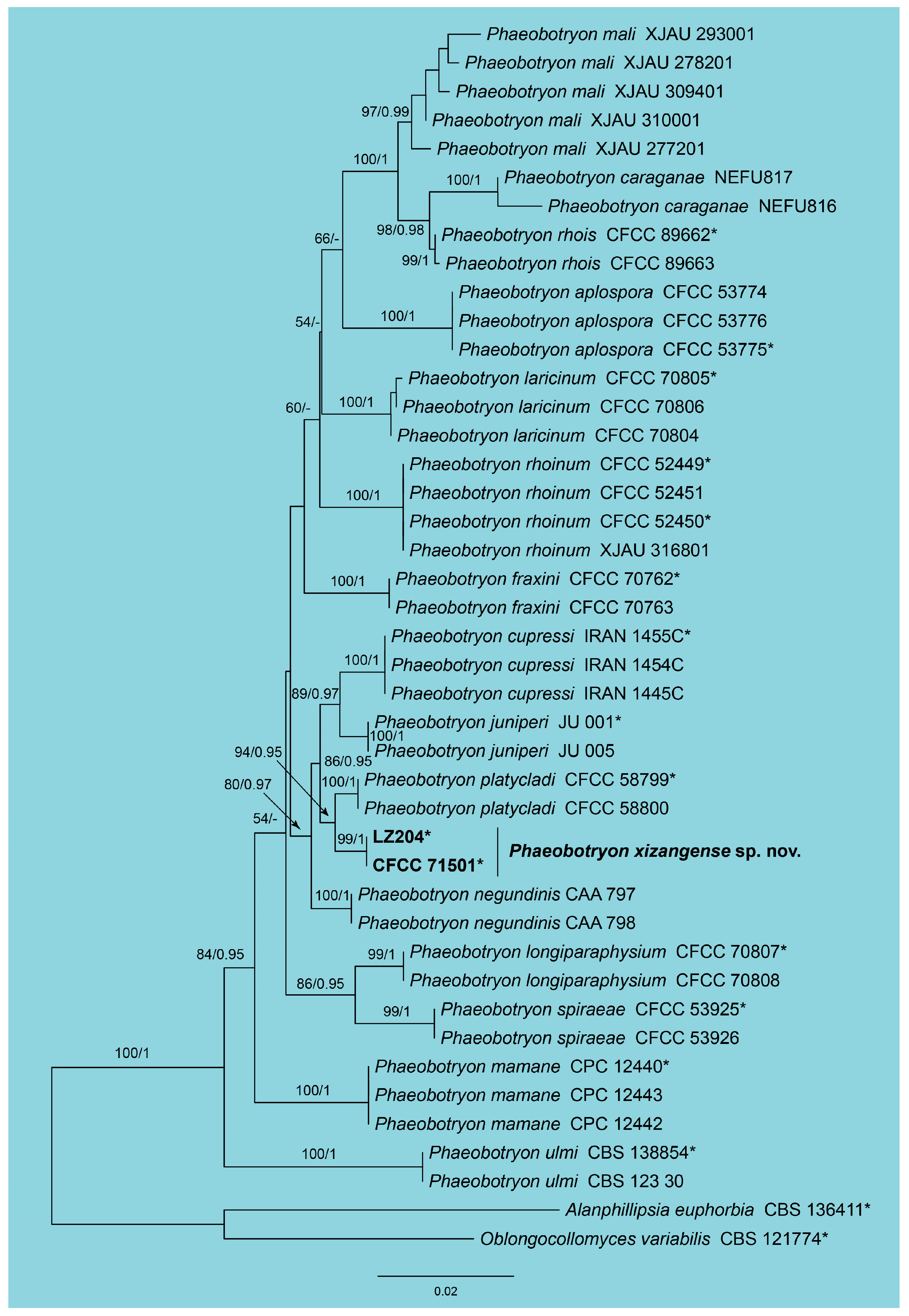
3.1. Phylogeny
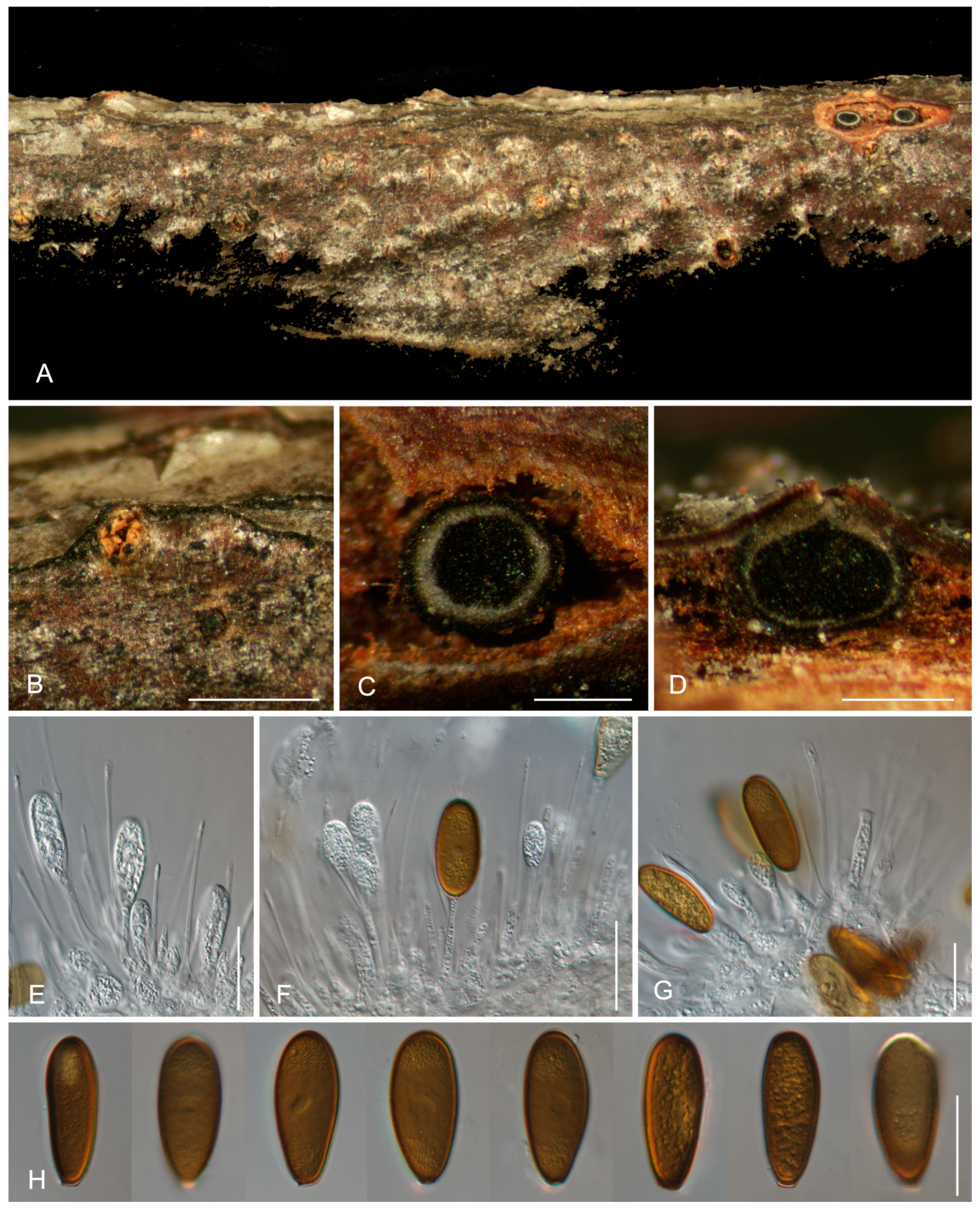
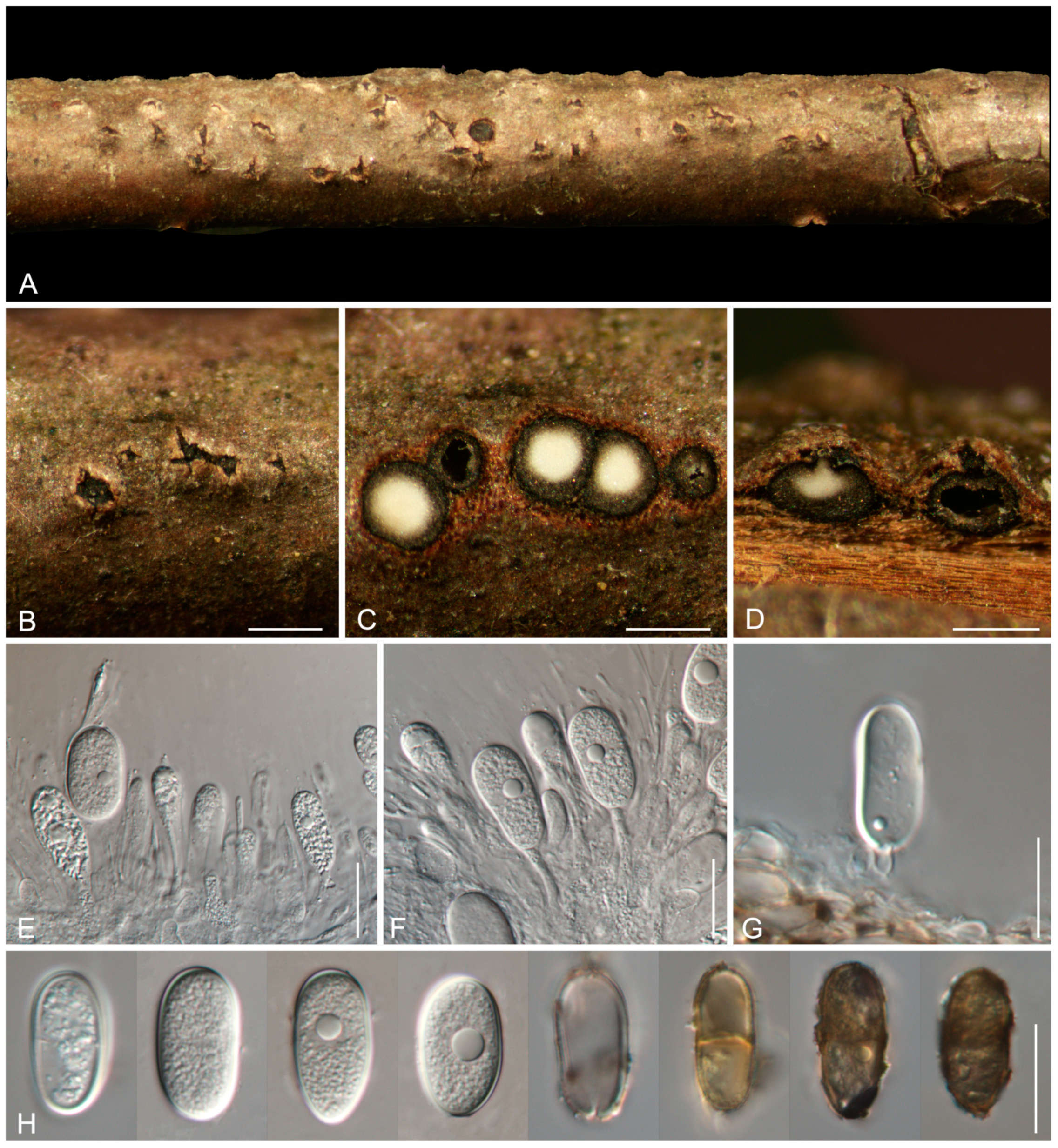
3.2. Taxonomy
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Fan, X.L.; Bezerra, J.D.P.; Tian, C.M.; Crous, P.W. Cytospora (Diaporthales) in China. Persoonia 2020, 45, 1–45. [Google Scholar] [CrossRef] [PubMed]
- Jiang, N.; Fan, X.L.; Crous, P.W.; Tian, C.M. Species of Dendrostoma (Erythrogloeaceae, Diaporthales) associated with chestnut and oak canker diseases in China. MycoKeys 2019, 48, 67–96. [Google Scholar] [CrossRef]
- Zhu, H.Y.; Tian, C.M.; Fan, X.L. Studies of botryosphaerialean fungi associated with canker and dieback of tree hosts in Dongling Mountain of China. Phytotaxa 2018, 348, 63–76. [Google Scholar] [CrossRef]
- Weiland, J.E.; Sniezko, R.A.; Wiseman, M.S.; Serdani, M.; Putnam, M.L. First report of Phaeobotryon cupressi causing canker of Calocedrus decurrens (incense-cedar) in Oregon. Plant Dis. 2016, 100, 1793. [Google Scholar] [CrossRef]
- Jiang, N.; Voglmayr, H.; Ma, C.Y.; Xue, H.; Piao, C.G.; Li, Y. A new Arthrinium-like genus of Amphisphaeriales in China. MycoKeys 2022, 92, 27–43. [Google Scholar] [CrossRef]
- Wen, Y.; Li, X.; Xie, K.Z.; Deng, W.Q.; Zhuang, W.Y. Systematics and species diversity of botryosphaeriaceous fungi. Biodivers. Sci. 2017, 25, 874–885. [Google Scholar]
- Jiang, N.; Fan, X.L.; Tian, C.M. Identification and pathogenicity of Cryphonectriaceae species associated with chestnut canker in China. Plant Pathol. 2019, 68, 1132–1145. [Google Scholar] [CrossRef]
- Jiang, N.; Yang, Q.; Fan, X.L.; Tian, C.M. Identification of six Cytospora species on Chinese chestnut in China. MycoKeys 2020, 62, 1–25. [Google Scholar] [CrossRef]
- Pan, M.; Zhu, H.; Bezerra, J.D.P.; Bonthond, G.; Tian, C.; Fan, X. Botryosphaerialean fungi causing canker and dieback of tree hosts from Mount Yudu in China. Mycol. Progr. 2019, 18, 1341–1361. [Google Scholar] [CrossRef]
- Zlatković, M.; Keča, N.; Wingfield, M.J.; Jami, F.; Slippers, B. Botryosphaeriaceae associated with the die-back of ornamental trees in the Western Balkans. Anton. Leeuw. Int. J. G 2016, 109, 543–564. [Google Scholar] [CrossRef]
- Chethana, K.W.T.; Li, X.; Zhang, W.; Hyde, K.D.; Yan, J. Trail of decryption of molecular research on Botryosphaeriaceae in woody plants. Phytopathol. Mediterr. 2016, 55, 147–171. [Google Scholar]
- Manawasinghe, I.S.; Phillips, A.J.L.; Hyde, K.D.; Chethana, K.W.T.; Zhang, W.; Zhao, W.S.; Yan, J.Y.; Li, X.H. Mycosphere Essays 14: Assessing the aggressiveness of plant pathogenic Botryosphaeriaceae. Mycosphere 2016, 7, 883–892. [Google Scholar] [CrossRef]
- Ilyukhin, E.; Ellouze, W. First report of Phaeobotryon negundinis associated with twig and branch dieback of Malus domestica trees in southern Ontario, Canada and worldwide. J. Plant Pathol. 2022, 105, 355–356. [Google Scholar] [CrossRef]
- Slippers, B.; Wingfield, M.J. Botryosphaeriaceae as endophytes and latent pathogens of woody plants: Diversity, ecology and impact. Fungal Biol. Rev. 2007, 21, 90–106. [Google Scholar] [CrossRef]
- Jiang, N.; Fan, X.; Tian, C. Identification and characterization of leaf-inhabiting fungi from Castanea plantations in China. J. Fungi 2021, 7, 64. [Google Scholar] [CrossRef]
- Jiang, N.; Phillips, A.J.L.; Zhang, Z.X.; Tian, C.M. Morphological and molecular identification of two novel species of Melanops in China. Mycosphere 2018, 9, 1187–1196. [Google Scholar] [CrossRef]
- Jiang, N.; Wang, X.W.; Liang, Y.M.; Tian, C.M. Lasiodiplodia cinnamomi sp. nov. from Cinnamomum camphora in China. Mycotaxon 2018, 133, 249–259. [Google Scholar] [CrossRef]
- Schoch, C.L.; Shoemaker, R.A.; Seifert, K.A.; Hambleton, S.; Spatafora, J.W.; Crous, P.W. A multigene phylogeny of the Dothideomycetes using four nuclear loci. Mycologia 2006, 98, 1041–1052. [Google Scholar] [CrossRef] [PubMed]
- Slippers, B.; Boissin, E.; Phillips, A.J.L.; Groenewald, J.Z.; Lombard, L.; Wingfeld, M.J.; Postma, A.; Burgess, T.; Crous, P.W. Phylogenetic lineages in the Botryosphaeriales: A systematic and evolutionary framework. Stud. Mycol. 2013, 76, 31–49. [Google Scholar] [CrossRef]
- Slippers, B.; Smit, W.A.; Crous, P.W.; Coutinho, T.A.; Wingfield, B.D.; Wingfield, M.J. Taxonomy, phylogeny and identification of Botryosphaeriaceae associated with pome and stone fruit trees in South Africa and other regions of the world. Plant Pathol. 2007, 56, 128–139. [Google Scholar] [CrossRef]
- Phillips, A.J.L.; Hyde, K.D.; Alves, A.; Liu, J.K. Families in Botryosphaeriales: A phylogenetic, morphological and evolutionary perspective. Fungal Divers. 2019, 94, 1–22. [Google Scholar] [CrossRef]
- Liu, J.K.; Phookamsak, R.; Wikee, S.; Li, Y.M.; Ariyawansha, H.; Boonmee, S.; Chomnunti, P.; Dai, D.Q.; Bhat, J.D.; Romero, A.I.; et al. Towards a natural classification of Botryosphaeriales. Fungal Divers. 2012, 57, 149–210. [Google Scholar] [CrossRef]
- Mehl, J.W.; Slippers, B.; Roux, J.; Wingfield, M.J. Overlap of latent pathogens in the Botryosphaeriaceae on a native and agricultural host. Fungal Biol. 2017, 121, 405–419. [Google Scholar] [CrossRef] [PubMed]
- Hongsanan, S.; Hyde, K.D.; Phookamsak, R.; Wanasinghe, D.N.; McKenzie, E.H.C.; Sarma, V.V.; Boonmee, S.; Luecking, R.; Bhat, D.J.; Liu, N.G.; et al. Refined families of Dothideomycetes: Orders and families incertae sedis in Dothideomycetes. Fungal Divers. 2020, 105, 17–318. [Google Scholar]
- Abdollahzadeh, J.; Goltapeh, E.M.; Javadi, A.; Shams-Bakhsh, M.; Zare, R.; Phillips, A.J.L. Barriopsis iraniana and Phaeobotryon cupressi: Two new species of the Botryosphaeriaceae from trees in Iran. Persoonia 2009, 23, 1–8. [Google Scholar] [CrossRef]
- Abdollahzadeh, J.; Javadi, A.; Mohammadi, G.E.; Zare, R.; Phillips, A.J.L. Phylogeny and morphology of four new species of Lasiodiplodia from Iran. Persoonia 2010, 25, 1–10. [Google Scholar] [CrossRef]
- Chen, J.; Hao, X.; Liu, X.; Liu, Z.; Gao, F. Identification of Caragana arborescens shoot blight disease caused by Phaeobotryon caraganae sp. nov. (Botryosphaeriales) in China. Eur. J. Plant Pathol. 2019, 155, 537–544. [Google Scholar] [CrossRef]
- Fan, X.L.; Hyde, K.D.; Liu, J.; Liang, Y.; Tian, C.M. Multigene phylogeny and morphology reveal Phaeobotryon rhois sp. nov. (Botryosphaeriales, Ascomycota). Phytotaxa 2015, 205, 90–98. [Google Scholar] [CrossRef]
- Phillips, A.J.L.; Alves, A.; Abdollahzadeh, J.; Slippers, B.; Wingfield, M.J.; Groenewald, J.Z.; Crous, P.W. The Botryosphaeriaceae: Genera and species known from culture. Stud. Mycol. 2013, 76, 51–167. [Google Scholar] [CrossRef]
- Phillips, A.J.L.; Alves, A.; Correia, A.; Luque, J. Two new species of Botryosphaeria with brown, 1-septate ascospores and Dothiorella anamorphs. Mycologia 2005, 97, 513–529. [Google Scholar] [CrossRef]
- Alves, A.; Correia, A.; Luque, J.; Phillips, A. Botryosphaeria corticola, sp. nov. on Quercus species, with notes and description of Botryosphaeria stevensii and its anamorph, Diplodia mutila. Mycologia 2004, 96, 598–613. [Google Scholar] [CrossRef] [PubMed]
- Smith, D.R.; Stanosz, G.R. Occurrence of Diplodia corticola, including new oak host records, in Wisconsin, USA. For. Pathol. 2018, 48, e12427. [Google Scholar] [CrossRef]
- Lódolo, X.V.; Lutz, M.C.; Mondino, P.; Ousset, J.; Sosa, M.C. First report of Diplodia seriata, Diplodia mutila, and Dothiorella omnivora associated with apple cankers and dieback in Rio Negro, Argentina. Plant Dis. 2022, 106, 325. [Google Scholar] [CrossRef]
- Li, G.Q.; Liang, Y.H.; Lu, L.Q.; Liu, F.F. A new species and a new Chinese record of Diplodia associated with canker disease on loquat in China. Mycosystema 2024, 43, 240033. [Google Scholar]
- Dissanayake, A.J.; Camporesi, E.; Hyde, K.D.; Phillips, A.J.L.; Fu, C.Y.; Yan, J.Y.; Li, X.H. Dothiorella species associated with woody hosts in Italy. Mycosphere 2016, 7, 51–63. [Google Scholar] [CrossRef]
- Dissanayake, A.J.; Phillips, A.J.L.; Li, X.H.; Hyde, K.D. Botryosphaeriaceae: Current status of genera and species. Mycosphere 2016, 7, 1001–1073. [Google Scholar] [CrossRef]
- Batista, E.; Lopes, A.; Alves, A. Botryosphaeriaceae species on forest trees in Portugal: Diversity, distribution and pathogenicity. Eur. J. Plant Pathol. 2020, 158, 693–720. [Google Scholar] [CrossRef]
- Pitt, W.M.; Úrbez-Torres, J.R.; Trouillas, F.P. Dothiorella vidmadera, a novel species from grapevines in Australia and notes on Spencermartinsia. Fungal Divers. 2013, 61, 209–219. [Google Scholar] [CrossRef]
- You, C.J.; Liu, X.; Li, L.X.; Tsui, C.K.M.; Tian, C.M. Dothiorella magnoliae, a new species associated with dieback of Magnolia grandiflora from China. Mycosphere 2017, 8, 1031–1041. [Google Scholar] [CrossRef]
- Theissen, F.; Sydow, H. Die Dothideales. kritisch-systematisch originaluntersuchungen. Ann. Mycol. 1915, 13, 431–746. [Google Scholar]
- Li, W.L.; Liang, R.R.; Dissanayake, A.J.; Liu, J.K. Botryosphaerialean fungi associated with woody oil plants cultivated in Sichuan Province, China. MycoKeys 2023, 97, 71–116. [Google Scholar] [CrossRef]
- Lin, L.; Bai, Y.; Pan, M.; Tian, C.; Fan, X. Morphology and molecular analyses reveal three new species of Botryosphaeriales isolated from diseased plant branches in China. MycoKeys 2023, 97, 1–19. [Google Scholar] [CrossRef] [PubMed]
- Tennakoon, D.S.; Kuo, C.H.; Maharachchikumbura, S.S.; Thambugala, K.M.; Gentekaki, E.; Phillips, A.J.; Hyde, K.D. Taxonomic and phylogenetic contributions to Celtis formosana, Ficus ampelas, F. septica, Macaranga tanarius and Morus australis leaf litter inhabiting microfungi. Fungal Divers. 2021, 108, 1–215. [Google Scholar] [CrossRef]
- Wu, Y.; Peng, C.; Yuan, R.; Zhang, M.; Hu, Y.; Tian, C. New species and records of Botryosphaeriales (Dothideomycetes) associated with tree dieback in Beijing, China. MycoKeys 2024, 106, 225–250. [Google Scholar] [CrossRef]
- Zhu, Y.; Liang, Y.; Peng, C. New species and records of Phaeobotryon (Botryosphaeriales, Botryosphaeriaceae) from Larix in China. MycoKeys 2025, 112, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Doyle, J.J. Isolation of plant DNA from fresh tissue. Focus 1990, 12, 13–15. [Google Scholar]
- Carbone, I.; Kohn, L.M. A method for designing primer sets for speciation studies in filamentous ascomycetes. Mycologia 1999, 3, 553–556. [Google Scholar] [CrossRef]
- Glass, N.L.; Donaldson, G.C. Development of primer sets designed for use with the PCR to amplify conserved genes from filamentous ascomycetes. Appl. Environ. Microb. 1995, 61, 1323–1330. [Google Scholar] [CrossRef] [PubMed]
- White, T.J.; Bruns, T.; Lee, S.; Taylor, J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. PCR Protoc. Guide Methods Appl. 1990, 18, 315–322. [Google Scholar]
- Vilgalys, R.; Hester, M. Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. J. Bacteriol. 1990, 172, 4238–4246. [Google Scholar] [CrossRef]
- Katoh, K.; Toh, H. Parallelization of the MAFFT multiple sequence alignment program. Bioinformatics 2010, 26, 1899–1900. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef]
- Miller, M.A.; Pfeiffer, W.; Schwartz, T. Creating the CIPRES Science Gateway for Inference of Large Phylogenetic Trees; Institute of Electrical and Electronics Engineers: New Orleans, LA, USA, 2010. [Google Scholar]
- Ronquist, F.; Huelsenbeck, J.P. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 2003, 19, 1572–1574. [Google Scholar] [CrossRef]
- Stamatakis, A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 2014, 30, 1312–1313. [Google Scholar] [CrossRef] [PubMed]
- Phookamsak, R.; Hyde, K.D.; Jeewon, R.; Bhat, D.J.; Jones, E.G.; Maharachchikumbura, S.S.; Xu, J. Fungal diversity notes 929–1035: Taxonomic and phylogenetic contributions on genera and species of fungi. Fungal Divers. 2019, 95, 1–273. [Google Scholar] [CrossRef]
- Pan, M.; Tian, C.M.; Fan, X.L. Two fungal species associated with canker disease of Jujube tree in China. Mycoasia 2021, 3, 1–21. [Google Scholar] [CrossRef]
- Phillips, A.J.L.; Alves, A.; Pennycook, S.R.; Johnston, P.R.; Ramaley, A.; Akulov, A.; Crous, P.W. Resolving the phylogenetic and taxonomic status of dark-spored teleomorph genera in the Botryosphaeriaceae. Persoonia 2008, 21, 29–55. [Google Scholar] [CrossRef] [PubMed]
- Yang, T.; Groenewald, J.Z.; Cheewangkoon, R.; Jami, F.; Abdollahzadeh, J.; Lombard, L.; Crous, P.W. Families, genera, and species of Botryosphaeriales. Fungal Biol. 2017, 121, 322–346. [Google Scholar] [CrossRef]
- Zhang, M.; He, W.; Wu, J.R.; Zhang, Y. Two new species of Spencermartinsia (Botryosphaeriaceae, Botryosphaeriales) from China. Mycosphere 2016, 7, 942–949. [Google Scholar] [CrossRef]
- Wang, C.B.; Yang, J.; Li, Y.; Xue, H.; Piao, C.G.; Jiang, N. Multi-gene phylogeny and morphology of two new Phyllosticta (Phyllostictaceae, Botryosphaeriales) species from China. MycoKeys 2023, 95, 189–207. [Google Scholar] [CrossRef]
- Zhao, L.; Cai, J.; He, W.; Zhang, Y. Macrophomina vaccinii sp. nov. causing blueberry stem blight in China. MycoKeys 2019, 55, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.E.; Meng, C.R.; Phillips, A.J.; Wang, Y. Two new Botryosphaeria (Botryosphaeriales, Botryosphaeriaceae) species in China. MycoKeys 2022, 94, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Groenewald, J.Z.; Lombard, L.; Schumacher, R.K.; Phillips, A.J.L.; Crous, P.W. Evaluating species in Botryosphaeriales. Persoonia 2021, 46, 63–115. [Google Scholar] [CrossRef] [PubMed]
| Species | Isolates | GenBank Accession Number | ||
|---|---|---|---|---|
| ITS | tef1 | tub2 | ||
| Botryosphaeria dothidea | CBS 115476 * | AY236949 | AY236898 | AY236927 |
| Diplodia acerigena | CGMCC 3.24157 | OQ190518 | OQ241452 | NA |
| Diplodia acerigena | UESTCC 22.0074 | OQ190519 | OQ241453 | OQ338163 |
| Diplodia acerigena | UESTCC 22.0075 | OQ190520 | OQ241454 | OQ338164 |
| Diplodia africana | CBS 120835 * | KF766155 | KF766397 | KF766129 |
| Diplodia africana | STE-U 5946 | EF445344 | EF445383 | NA |
| Diplodia afrocarpi | CBS 131681 * | MT587333 | MT592035 | MT592471 |
| Diplodia agrifolia | CBS 124.30 | KX464087 | KX464557 | KX464783 |
| Diplodia alanphillipsii | IRAN 1508C | KF890208 | KF890190 | NA |
| Diplodia alanphillipsii | IRAN 1509C | KF890209 | KF890191 | NA |
| Diplodia alanphillipsii | IRAN 1574C | MT258875 | MT270153 | NA |
| Diplodia allocellula | CBS 130408 * | JQ239397 | JQ239384 | JQ239378 |
| Diplodia allocellula | CMW 36470 | JQ239399 | JQ239386 | JQ239380 |
| Diplodia arengae | MFLU 17-2769 * | MG762771 | MG762774 | MG783039 |
| Diplodia bulgarica | CBS 124254 | GQ923853 | GQ923821 | NA |
| Diplodia bulgarica | CBS 124135 * | GQ923852 | GQ923820 | NA |
| Diplodia citricarpa | CBS 124715 * | KF890207 | KF890189 | KX464784 |
| Diplodia corticola | CBS 112549 * | AY259100 | AY573227 | DQ458853 |
| Diplodia corticola | CBS 112546 | AY259090 | EU673310 | EU673117 |
| Diplodia corticola | CFCC 71193 | PV264850 | NA | PV339814 |
| Diplodia corticola | N183 | PV264851 | NA | PV339815 |
| Diplodia crataegicola | MFLU 15-1311 * | KT290244 | KT290248 | KT290246 |
| Diplodia cupressi | CBS 168.87 * | DQ458893 | DQ458878 | DQ458861 |
| Diplodia cupressi | CBS 261.85 | DQ458894 | DQ458879 | DQ458862 |
| Diplodia eriobotryicola | CBS 140851 * | KT240355 | KT240193 | MG015806 |
| Diplodia estuarina | CMW 41363 | KP860829 | KP860674 | KP860752 |
| Diplodia estuarina | CMW 41230 | KP860830 | KP860675 | KP860753 |
| Diplodia fici-septicae | MFLUCC 20-0037 | MW063180 | MW183802 | NA |
| Diplodia fici-septicae | NCYUCC 19-0007 | MW063181 | MW183803 | NA |
| Diplodia fraxini | CBS 136010 * | KF307700 | KF318747 | MG015807 |
| Diplodia fraxini | CBS 136011 | KF307711 | KF318748 | MG015808 |
| Diplodia galiicola | MFLU15-1310 * | KT290245 | KT290249 | MT592471 |
| Diplodia gallae | CBS 211.25 | KX464090 | KX464564 | KX464795 |
| Diplodia gallae | CBS 212.25 | KX464091 | KX464565 | KX464796 |
| Diplodia malorum | CBS 124130 * | GQ923865 | GQ923833 | NA |
| Diplodia malorum | BN-37 | KT240360 | KT240198 | NA |
| Diplodia mutila | CBS 112553 * | AY259093 | AY573219 | KY554743 |
| Diplodia mutila | UESTCC 22.0064 | OQ190521 | OQ241455 | OQ338165 |
| Diplodia mutila | UESTCC 22.0065 | OQ190522 | OQ241456 | OQ338166 |
| Diplodia mutila | UESTCC 22.0069 | OQ190523 | OQ241457 | OQ338167 |
| Diplodia mutila | UESTCC 22.0068 | OQ190524 | OQ241458 | OQ338168 |
| Diplodia mutila | UESTCC 22.0067 | OQ190525 | OQ241459 | OQ338169 |
| Diplodia mutila | UESTCC 22.0063 | OQ190526 | OQ241460 | OQ338170 |
| Diplodia mutila | CFCC 71473 | PV264852 | PV268109 | PV339816 |
| Diplodia neojuniperi | CPC 22753 * | KM006431 | KM006462 | NA |
| Diplodia neojuniperi | CPC 22754 | KM006432 | KM006463 | NA |
| Diplodia olivarum | CBS 121887 * | EU392302 | EU392279 | HQ660079 |
| Diplodia olivarum | IMI 390972 | HM028640 | HQ660078 | HQ660080 |
| Diplodia parva | KNU16-007 * | LC417238 | LC435495 | LC522938 |
| Diplodia pipa | CGMCC 3.27062 * | PP192032 | PP197939 | PP197952 |
| Diplodia pistaciicola | CGMCC 3.24156 | OQ190527 | OQ241461 | OQ338171 |
| Diplodia pistaciicola | UESTCC 22.0071 | OQ190528 | OQ241462 | OQ275062 |
| Diplodia pseudoseriata | CBS 124906 * | EU080927 | EU863181 | MG015820 |
| Diplodia quercivora | CBS 133852 * | JX894205 | JX894229 | MG015821 |
| Diplodia quercivora | MEAN 1017 | KU311198 | KU311201 | NA |
| Diplodia rosulata | CBS 116470 * | EU430265 | EU430267 | EU673132 |
| Diplodia rosulata | CBS 116472 | EU430266 | EU430268 | EU673131 |
| Diplodia salicicola sp. nov. | CFCC 71412 * | PV264853 | PV268110 | PV339817 |
| Diplodia salicicola sp. nov. | LZ100 * | PV264854 | PV268111 | PV339818 |
| Diplodia sapinea | CBS 393.84 * | DQ458895 | DQ458880 | DQ458863 |
| Diplodia sapinea | CBS 109726 | KX464094 | KX464568 | KX464800 |
| Diplodia scrobiculata | CBS 118110 * | AY253292 | AY624253 | AY624258 |
| Diplodia seriata | CBS 112555 * | AY259094 | AY573220 | DQ458856 |
| Diplodia subglobosa | CBS 124133 * | GQ923856 | GQ923824 | MT592576 |
| Diplodia subglobosa | CBS 124132 | DQ458887 | DQ458871 | DQ458852 |
| Diplodia tsugae | CBS 418.64 * | DQ458888 | DQ458873 | DQ458855 |
| Dothiorella dulcispinae | CBS 130413 | JQ239400 | JQ239387 | JQ239373 |
| Dothiorella dulcispinae | CMW 36462 | JQ239402 | JQ239389 | JQ239375 |
| Species | Isolates | GenBank Accession Number | ||
|---|---|---|---|---|
| ITS | tef1 | tub2 | ||
| Dothiorella acacicola | CPC 26349 * | NR_145255 | KX228376 | NA |
| Dothiorella acericola | KUMCC 18-0137 | MK359449 | MK361182 | NA |
| Dothiorella acericola | CFCC 71537 | PV264855 | PV268112 | PV339819 |
| Dothiorella acericola | N593 | PV264856 | PV268113 | PV339820 |
| Dothiorella albiziae | MFLUCC 22-0057 * | ON751762 | ON799588 | ON799590 |
| Dothiorella albiziae | MFLU 22-0093 | ON707683 | NA | ON677453 |
| Dothiorella alpina | CGMCC 3.18001 | KX499645 | KX499651 | NA |
| Dothiorella americana | UCD2272MO * | HQ288219 | HQ288263 | HQ288298 |
| Dothiorella americana | UCD2252MO | HQ288218 | HQ288262 | HQ288297 |
| Dothiorella brevicollis | CMW 36463 * | NR_111703 | JQ239390 | JQ239371 |
| Dothiorella camelliae | UESTCC 22.0080 | OQ190530 | NA | OQ275063 |
| Dothiorella camelliae | UESTCC 22.0079 | OQ190532 | OQ241465 | OQ275065 |
| Dothiorella camelliae | UESTCC 22.0078 | OQ190533 | OQ241466 | OQ275066 |
| Dothiorella camelliae | CGMCC 3.24158 | OQ190531 | OQ241464 | OQ275064 |
| Dothiorella capri-amiss | CBS 121763 | EU101323 | EU101368 | KX464850 |
| Dothiorella casuarini | CBS 120688 | DQ846773 | DQ875331 | NA |
| Dothiorella chiangmaiensis | YW177 | NA | NA | NA |
| Dothiorella citricola | ICMP16828 | EU673323 | EU673290 | EU673145 |
| Dothiorella diospyricola | CBS 145972 | MT587398 | MT592110 | MT592581 |
| Dothiorella dulcispinae | CMW 36460 | JQ239400 | JQ239387 | JQ239373 |
| Dothiorella dulcispinae | CMW 25407 * | EU101300 | MT592120 | KX464862 |
| Dothiorella guttulata | MFLUCC 17-0242 * | KY797637 | NA | NA |
| Dothiorella heterophyllae | CMW 46458 * | MN103794 | MH548348 | MH548324 |
| Dothiorella iberica | CBS 115041 * | AY573202 | AY573222 | EU673096 |
| Dothiorella iberica | CBS 113189 | AY573199 | AY573230 | KX464855 |
| Dothiorella iranica | IRAN1587C | KC898231 | KC898214 | NA |
| Dothiorella italica | MFLUCC 17-0951 * | MG828897 | MG829267 | MT592592 |
| Dothiorella juglandis | CBS 188.87 | EU673316 | EU673283 | EU673119 |
| Dothiorella koae | CMW 48017 * | MH447652 | MH548338 | MH548327 |
| Dothiorella lampangensis | MFLUCC 18-0232 | MK347758 | MK340869 | MK412874 |
| Dothiorella longicollis | CBS 122068 | EU144054 | EU144069 | NA |
| Dothiorella magnoliae | CFCC 51563 * | KY111247 | KY213686 | NA |
| Dothiorella magnoliae | CFCC 71215 | PV264857 | NA | PV339821 |
| Dothiorella mangifericola | CBS 121760 * | EU101290 | EU101335 | KX464877 |
| Dothiorella mangifericola | IRAN1584C | KC898221 | KC898204 | NA |
| Dothiorella moneti | MUCC505 * | EF591920 | EF591971 | EF591954 |
| Dothiorella obovata | MFLUCC 22-0058 * | ON751763 | ON799589 | ON799591 |
| Dothiorella obovata | MFLU 22-0094 | ON707682 | NA | ON677452 |
| Dothiorella omnivora | CBS 140349 * | KP205497 | KP205470 | NA |
| Dothiorella parva | CBS 124720 * | KC898234 | KC898217 | KX464866 |
| Dothiorella parva | CBS 124721 | KX464123 | KX464615 | KX464867 |
| Dothiorella plurivora | IRAN1557C | KC898225 | KC898208 | NA |
| Dothiorella pretoriensis | CMW 36480 * | JQ239405 | JQ239392 | JQ239376 |
| Dothiorella prunicola | CAP187 | EU673313 | EU673280 | EU673100 |
| Dothiorella rhamni | MFLUCC 14-0902 * | KT240287 | MT592111 | MT592582 |
| Dothiorella santali | MUCC 509 * | EF591924 | EF591975 | EF591958 |
| Dothiorella sarmentorum | CBS 115038 | AY573206 | AY573223 | EU673101 |
| Dothiorella sarmentorum | IMI 63581b * | AY573212 | AY573235 | NA |
| Dothiorella sarmentorum | UESTCC 22.0076 | OQ190534 | NA | OQ275067 |
| Dothiorella sarmentorum | UESTCC 22.0077 | OQ190535 | OQ241467 | OQ275068 |
| Dothiorella sempervirentis | IRAN1581C | KC898237 | KC898219 | KX464885 |
| Dothiorella sempervirentis | IRAN1583C | KC898236 | KC898220 | KX464884 |
| Dothiorella striata | ICMP 16819 | EU673320 | EU673287 | EU673142 |
| Dothiorella striata | DAR80992 * | KJ573643 | KJ573640 | NA |
| Dothiorella symphoricarposicola | MFLUCC 13-0498 | KJ742379 | KJ742382 | NA |
| Dothiorella symphoricarposicola | MFLUCC 13-0497 * | KJ742378 | KJ742381 | NA |
| Dothiorella tectonae | MFLUCC12-0382 | KM396899 | KM409637 | KM510357 |
| Dothiorella thailandica | MFLUCC 11-0438 * | NR_111794 | JX646861 | JX646844 |
| Dothiorella thripsita | BRIP 51876 | KJ573642 | KJ573639 | KJ577550 |
| Dothiorella uruguayensis | CBS 124908 * | NR_156208 | KX464886 | |
| Dothiorella vidmadera | DAR 78992 * | EU768874 | EU768881 | HM800522 |
| Dothiorella vidmadera | CBS 621.74 | KX464129 | KX464621 | KX464887 |
| Dothiorella vidmadera | CBS 725.79 | KX464130 | KX464622 | KX464888 |
| Dothiorella vidmadera | CFCC 71191 | PV264858 | PV268114 | PV339822 |
| Dothiorella vidmadera | N282 | PV264859 | PV268115 | PV339823 |
| Dothiorella vinea-gemmae | B116-3 * | KJ573644 | KJ573641 | KJ577552 |
| Dothiorella viticola | WA10NO01 * | HM009376 | HM800511 | HM800519 |
| Dothiorella viticola | WA10NO02 | HM009377 | HM800512 | HM800520 |
| Dothiorella viticola | IRNBS28 | MN634039 | MN633993 | NA |
| Dothiorella viticola | MFLUCC 22-0059 | ON707685 | ON720571 | ON677455 |
| Dothiorella yunnana comb. nov. | CGMCC 3.18000 * | KX499644 | KX499650 | NA |
| Dothiorella yunnana comb. nov. | CFCC 71177 | PV264860 | NA | PV339824 |
| Dothiorella zanthoxyli | UESTCC 22.0083 | OQ190537 | OQ241469 | OQ275070 |
| Dothiorella zanthoxyli | UESTCC 22.0084 | OQ190538 | OQ241470 | OQ275071 |
| Dothiorella zanthoxyli | CGMCC 3.24159 * | OQ190536 | OQ241468 | OQ275069 |
| Dothiorella zanthoxyli | N280 | PV264861 | NA | PV339825 |
| Dothiorella zanthoxyli | N280B | PV264862 | NA | PV339826 |
| Neofusicoccum luteum | CBS 562.92 * | KX464170 | KX464690 | KX464968 |
| Neofusicoccum luteum | CMW 41365 | NR_147360 | KP860702 | KP860779 |
| Species | Isolates | GenBank Accession Number | ||
|---|---|---|---|---|
| LSU | ITS | tef1 | ||
| Alanphillipsia euphorbia | CBS 136411 * | KF777196 | KF777140 | MT592029 |
| Phaeobotryon aplospora | CFCC 53774 | MN215871 | MN215836 | MN205996 |
| Phaeobotryon aplospora | CFCC 53775 * | MN215872 | MN215837 | NA |
| Phaeobotryon aplospora | CFCC 53776 | MN215873 | MN215838 | MN205997 |
| Phaeobotryon caraganae | NEFU817 | NA | MH014076 | MH036714 |
| Phaeobotryon caraganae | NEFU816 | NA | MF193891 | MF509765 |
| Phaeobotryon cupressi | IRAN 1455C * | KX464539 | FJ919672 | FJ919661 |
| Phaeobotryon cupressi | IRAN 1454C | KX464538 | FJ919673 | FJ919662 |
| Phaeobotryon cupressi | IRAN 1445C | NA | KF766208 | KF766428 |
| Phaeobotryon fraxini | CFCC 70762 * | PP177348 | PP188527 | NA |
| Phaeobotryon fraxini | CFCC 70763 | PP177349 | PP188528 | NA |
| Phaeobotryon juniperi | JU 001 * | OP941644 | OP941637 | OP948218 |
| Phaeobotryon juniperi | JU 005 | OP941645 | OP941638 | OP948219 |
| Phaeobotryon laricinum | CFCC 70804 | PP960198 | PP960188 | PQ046941 |
| Phaeobotryon laricinum | CFCC 70805 * | PP960199 | PP960189 | PQ046942 |
| Phaeobotryon laricinum | CFCC 70806 | PP960200 | PP960190 | PQ046943 |
| Phaeobotryon longiparaphysium | CFCC 70807 * | PP960203 | PP960193 | PQ046946 |
| Phaeobotryon longiparaphysium | CFCC 70808 | PP960204 | PP960194 | PQ046947 |
| Phaeobotryon mali | XJAU 293001 | MW367101 | MW326854 | MW509519 |
| Phaeobotryon mali | XJAU 277201 | MW367094 | MW326853 | MW509520 |
| Phaeobotryon mali | XJAU 278201 | MW367092 | MW326852 | MW509516 |
| Phaeobotryon mali | XJAU 309401 | MW367100 | MW326858 | MW509517 |
| Phaeobotryon mali | XJAU 310001 | MW367093 | MW326878 | MW509518 |
| Phaeobotryon mamane | CPC 12442 | DQ377899 | EU673333 | EU673299 |
| Phaeobotryon mamane | CPC 12440 * | EU673248 | KF766209 | EU673298 |
| Phaeobotryon mamane | CPC 12443 | EU673249 | EU673334 | EU673300 |
| Phaeobotryon negundinis | CAA 797 | KU820971 | KX061513 | KX061507 |
| Phaeobotryon negundinis | CAA 798 | NG_069332 | KX061514 | KX061508 |
| Phaeobotryon platycladi | CFCC 58799 * | OQ652543 | OQ651172 | OQ692930 |
| Phaeobotryon platycladi | CFCC 58800 | OQ652544 | OQ651173 | OQ692931 |
| Phaeobotryon rhoinum | CFCC 52449 | MH133940 | MH133923 | MH133957 |
| Phaeobotryon rhoinum | CFCC 52450 * | MH133941 | MH133924 | MH133958 |
| Phaeobotryon rhoinum | CFCC 52451 | MH133942 | MH133925 | MH133959 |
| Phaeobotryon rhoinum | XJAU 146801 | MW367102 | MW326857 | MW509522 |
| Phaeobotryon rhoinum | XJAU 276401 | MW367095 | MW326855 | MW509524 |
| Phaeobotryon rhoinum | XJAU 304901 | MW367096 | MW326856 | MW509523 |
| Phaeobotryon rhoinum | XJAU 316801 | MW367097 | MW326877 | MW509521 |
| Phaeobotryon rhois | CFCC 89662 * | KM030591 | KM030584 | KM030598 |
| Phaeobotryon rhois | CFCC 89663 | KM030592 | KM030585 | KM030599 |
| Phaeobotryon spiraeae | CFCC 53925 * | OM049432 | OM049420 | NA |
| Phaeobotryon spiraeae | CFCC 53926 | OM049433 | OM049421 | NA |
| Phaeobotryon ulmi | CBS 138854 * | MT587321 | MT587540 | MT592274 |
| Phaeobotryon ulmi | CBS 123.30 | DQ377861 | KX464232 | KX464766 |
| Phaeobotryon xizangense sp. nov. | CFCC 71501 * | PV264865 | PV264863 | PV268116 |
| Phaeobotryon xizangense sp. nov. | LZ204 * | PV264866 | PV264864 | PV268117 |
| Oblongocollomyces variabilis | CBS 121774 * | KX464536 | NR_136994 | EU101357 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhou, J.; Li, A.; Jiang, N. Morphology and Phylogeny Reveal New Species and Records of Diplodia, Dothiorella, and Phaeobotryon Associated with Tree Cankers in Xizang, China. J. Fungi 2025, 11, 331. https://doi.org/10.3390/jof11050331
Zhou J, Li A, Jiang N. Morphology and Phylogeny Reveal New Species and Records of Diplodia, Dothiorella, and Phaeobotryon Associated with Tree Cankers in Xizang, China. Journal of Fungi. 2025; 11(5):331. https://doi.org/10.3390/jof11050331
Chicago/Turabian StyleZhou, Jia, Aining Li, and Ning Jiang. 2025. "Morphology and Phylogeny Reveal New Species and Records of Diplodia, Dothiorella, and Phaeobotryon Associated with Tree Cankers in Xizang, China" Journal of Fungi 11, no. 5: 331. https://doi.org/10.3390/jof11050331
APA StyleZhou, J., Li, A., & Jiang, N. (2025). Morphology and Phylogeny Reveal New Species and Records of Diplodia, Dothiorella, and Phaeobotryon Associated with Tree Cankers in Xizang, China. Journal of Fungi, 11(5), 331. https://doi.org/10.3390/jof11050331







