Genotype, but Not Climate, Affects the Resistance of Honey Bees (Apis mellifera) to Viral Infections and to the Mite Varroa destructor
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Samples and Locations
2.2. Sample Analyses
2.3. Prevalence and Intensity of Varroa destructor Parasitism
2.4. Prevalence and Intensity of Nosema spp. Infection
2.5. Viral Diagnosis
2.6. Viral Quantification
2.7. Mitotype of the Samples
2.8. Morphotype of the Samples
2.9. Genotype of the Samples
2.10. Statistical Analyses
3. Results
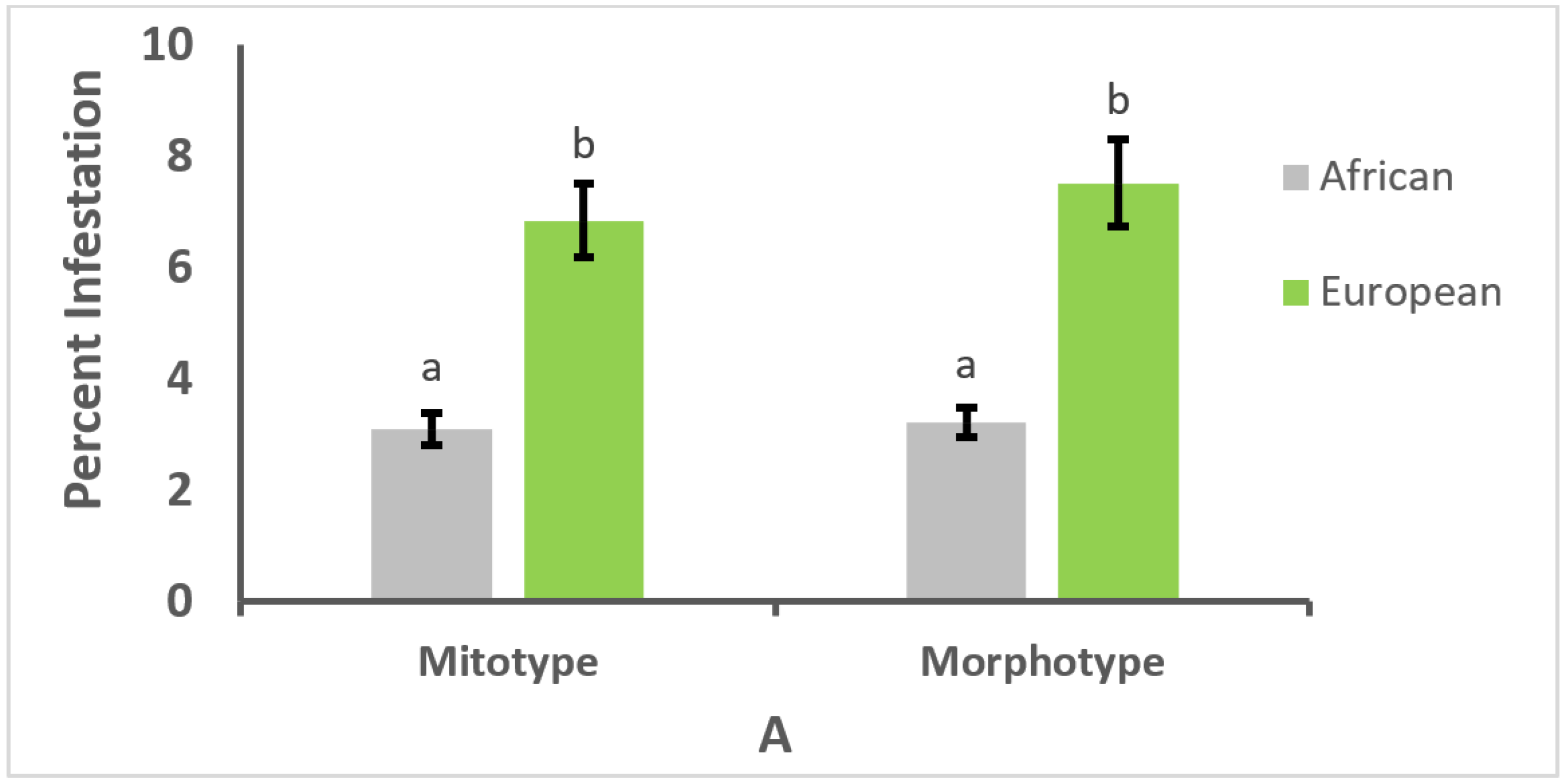
3.1. Effect of Mitotype and Morphotype on Pathogen Prevalence
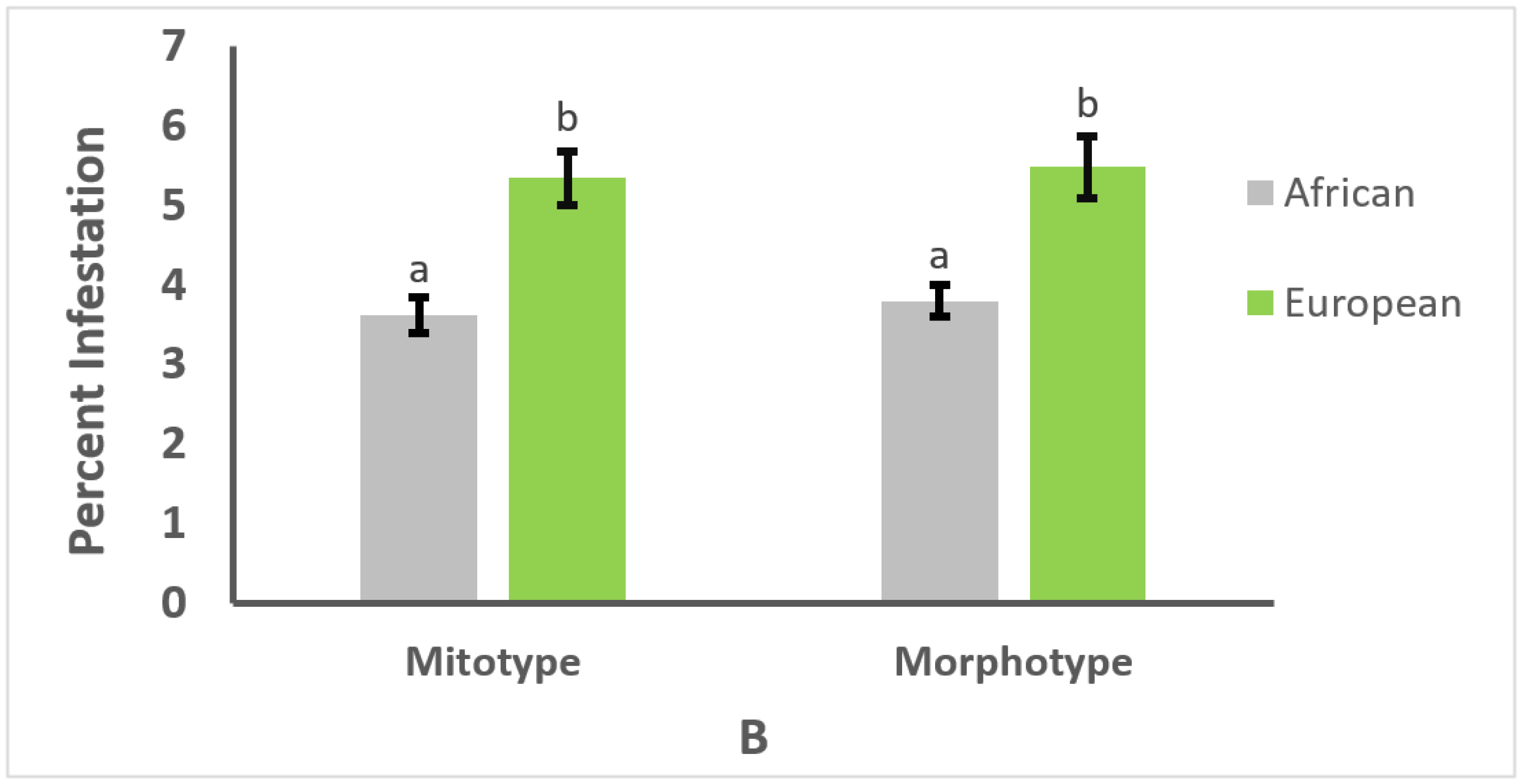
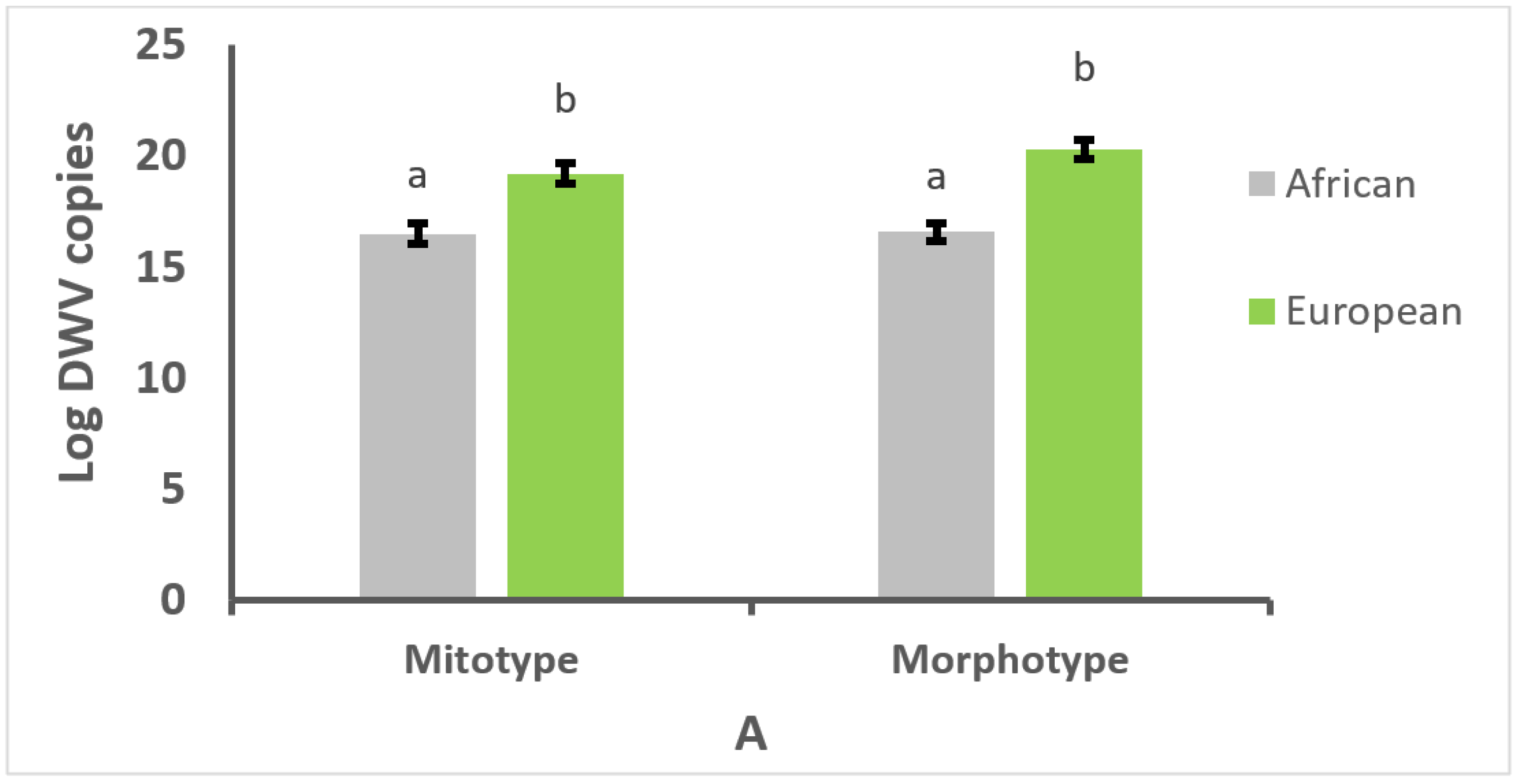
3.2. Effect of Mitotype and Morphotype on Parasitism or Infection Intensity
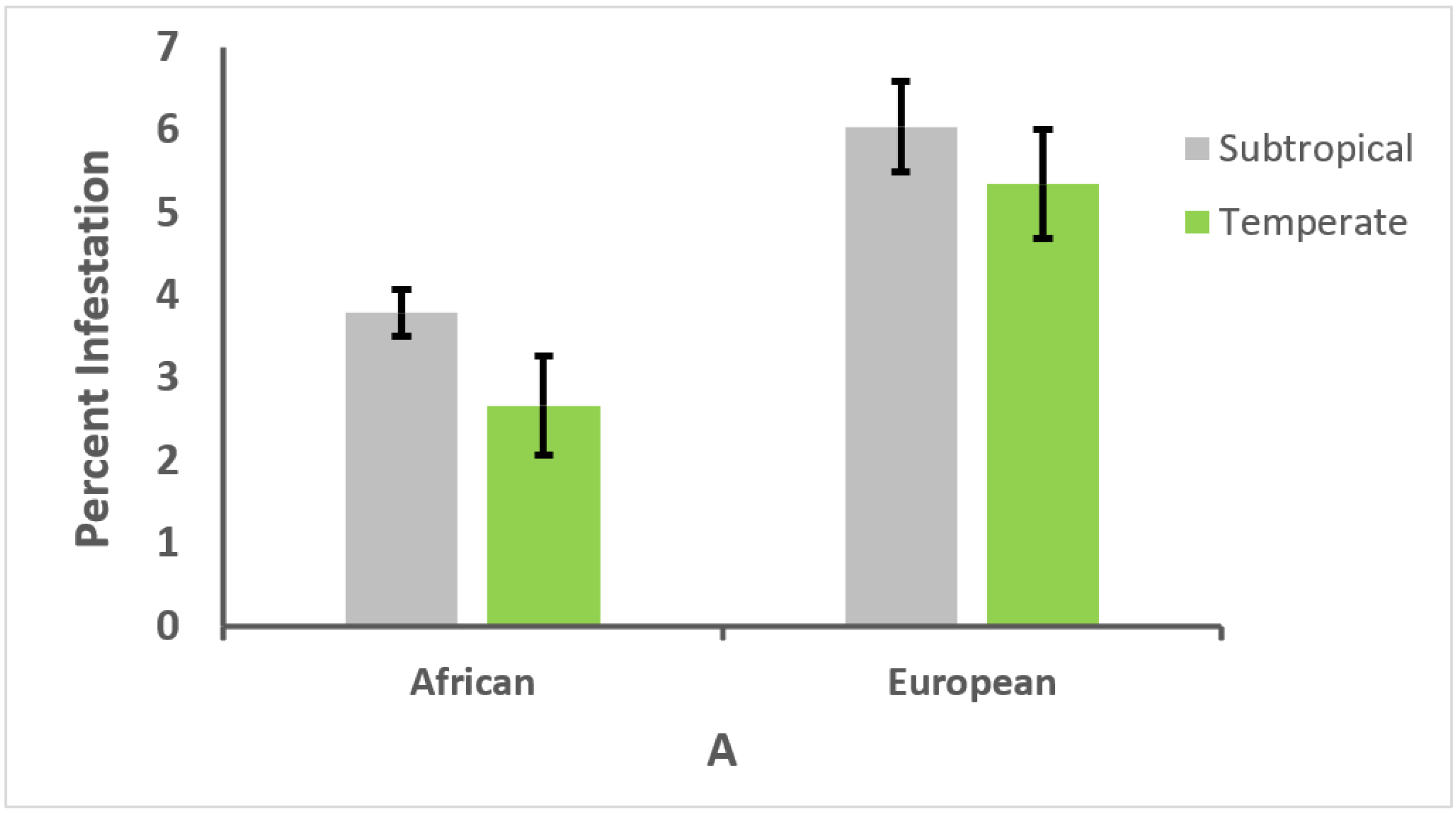
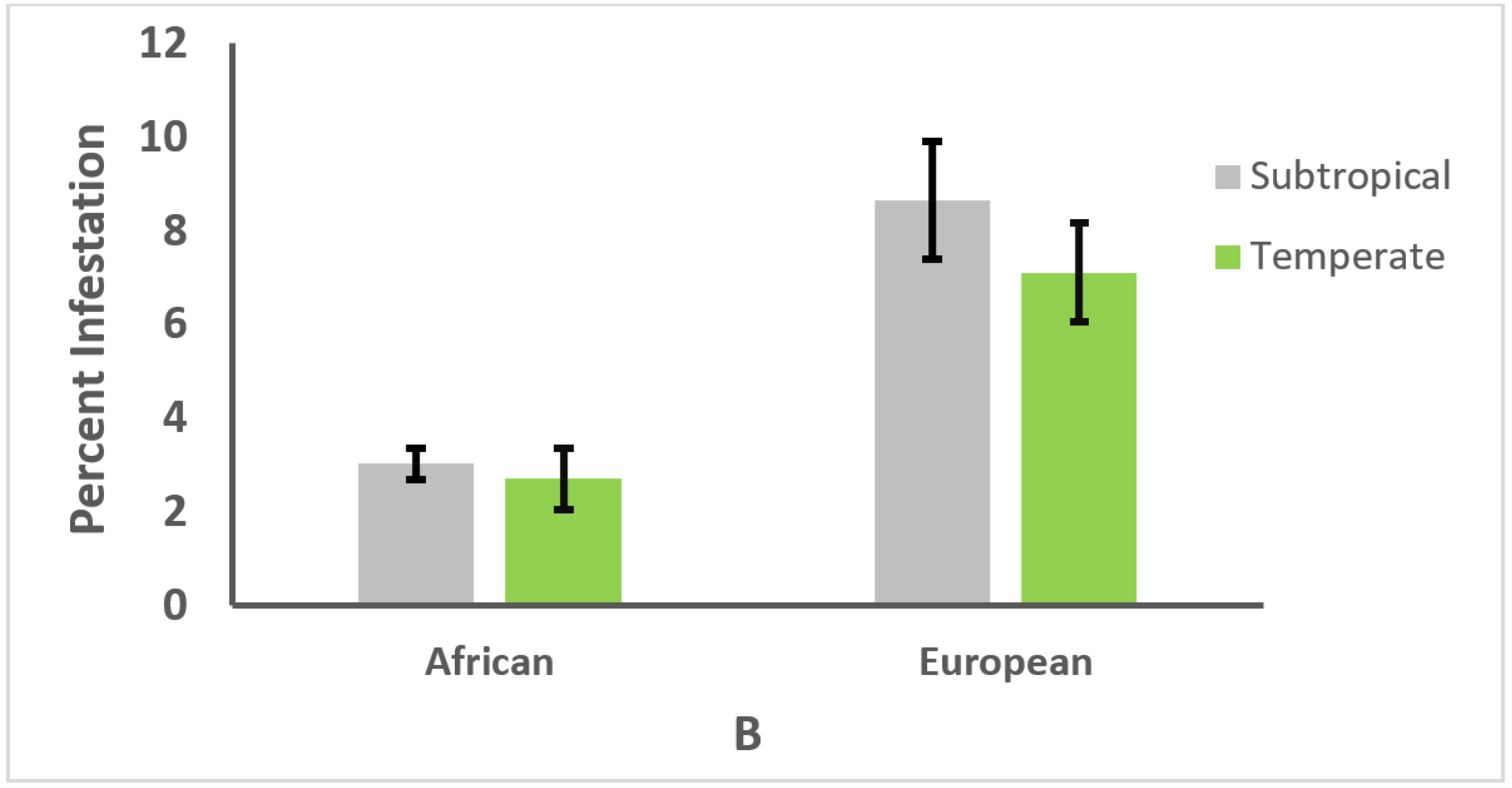
3.3. Effect of Climate on Genotype and Parasitism or Infection Intensity
3.4. Relationships between Honey Bee Type, Climate, and Parasitism or Infection Intensity
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kevan, P.G.; Guzman-Novoa, E.; Skinner, A.; Van Englesdorp, D. Colony collapse disorder in Canada: Do we have a problem? Hive Lights 2007, 20, 14–16. [Google Scholar]
- Guzman-Novoa, E.; Eccles, L.; Calvete, Y.; McGowan, J.; Kelly, P.G.; Correa-Benítez, A. Varroa destructor is the main culprit for the death and reduced populations of overwintered honey bee (Apis mellifera) colonies in Ontario, Canada. Apidologie 2010, 41, 443–450. [Google Scholar] [CrossRef]
- Le Conte, Y.; Ellis, M.; Ritter, W. Varroa mites and honey bee health: Can Varroa explain part of the colony losses? Apidologie 2010, 41, 353–363. [Google Scholar] [CrossRef]
- Neumann, P.; Carreck, N.L. Honey bee colony losses. J. Apic. Res. 2010, 49, 1–6. [Google Scholar] [CrossRef]
- McMenamin, A.J.; Genersch, E. Honey bee colony losses and associated viruses. Curr. Opin. Insect Sci. 2015, 8, 121–129. [Google Scholar] [CrossRef]
- Yang, X.; Cox-Foster, D. Effects of parasitization by Varroa destructor on survivorship and physiological traits of Apis mellifera in correlation with viral incidence and microbial challenge. Parasitology 2007, 134, 405–412. [Google Scholar] [CrossRef]
- Koleoglu, G.; Goodwin, P.H.; Reyes-Quintana, M.; Hamiduzzaman, M.M.; Guzman-Novoa, E. Effect of Varroa destructor, wounding and Varroa homogenate on gene expression in brood and adult honey bees. PLoS ONE 2017, 12, e0169669. [Google Scholar] [CrossRef] [PubMed]
- Koleoglu, G.; Goodwin, P.H.; Reyes-Quintana, M.; Hamiduzzaman, M.M.; Guzman-Novoa, E. Varroa destructor parasitism reduces hemocyte concentrations and prophenol oxidase gene expression in bees from two populations. Parasitol. Res. 2018, 117, 1175–1183. [Google Scholar] [CrossRef]
- Emsen, B.; Guzman-Novoa, E.; Kelly, P.G. Honey production of honey bee (Hymenoptera: Apidae) colonies with high and low Varroa destructor (Acari: Varroidae) infestation rates in eastern Canada. Can. Entomol. 2014, 146, 236–240. [Google Scholar] [CrossRef]
- Williams, G.R.; Shutler, D.; Burgher-MacLellan, K.L.; Rogers, R.E.L. Infra-population and community dynamics of the parasites Nosema apis and Nosema ceranae, and consequences for honey bee (Apis mellifera) hosts. PLoS ONE 2014, 9, e99465. [Google Scholar] [CrossRef]
- Martín-Hernández, R.; Bartolomé, C.; Chejanovsky, N.; Le Conte, Y.; Dalmon, A.; Dussaubat, C.; García-Palencia, P.; Meana, A.; Pinto, A.; Soroker, V.; et al. Nosema ceranae in Apis mellifera: A 12 years postdetection perspective. Environ. Microbiol. 2018, 20, 1302–1329. [Google Scholar] [CrossRef] [PubMed]
- Valizadeh, P.; Guzman-Novoa, E.; Goodwin, P.H. Effect of immune inducers on Nosema ceranae multiplication and their impact on honey bee (Apis mellifera L.) survivorship and behaviors. Insects 2020, 11, 572. [Google Scholar] [CrossRef] [PubMed]
- Emsen, B.; Guzman-Novoa, E.; Hamiduzzaman, M.M.; Eccles, L.; Lacey, B.; Ruiz-Pérez, R.A.; Nasr, M. Higher prevalence and levels of Nosema ceranae than Nosema apis infections in Canadian honey bee colonies. Parasitol. Res. 2016, 115, 175–181. [Google Scholar] [CrossRef] [PubMed]
- Emsen, B.; De la Mora, A.; Lacey, B.; Eccles, L.; Kelly, P.G.; Medina-Flores, C.A.; Petukhova, T.; Morfin, N.; Guzman-Novoa, E. Seasonality of Nosema ceranae infections and their relationship with honey bee populations, food stores, and survivorship in a North American region. Vet. Sci. 2020, 7, 131. [Google Scholar] [CrossRef]
- Beaurepaire, A.; Piot, N.; Doublet, B.; Antúnez, K.; Campbell, B.; Chantawannakul, P.; Chejanovsky, N.; Gajda, A.; Heerman, M.; Panziera, D.; et al. Diversity and global distribution of viruses of the western honey bee, Apis mellifera. Insects 2020, 11, 239. [Google Scholar] [CrossRef]
- Cox-Foster, D.L.; Conlan, S.; Holmes, E.C.; Palacios, G.; Evans, J.D.; Moran, N.A.; Quan, P.L.; Briese, T.; Hornig, M.; Geiser, D.M.; et al. A metagenomic survey of microbes in honey bee colony collapse disorder. Science 2007, 318, 283–287. [Google Scholar] [CrossRef]
- Van Engelsdorp, D.; Evans, J.D.; Saegerman, C.; Mullin, C.; Haubruge, E.; Nguyen, B.K.; Frazier, M.; Frazier, J.; Cox-Foster, D.; Chen, Y.; et al. Colony collapse disorder: A descriptive study. PLoS ONE 2009, 4, e6481. [Google Scholar]
- Gisder, S.; Genersch, E. Viruses of commercialized insect pollinators. J. Invertebr. Pathol. 2017, 147, 51–59. [Google Scholar] [CrossRef]
- Gisder, S.; Aumeier, P.; Genersch, E. Deformed wing virus: Replication and viral load in mites (Varroa destructor). J. Gen. Virol. 2009, 90, 463–467. [Google Scholar] [CrossRef]
- Dainat, B.; Evans, J.D.; Chen, Y.P.; Gauthier, L.; Neumann, P. Predictive markers of honey bee colony collapse. PLoS ONE 2012, 7, e32151. [Google Scholar]
- Natsopoulou, M.E.; McMahon, D.P.; Doublet, V.; Frey, E.; Rosenkranz, P.; Paxton, R.J. The virulent, emerging genotype B of Deformed wing virus is closely linked to overwinter honeybee worker loss. Sci. Rep. 2017, 7, 5242. [Google Scholar] [CrossRef] [PubMed]
- Tentcheva, D.; Gauthier, L.; Zappulla, N.; Dainat, B.; Cousserans, F.; Colin, M.E.; Bergoin, M. Prevalence and seasonal variations of six bee viruses in Apis mellifera L. and Varroa destructor mite populations in France. App. Environ. Microbiol. 2004, 70, 7185–7191. [Google Scholar] [CrossRef] [PubMed]
- Kevan, P.G.; Hannan, M.A.; Ostiguy, N.; Guzman-Novoa, E. A summary of the varroa-virus disease complex in honey bees. Am. Bee J. 2006, 146, 694–697. [Google Scholar]
- Di Prisco, G.; Pennacchio, F.; Caprio, E.; Boncristiani, H.F., Jr.; Evans, J.D.; Chen, Y. Varroa destructor is an effective vector of Israeli acute paralysis virus in the honeybee, Apis mellifera. J. Gen. Virol. 2011, 92, 151–155. [Google Scholar] [CrossRef]
- Yue, C.; Genersch, E. RT-PCR analysis of Deformed wing virus in honeybees (Apis mellifera) and mites (Varroa destructor). J. Gen. Virol. 2005, 88, 2329–2336. [Google Scholar] [CrossRef]
- Di Prisco, G.; Annoscia, D.; Margiotta, M.; Ferrara, R.; Varricchio, P.; Zanni, V.; Caprio, E.; Nazzi, F.; Pennacchio, F. A mutualistic symbiosis between a parasitic mite and a pathogenic virus undermines honey bee immunity and health. Proc. Natl. Acad. Sci. USA 2016, 113, 3203–3208. [Google Scholar] [CrossRef]
- Reyes-Quintana, M.; Espinosa-Montaño, L.G.; Prieto-Merlos, D.; Koleoglu, G.; Petukhova, T.; Correa-Benítez, A.; Guzman-Novoa, E. Impact of Varroa destructor and deformed wing virus on emergence, cellular immunity, wing integrity and survivorship of Africanized honey bees in Mexico. J. Invert. Pathol. 2019, 164, 43–48. [Google Scholar] [CrossRef]
- Sabahi, Q.; Morfin, N.; Nehzati-Paghaleh, G.; Guzman-Novoa, E. Detection and replication of deformed wing virus and black queen cell virus in parasitic mites, Varroa destructor, from Iranian honey bee (Apis mellifera) colonies. J. Apic. Res. 2020, 59, 211–217. [Google Scholar] [CrossRef]
- Mondet, F.; De Miranda, J.R.; Kretzschmar, A.; Leconte, Y.; Mercer, A. On the front line: Quantitative virus dynamics in honeybee (Apis mellifera L.) colonies along a new expansion front of the parasite Varroa destructor. PLoS Pathog. 2014, 10, e1004323. [Google Scholar] [CrossRef]
- Gajda, A.M.; Manzur, E.D.; Bober, A.M.; Czopowics, M. Nosema ceranae interactions with Nosema apis and black queen cell virus. Agriculture 2021, 11, 963. [Google Scholar] [CrossRef]
- Guzman-Novoa, E.; Morfin, N. Disease resistance in honey bees (Apis mellifera L.) at the colony and individual levels. In Comprehensive Biotechnology, 3rd ed.; Moo-Young, M., Ed.; Pergamon Press: Oxford, UK, 2019; Volume 4, pp. 811–817. [Google Scholar]
- Moretto, G.; Gonçalves, L.S.; De Jong, D.; Bichuette, M.Z. The effects of climate and bee race on Varroa jacobsoni Oud. infestations in Brazil. Apidologie 1991, 22, 197–203. [Google Scholar] [CrossRef]
- Moretto, G.; Gonçalves, L.S.; De Jong, D. Heritability of Africanized and European honey bee defensive behavior against the mite Varroa jacobsoni. Rev. Bras. Genet. 1993, 16, 71–77. [Google Scholar]
- Guzman-Novoa, E.; Sánchez, A.; Page, R.E., Jr.; García, T. Susceptibility of European and Africanized honeybees (Apis mellifera L.) and their hybrids to Varroa jacobsoni Oud. Apidologie 1996, 27, 93–103. [Google Scholar] [CrossRef]
- Guzman-Novoa, E.; Vandame, R.; Arechavaleta, M.E. Susceptibility of European and Africanized honey bees (Apis mellifera L.) to Varroa jacobsoni Oud. in Mexico. Apidologie 1999, 30, 173–182. [Google Scholar] [CrossRef]
- Guzman-Novoa, E.; Emsen, B.; Unger, P.; Espinoza-Montaño, L.G.; Petukhova, T. Genotypic variability and relationships between mite infestation levels, mite damage, grooming intensity, and removal of Varroa destructor mites in selected strains of worker honey bees (Apis mellifera L.). J. Invertebr. Pathol. 2012, 110, 314–320. [Google Scholar] [CrossRef]
- Arechavaleta-Velasco, M.E.; Guzman-Novoa, E. Relative effect of four characteristics that restrain the population growth of the mite Varroa destructor in honey bee (Apis mellifera) colonies. Apidologie 2001, 32, 157–174. [Google Scholar] [CrossRef]
- Mondragón, L.; Spivak, M.; Vandame, R. A multifactorial study of the resistance of honeybees Apis mellifera to the mite Varroa destructor over one year in Mexico. Apidologie 2005, 36, 345–358. [Google Scholar] [CrossRef]
- Mondragón, L.; Martin, S.; Vandame, R. Mortality of mite offspring: A major component of Varroa destructor resistance in a population of Africanized bees. Apidologie 2006, 37, 67–74. [Google Scholar] [CrossRef]
- Calderón, R.A.; Van Veen, J.W.; Sommeijer, M.J.; Sánchez, L.A. Reproductive biology of Varroa destructor in Africanized honey bees (Apis mellifera). Exp. Appl. Acarol. 2010, 50, 281–297. [Google Scholar] [CrossRef]
- Medina-Flores, C.A.; Guzman-Novoa, E.; Hamiduzzaman, M.M.; Aréchiga-Flores, C.F.; López-Carlos, M.A. Africanized honey bees (Apis mellifera) have low infestation levels of the mite Varroa destructor in different ecological regions in Mexico. Genet. Mol. Res. 2014, 13, 7282–7293. [Google Scholar] [CrossRef]
- Invernizzi, C.; Zefferino, I.; Santos, E.; Sánchez, L.; Mendoza, Y. Multilevel assessment of grooming behavior against Varroa destructor in Italian and Africanized honey bees. J. Apic. Res. 2015, 54, 321–327. [Google Scholar] [CrossRef]
- Sánchez, H.U.R.; Peña, A.R.M.; Reyna, J.A.G. Actualización del Atlas Bioclimático del estado de Jalisco. Investig. Geográficas Boletín Inst. Geogr. 2013, 82, 66–92. [Google Scholar]
- De Jong, D.; Roma, D.A.; Gonçalves, L.S. A comparative analysis of shaking solutions for the detection of Varroa jacobsoni on adult honeybees. Apidologie 1982, 13, 297–306. [Google Scholar] [CrossRef]
- Fries, I.; Chauzat, M.P.; Chen, Y.P.; Doublet, V.; Genersch, E.; Gisder, S.; Higes, M.; Mcmahon, D.; Martín-Hernández, R.; Natsopoulou, M.; et al. Standard methods for Nosema research. J. Apic. Res. 2013, 52, 1–28. [Google Scholar] [CrossRef]
- Hamiduzzaman, M.M.; Guzman-Novoa, E.; Goodwin, P.H. A multiplex PCR assay to diagnose and quantify Nosema infections in honey bees (Apis mellifera). J. Invertebr. Pathol. 2010, 105, 151–155. [Google Scholar]
- Guzman-Novoa, E.; Hamiduzzaman, M.M.; Espinosa-Montaño, L.G.; Correa-Benítez, A.; Anguiano-Baez, R.; Ponce-Vázquez, R. First detection of four viruses in honey bee (Apis mellifera) workers with and without deformed wings and Varroa destructor in Mexico. J. Apic. Res. 2012, 51, 342–346. [Google Scholar] [CrossRef]
- Guzman-Novoa, E.; Hamiduzzaman, M.M.; Correa-Benítez, A.; Espinosa-Montaño, L.G.; Uribe-Rubio, J.L. A scientific note on the first detection of black queen cell virus in honey bees (Apis mellifera) in Mexico. Apidologie 2013, 44, 382–384. [Google Scholar] [CrossRef]
- García-Anaya, M.C.; Romo-Chacón, A.; Zamudio-Flores, P.B.; Ríos-Velasco, C.; Acosta-Muñiz, C.H. Detection of viruses in colonies of honey bees (Apis mellifera L.) in the state of Chihuahua, México. J. Apic. Res. 2016, 55, 240–242. [Google Scholar] [CrossRef]
- García-Anaya, M.C.; Romo-Chacón, A.; Sáenz-Mendoza, A.I.; Pérez-Ordoñez, G.; Acosta-Muñiz, C.H. Detection of Israeli acute paralysis virus (IAPV) and Apis mellifera filamentous virus (AmFV) in honey bees in México. J. Apic. Sci. 2018, 62, 141–144. [Google Scholar]
- Di Prisco, G.; Cavaliere, V.; Annoscia, D.; Varricchio, P.; Caprio, E.; Nazzi, F.; Gargiulo, G.; Pennacchio, F. Neonicotinoid clothianidin adversely affects insect immunity and promotes replication of a viral pathogen in honey bees. Proc. Natl. Acad. Sci. USA 2013, 110, 18466–18471. [Google Scholar] [CrossRef]
- Chantawannakul, P.; Ward, L.; Boonham, N.; Brown, M. A scientific note on the detection of honeybee viruses using real-time PCR (TaqMan) in Varroa mites collected from a Thai honeybee (Apis mellifera) apiary. J. Invertebr. Pathol. 2006, 91, 69–73. [Google Scholar] [CrossRef] [PubMed]
- Morfin, N.; Gashout, H.A.; Macías-Macías, J.O.; De la Mora, A.; Tapia-Rivera, J.C.; Tapia-González, J.M.; Contreras-Escareño, F.; Guzman-Novoa, E. Detection, replication and quantification of deformed wing virus-A, deformed wing virus-B, and black queen cell virus in the endemic stingless bee, Melipona colimana, from Jalisco, Mexico. Int. J. Trop. Insect Sci. 2020, 41, 1285–1292. [Google Scholar] [CrossRef]
- Morfin, N.; Given, K.; Evans, M.; Guzman-Novoa, E.; Hunt, G.J. Grooming behavior and gene expression of the Indiana “mite-bitter” honey bee stock. Apidologie 2020, 51, 267–275. [Google Scholar]
- Nielsen, D.I.; Ebert, P.R.; Page, R.E.; Hunt, G.J.; Guzman-Novoa, E. Improved polymerase chain reaction-based mitochondrial genotype assay for identification of the Africanized honey bee (Hymenoptera: Apidae). Ann. Entomol. Soc. Am. 2000, 93, 1–6. [Google Scholar] [CrossRef]
- Pinto, M.A.; Johnston, J.S.; Rubink, W.L.; Coulson, R.N.; Patton, J.C.; Sheppard, W.S. Identification of Africanized honey bee (Hymenoptera: Apidae) mitochondrial DNA: Validation of a rapid polymerase chain reaction-based assay. Ann. Entomol. Soc. Am. 2003, 96, 679–684. [Google Scholar]
- Sylvester, H.A.; Rinderer, T.E. Fast Africanized bee identification system (FABIS) manual. Am. Bee J. 1987, 27, 511–516. [Google Scholar]
- Tibatá, V.M.; Sánchez, A.; Palmer-Young, E.; Junca, H.; Solarte, V.M.; Madella, S.; Ariza, F.; Figueroa, J.; Corona, M. Africanized honey bees in Colombia exhibit high prevalence but low level of infestation of Varroa mites and low prevalence of pathogenic viruses. PLoS ONE 2021, 16, e0244906. [Google Scholar] [CrossRef]
- Rosenkranz, P. Honey bee (Apis mellifera L.) tolerance to Varroa jacobsoni Oud. in South America. Apidologie 1999, 30, 159–172. [Google Scholar] [CrossRef]
- Martin, S.J.; Medina, L.M. Africanized honeybees have unique tolerance to Varroa mites. Trends Parasitol. 2004, 20, 112–114. [Google Scholar] [CrossRef]
- Ritter, W.; De Jong, D. Reproduction of Varroa jacobsoni O. in Europe, the middle East and tropical South America. J. Appl. Entomol. 1984, 98, 55–57. [Google Scholar] [CrossRef]
- Medina, L.M.; Martin, S.J. A comparative study of Varroa jacobsoni reproduction in worker cells of honey bees (Apis mellifera) in England and Africanized bees in Yucatan, Mexico. Exp. Appl. Acarol. 1999, 23, 659–667. [Google Scholar] [CrossRef]
- Guzman-Novoa, E.; Morfin, N.; De la Mora, A.; Macías-Macías, J.O.; Tapia-González, J.M.; Contreras-Escareño, F.; Medina-Flores, C.A.; Correa-Benítez, A.; Quezada-Euán, J.J.G. The process and outcome of the Africanization of honey bees in Mexico: Lessons and future directions. Front. Ecol. Evol. 2020, 8, 404. [Google Scholar] [CrossRef]
- Guerra, J.C.V., Jr.; Gonçalves, L.S.; De Jong, D. Africanized honey bees (Apis mellifera L.) are more efficient at removing worker brood artificially infested with the parasitic mite Varroa jacobsoni Oudemans than are Italian bees or Italian/Africanized hybrids. Genet. Mol. Biol. 2000, 23, 89–92. [Google Scholar]
- Vandame, R.; Morand, S.; Colin, M.E.; Belzunces, L.P. Parasitism in the social bee Apis mellifera: Quantifying costs and benefits of behavioral resistance to Varroa destructor mites. Apidologie 2002, 33, 433–445. [Google Scholar]
- Medina-Flores, C.A.; Medina-Medina, L.A.; Guzman-Novoa, E. Effect of hygienic behavior on resistance to chalkbrood disease (Ascosphaera apis) in Africanized bee colonies (Apis mellifera). Rev. Mex. Cienc. Pecu. 2022, 13, 225–239. [Google Scholar]
- Guerrero-Molina, C.; Correa-Benítez, A.; Hamiduzzaman, M.M.; Guzman-Novoa, E. Nosema ceranae is an old resident of honey bee (Apis mellifera) colonies in Mexico, causing infection levels of one million spores per bee or higher during summer and fall. J. Invertebr. Pathol. 2016, 141, 38–40. [Google Scholar] [CrossRef]
- Antúnez, K.; D’Alessandro, B.; Corbella, E.; Ramallo, G.; Zunino, P. Honeybee viruses in Uruguay. J. Invertebr. Pathol. 2006, 93, 67–70. [Google Scholar] [CrossRef]
- Teixeira, E.W.; Chen, Y.; Message, D.; Pettis, J.; Evans, J.D. Virus infections in Brazilian honey bees. J. Invertebr. Pathol. 2008, 99, 117–119. [Google Scholar] [CrossRef]
- Mendoza, Y.; Tomasco, I.H.; Antúnez, K.; Castelli, L.; Branchiccela, B.; Santos, E.; Invernizzi, C. Unraveling honey bee–Varroa destructor interaction: Multiple factors involved in differential resistance between two Uruguayan populations. Vet. Sci. 2020, 7, 116. [Google Scholar]
- Hamiduzzaman, M.M.; Guzman-Novoa, E.; Goodwin, P.H.; Reyes-Quintana, M.; Koleoglu, G.; Correa-Benítez, A.; Petukhova, T. Differential responses of Africanized and European honey bees (Apis mellifera) to viral replication following mechanical transmission or Varroa destructor parasitism. J. Invertebr. Pathol. 2015, 126, 12–20. [Google Scholar] [CrossRef]
- Emsen, B.; Hamiduzzaman, M.M.; Goodwin, P.H.; Guzman-Novoa, E. Lower virus infections in Varroa destructor-infested and uninfested brood and adult honey bees (Apis mellifera) of a low mite population growth colony compared to a high mite population growth colony. PLoS ONE 2015, 10, e0118885. [Google Scholar]
- Anguiano-Baez, R.; Guzman-Novoa, E.; Hamiduzzaman, M.; Espinosa-Montaño, L.G.; Correa-Benítez, A. Varroa destructor (Mesostigmata: Varroidae) parasitism and climate differentially influence the prevalence, levels, and overt infections of deformed wing virus in honey bees (Hymenoptera: Apidae). J. Insect Sci. 2016, 16, 44. [Google Scholar] [CrossRef] [PubMed]
- Mendoza, Y.; Antúnez, K.; Branchiccela, B.; Anido, M.; Santos, E.; Invernizzi, C. Nosema ceranae and RNA viruses in European and Africanized honeybee colonies (Apis mellifera) in Uruguay. Apidologie 2014, 45, 224–234. [Google Scholar] [CrossRef][Green Version]
- Tapia-González, J.M.; Alcazar-Oceguera, G.; Macías-Macías, J.O.; Contreras-Escareño, F.; Tapia-Rivera, J.C.; Petukhova, T.; Guzman-Novoa, E. Varroosis in honey bees in different environmental and regional conditions of Jalisco, Mexico. Ecosist. Recur. Agropec. 2019, 6, 243–251. [Google Scholar]
- Dalmon, A.; Peruzzi, M.; Le Conte, Y.; Alaux, C.; Pioz, M. Temperature-driven changes in viral loads in the honey bee Apis mellifera. J. Invertebr. Pathol. 2019, 160, 87–94. [Google Scholar]
- Penn, H.J.; Simone-Finstrom, M.D.; Chen, I.; Healy, K.B. Honey bee genetic stock determines deformed wing virus symptom severity but not viral load or dissemination following pupal exposure. Front. Genet. 2022, 13, 909392. [Google Scholar] [CrossRef]
- Larsen, A.; Reynaldi, F.J.; Guzman-Novoa, E. Fundaments of the honey bee (Apis mellifera) immune system. Review. Rev. Mex. Cienc. Pecu. 2019, 10, 705–728. [Google Scholar]
- Bronkhorst, A.W.; Van Rij, R.P. The long and short antiviral defense: Small RNA-based immunity in insects. Curr. Opin. Virol. 2014, 7, 19–28. [Google Scholar] [CrossRef]
- Brutscher, L.M.; Daughenbaugh, K.F.; Flenniken, M.L. Antiviral defense mechanisms in honey bees. Curr. Opin. Insect. Sci. 2015, 10, 71–82. [Google Scholar] [CrossRef]
- Chen, Y.P.; Higgins, J.A.; Feldlaufer, M.F. Quantitative real-time reverse transcription-PCR analysis of deformed wing virus infection in the honeybee (Apis mellifera L.). Appl. Environ. Microbiol. 2005, 71, 436–441. [Google Scholar] [CrossRef]
- Crozier, Y.C.; Koulianos, S.; Crozier, R.H. An improved test for Africanized honeybee mitochondrial DNA. Experientia 1991, 47, 968–969. [Google Scholar] [CrossRef]
- Li, B.; Deng, S.; Yang, D.; Hou, C.; Diao, Q. Complete sequences of the RNA 1 and RNA 2 segments of chronic bee paralysis virus strain CBPV-BJ detected in China. Arch. Virol. 2017, 162, 2451–2456. [Google Scholar] [CrossRef]
- Maori, E.; Paldi, N.; Shafir, S.; Kalev, H.; Tsur, E.; Glick, E.; Sela, I. IAPV, a bee‐affecting virus associated with Colony Collapse Disorder can be silenced by dsRNA ingestion. Insect Mol. Biol. 2009, 18, 55–60. [Google Scholar] [CrossRef]
- Thompson, G.J.; Yockey, H.; Lim, J.; Oldroyd, B.P. Experimental manipulation of ovary activation and gene expression in honey bee (Apis mellifera) queens and workers: Testing hypotheses of reproductive regulation. J. Exp. Zool. 2007, 307, 600–610. [Google Scholar] [CrossRef]


| Pathogen | N | % MitA | % MitE | p | % MorA | % MorE | p |
|---|---|---|---|---|---|---|---|
| V. Brood | 334 | 76.7 | 84.2 | 0.09 | 75.0 | 88.0 | 0.003 * |
| V. Adults | 334 | 93.8 | 97.5 | 0.11 | 93.9 | 98.4 | 0.048 * |
| N. ceranae | 55 | 13.6 | 18.4 | 0.29 | 16.5 | 15.8 | 0.88 |
| DWV | 76 | 30.5 | 45.0 | 0.24 | 32.6 | 48.2 | 0.22 |
| BQCV | 76 | 55.5 | 72.5 | 0.15 | 61.2 | 70.4 | 0.46 |
| Correlated Variables | N | r | p |
|---|---|---|---|
| W. L.—V. d. brood | 339 | 0.32 | <0.001 |
| W. L.—V. d. adults | 339 | 0.27 | <0.001 |
| W. L.—DWV | 76 | 0.39 | <0.01 |
| Masl—V. d. brood | 339 | 0.14 | <0.01 |
| Masl—W. L. V. d.brood—DWV V. d.adults—DWV | 339 76 76 | 0.51 0.92 0.71 | <0.01 <0.01 <0.05 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ramos-Cuellar, A.K.; De la Mora, A.; Contreras-Escareño, F.; Morfin, N.; Tapia-González, J.M.; Macías-Macías, J.O.; Petukhova, T.; Correa-Benítez, A.; Guzman-Novoa, E. Genotype, but Not Climate, Affects the Resistance of Honey Bees (Apis mellifera) to Viral Infections and to the Mite Varroa destructor. Vet. Sci. 2022, 9, 358. https://doi.org/10.3390/vetsci9070358
Ramos-Cuellar AK, De la Mora A, Contreras-Escareño F, Morfin N, Tapia-González JM, Macías-Macías JO, Petukhova T, Correa-Benítez A, Guzman-Novoa E. Genotype, but Not Climate, Affects the Resistance of Honey Bees (Apis mellifera) to Viral Infections and to the Mite Varroa destructor. Veterinary Sciences. 2022; 9(7):358. https://doi.org/10.3390/vetsci9070358
Chicago/Turabian StyleRamos-Cuellar, Ana K., Alvaro De la Mora, Francisca Contreras-Escareño, Nuria Morfin, José M. Tapia-González, José O. Macías-Macías, Tatiana Petukhova, Adriana Correa-Benítez, and Ernesto Guzman-Novoa. 2022. "Genotype, but Not Climate, Affects the Resistance of Honey Bees (Apis mellifera) to Viral Infections and to the Mite Varroa destructor" Veterinary Sciences 9, no. 7: 358. https://doi.org/10.3390/vetsci9070358
APA StyleRamos-Cuellar, A. K., De la Mora, A., Contreras-Escareño, F., Morfin, N., Tapia-González, J. M., Macías-Macías, J. O., Petukhova, T., Correa-Benítez, A., & Guzman-Novoa, E. (2022). Genotype, but Not Climate, Affects the Resistance of Honey Bees (Apis mellifera) to Viral Infections and to the Mite Varroa destructor. Veterinary Sciences, 9(7), 358. https://doi.org/10.3390/vetsci9070358









