Colonisation Patterns of Nosema ceranae in the Azores Archipelago
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Survey and Sampling Collection
2.2. Nosema spp. Extraction and Detection for the Prevalence Study
2.3. N. ceranae Extraction and Load Determination
2.4. Statistical Analysis
3. Results
3.1. Quality Control
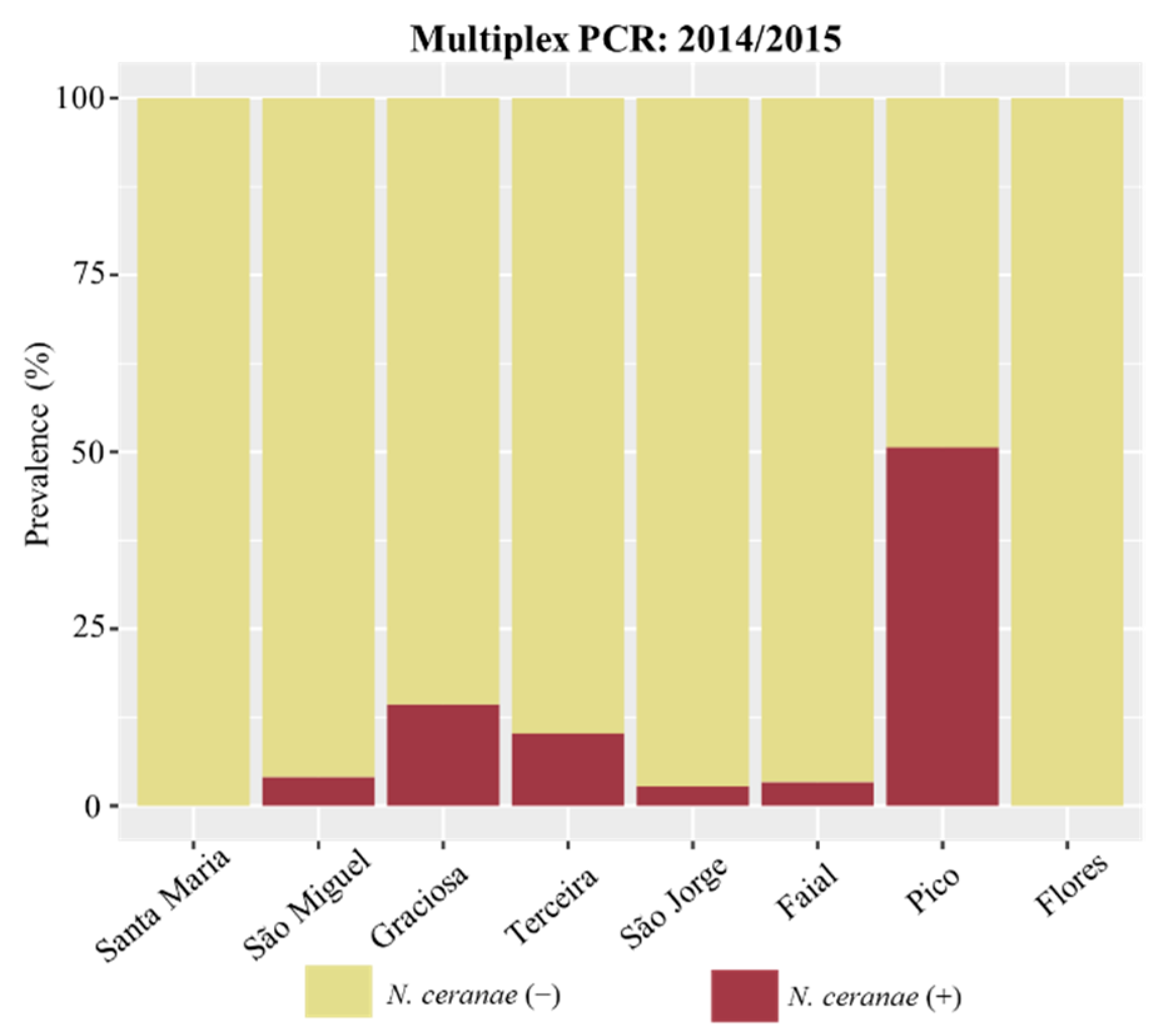
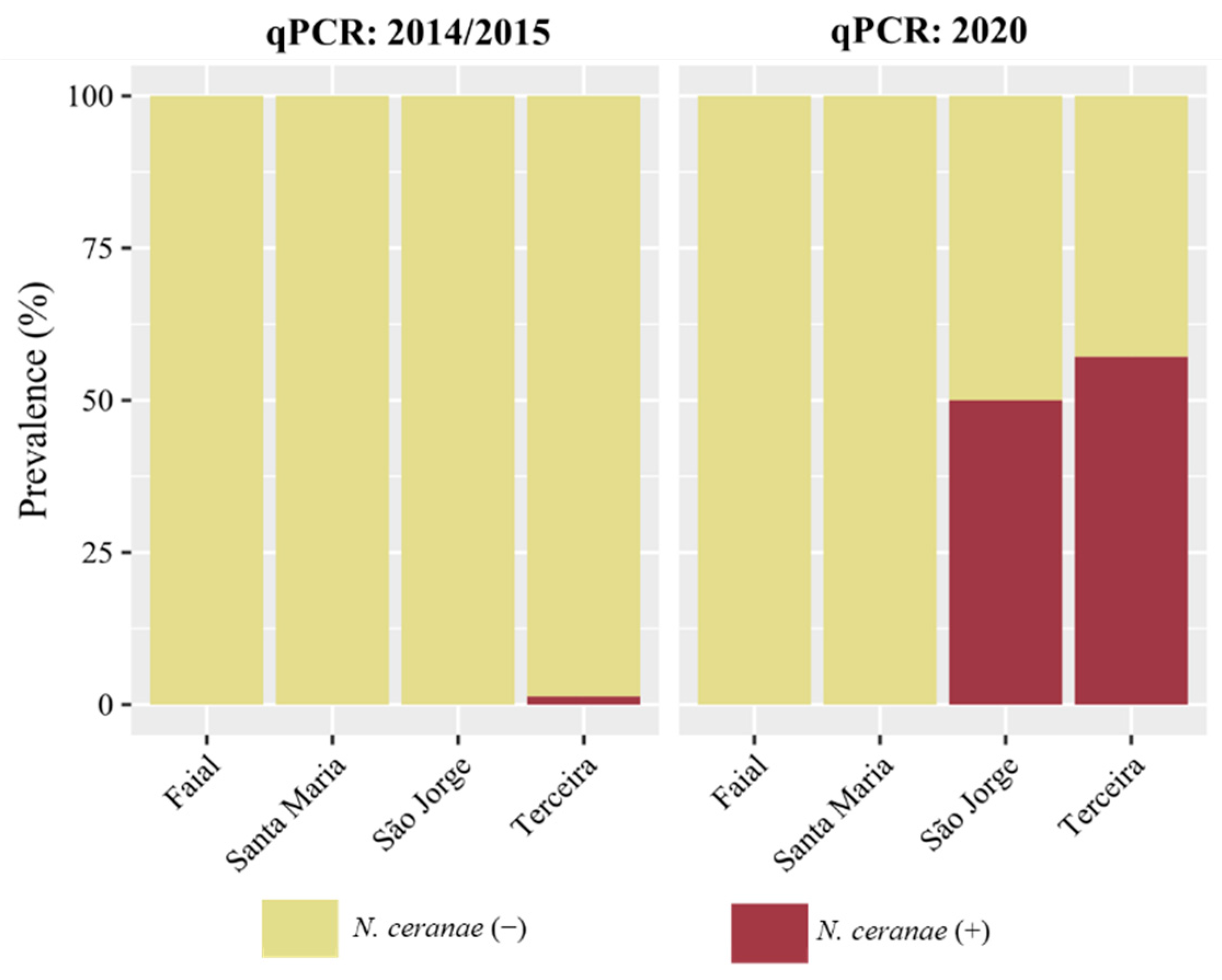
3.2. Detection and Prevalence of Nosema spp. across the Azores
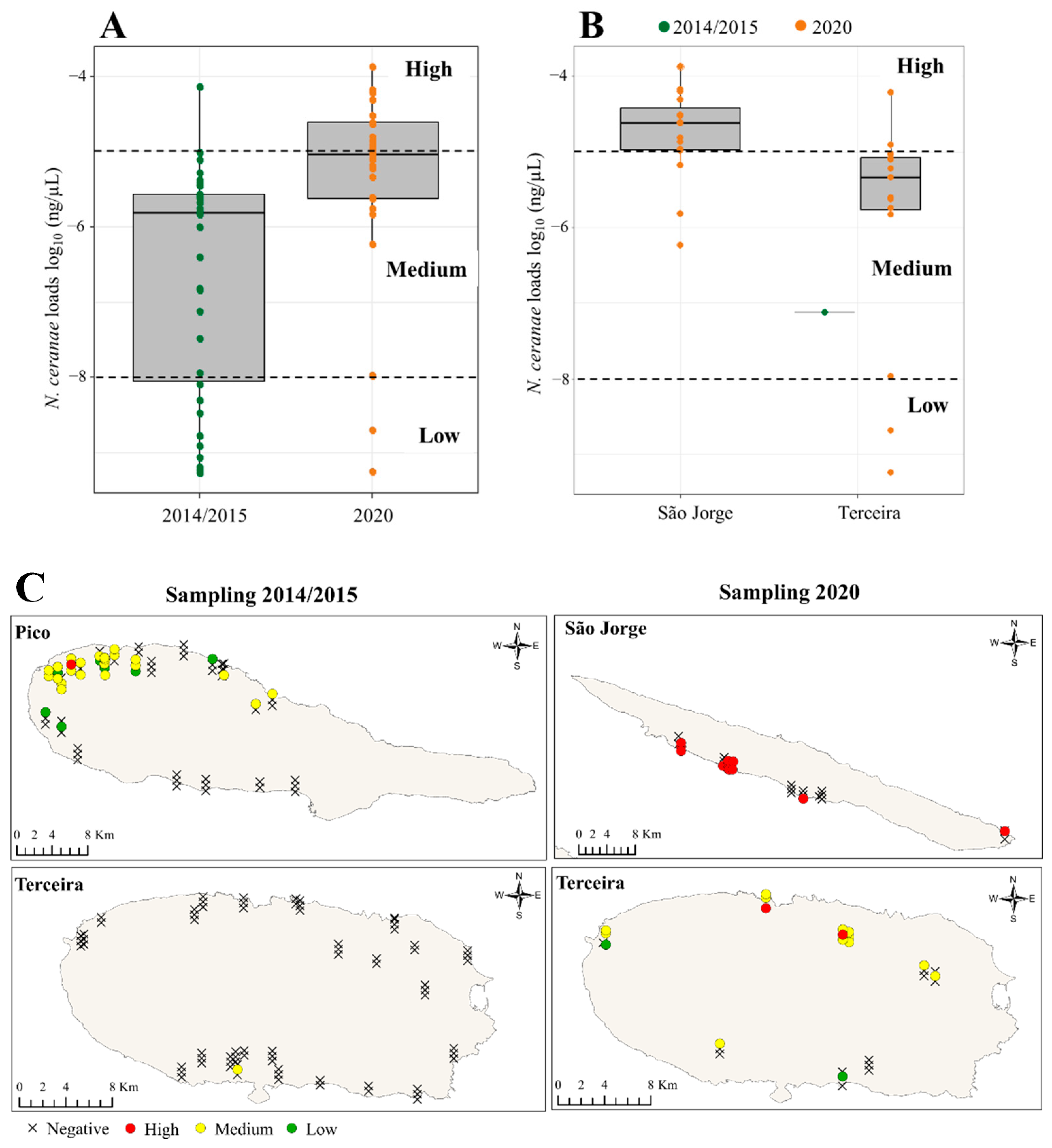
3.3. N. ceranae Loads across the Azores
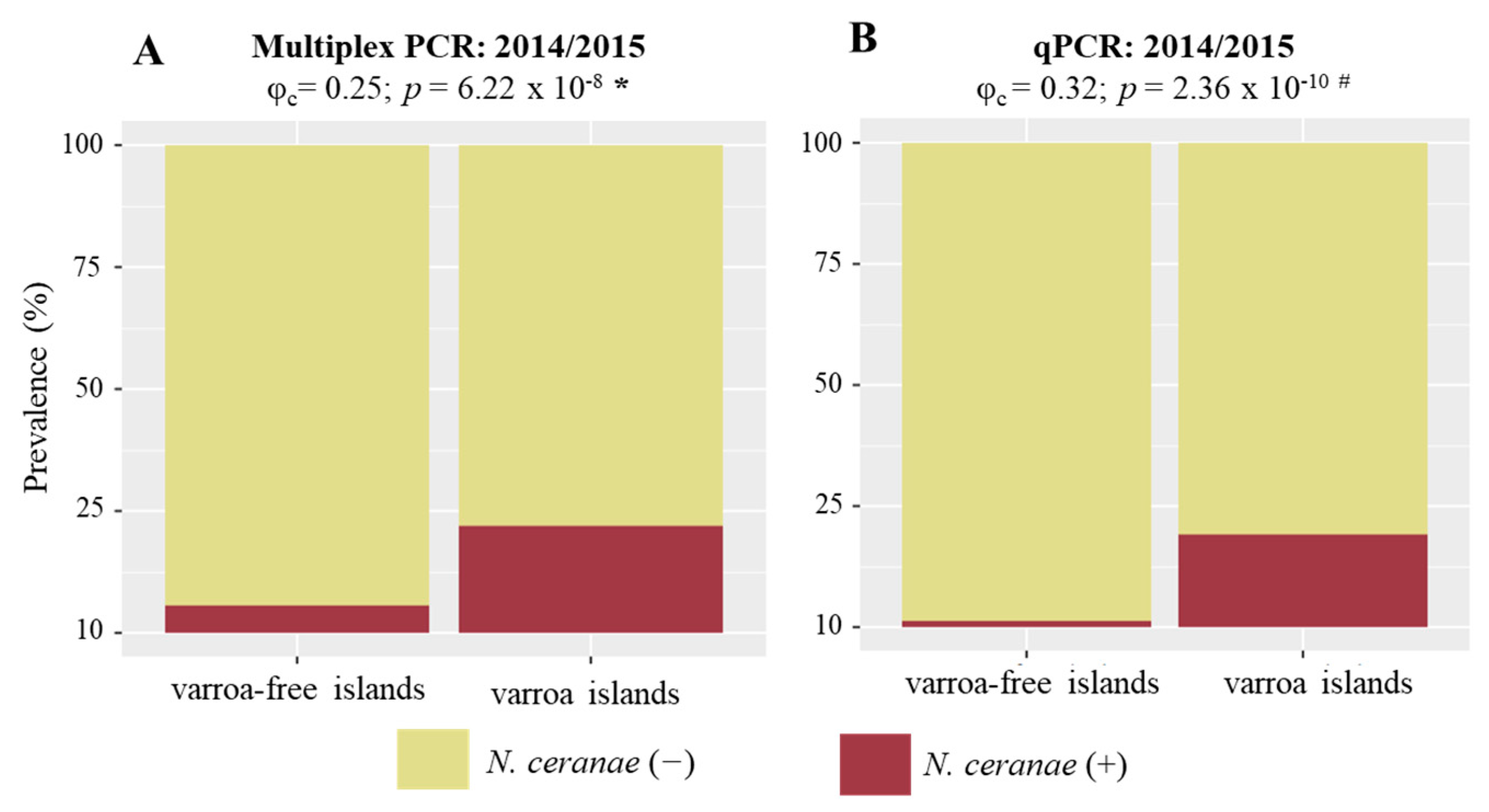
3.4. N. ceranae and V. destructor Associations
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Goulson, D.; Nicholls, E.; Botías, C.; Rotheray, E.L. Bee declines driven by combined stress from parasites, pesticides, and lack of flowers. Science 2015, 347, 1255957. [Google Scholar] [CrossRef]
- Potts, S.G.; Biesmeijer, J.C.; Kremen, C.; Neumann, P.; Schweiger, O.; Kunin, W.E. Global pollinator declines: Trends, impacts and drivers. Trends Ecol. Evol. 2010, 25, 345–353. [Google Scholar] [CrossRef]
- Gallai, N.; Salles, J.M.; Settele, J.; Vaissiere, B.E. Economic valuation of the vulnerability of world agriculture confronted with pollinator decline. Ecol. Econ. 2009, 68, 810–821. [Google Scholar] [CrossRef]
- Hristov, P.; Shumkova, R.; Palova, N.; Neov, B. Honey bee colony losses: Why are honey bees disappearing? Sociobiology 2021, 68, e5851. [Google Scholar] [CrossRef]
- Evans, J.D.; Schwarz, R.S. Bees brought to their knees: Microbes affecting honey bee health. Trends Microbiol. 2011, 19, 614–620. [Google Scholar] [CrossRef] [PubMed]
- Higes, M.; Martín-Hernández, R.; Botías, C.; Bailón, E.G.; González-Porto, A.V.; Barrios, L.; del Nozal, M.J.; Bernal, J.L.; Jiménez, J.J.; Palencia, P.G.; et al. How natural infection by Nosema ceranae causes honeybee colony collapse. Environ. Microbiol. 2008, 10, 2659–2669. [Google Scholar] [CrossRef] [PubMed]
- Emsen, B.; De la Mora, A.; Lacey, B.; Eccles, L.; Kelly, P.G.; Medina-Flores, C.A.; Petukhova, T.; Morfin, N.; Guzman-Novoa, E. Seasonality of Nosema ceranae infections and their relationship with honey bee populations, food stores, and survivorship in a North American region. Vet. Sci. 2020, 7, 131. [Google Scholar] [CrossRef]
- Tokarev, Y.S.; Huang, W.F.; Solter, L.F.; Malysh, J.M.; Becnel, J.J.; Vossbrinck, C.R. A formal redefinition of the genera Nosema and Vairimorpha (Microsporidia: Nosematidae) and reassignment of species based on molecular phylogenetics. J. Invertebr. Pathol. 2020, 169, 7. [Google Scholar] [CrossRef]
- Plischuk, S.; Martín-Hernández, R.; Prieto, L.; Lucía, M.; Botías, C.; Meana, A.; Abrahamovich, A.H.; Lange, C.; Higes, M. South American native bumblebees (Hymenoptera: Apidae) infected by Nosema ceranae (Microsporidia), an emerging pathogen of honeybees (Apis mellifera). Env. Microbiol. Rep. 2009, 1, 131–135. [Google Scholar] [CrossRef]
- Bravi, M.E.; Alvarez, L.J.; Lucia, M.; Pecoraro, M.R.I.; García, M.L.G.; Reynaldi, F.J. Wild bumble bees (Hymenoptera: Apidae: Bombini) as a potential reservoir for bee pathogens in northeastern Argentina. J. Apic. Res. 2019, 58, 710–713. [Google Scholar] [CrossRef]
- Fries, I. Nosema apis—A parasite in the honey bee colony. Bee World 1993, 74, 5–19. [Google Scholar] [CrossRef]
- Fries, I.; Feng, F.; daSilva, A.; Slemenda, S.B.; Pieniazek, N.J. Nosema ceranae n sp (Microspora, Nosematidae), morphological and molecular characterization of a microsporidian parasite of the Asian honey bee Apis cerana (Hymenoptera, Apidae). Eur. J. Protistol. 1996, 32, 356–365. [Google Scholar] [CrossRef]
- Chemurot, M.; De Smet, L.; Brunain, M.; De Rycke, R.; de Graaf, D.C. Nosema neumanni n. sp (Microsporidia, Nosematidae), a new microsporidian parasite of honeybees, Apis mellifera in Uganda. Eur. J. Protistol. 2017, 61, 13–19. [Google Scholar] [CrossRef]
- Higes, M.; Martín-Hernández, R.; Meana, A. Nosema ceranae, a new microsporidian parasite in honeybees in Europe. J. Invertebr. Pathol. 2006, 92, 93–95. [Google Scholar] [CrossRef] [PubMed]
- Huang, W.F.; Jiang, J.H.; Chen, Y.W.; Wang, C.H. A Nosema ceranae isolate from the honeybee Apis mellifera. Apidologie 2007, 38, 30–37. [Google Scholar] [CrossRef]
- Klee, J.; Besana, A.M.; Genersch, E.; Gisder, S.; Nanetti, A.; Tam, D.Q.; Chinh, T.X.; Puerta, F.; Ruz, J.M.; Kryger, P.; et al. Widespread dispersal of the microsporidian Nosema ceranae, an emergent pathogen of the western honey bee, Apis mellifera. J. Invertebr. Pathol. 2007, 96, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Evans, J.D.; Smith, I.B.; Pettis, J.S. Nosema ceranae is a long-present and wide-spread microsporidian infection of the European honey bee (Apis mellifera) in the United States. J. Invertebr. Pathol. 2008, 97, 186–188. [Google Scholar] [CrossRef]
- Giersch, T.; Berg, T.; Galea, F.; Hornitzky, M. Nosema ceranae infects honey bees (Apis mellifera) and contaminates honey in Australia. Apidologie 2009, 40, 117–123. [Google Scholar] [CrossRef]
- Williams, G.R.; Shafer, A.B.A.; Rogers, R.E.L.; Shutler, D.; Stewart, D.T. First detection of Nosema ceranae, a microsporidian parasite of European honey bees (Apis mellifera), in Canada and central USA. J. Invertebr. Pathol. 2008, 97, 189–192. [Google Scholar] [CrossRef]
- Stevanovic, J.; Stanimirovic, Z.; Genersch, E.; Kovacevic, S.R.; Ljubenkovic, J.; Radakovic, M.; Aleksic, N. Dominance of Nosema ceranae in honey bees in the Balkan countries in the absence of symptoms of colony collapse disorder. Apidologie 2011, 42, 49–58. [Google Scholar] [CrossRef]
- Fries, I. Nosema ceranae in European honey bees (Apis mellifera). J. Invertebr. Pathol. 2010, 103, S73–S79. [Google Scholar] [CrossRef] [PubMed]
- Bacandritsos, N.; Granato, A.; Budge, G.; Papanastasiou, I.; Roinioti, E.; Caldon, M.; Falcaro, C.; Gallina, A.; Mutinelli, F. Sudden deaths and colony population decline in Greek honey bee colonies. J. Invertebr. Pathol. 2010, 105, 335–340. [Google Scholar] [CrossRef] [PubMed]
- Papini, R.; Mancianti, F.; Canovai, R.; Cosci, F.; Rocchigiani, G.; Benelli, G.; Canale, A. Prevalence of the microsporidian Nosema ceranae in honeybee (Apis mellifera) apiaries in Central Italy. Saudi J. Biol. Sci. 2017, 24, 979–982. [Google Scholar] [CrossRef]
- Soroker, V.; Hetzroni, A.; Yakobson, B.; David, D.; David, A.; Voet, H.; Slabezki, Y.; Efrat, H.; Levski, S.; Kamer, Y.; et al. Evaluation of colony losses in Israel in relation to the incidence of pathogens and pests. Apidologie 2011, 42, 192–199. [Google Scholar] [CrossRef]
- Higes, M.; García-Palencia, P.; Martín-Hernández, R.; Meana, A. Experimental infection of Apis mellifera honeybees with Nosema ceranae (Microsporidia). J. Invertebr. Pathol. 2007, 94, 211–217. [Google Scholar] [CrossRef]
- Goblirsch, M.; Huang, Z.Y.; Spivak, M. Physiological and behavioral changes in honey bees (Apis mellifera) induced by Nosema ceranae infection. PLoS ONE 2013, 8, 8. [Google Scholar] [CrossRef]
- Mayack, C.; Naug, D. Energetic stress in the honeybee Apis mellifera from Nosema ceranae infection. J. Invertebr. Pathol. 2009, 100, 185–188. [Google Scholar] [CrossRef]
- Lecocq, A.; Jensen, A.B.; Kryger, P.; Nieh, J.C. Parasite infection accelerates age polyethism in young honey bees. Sci Rep. 2016, 6, 11. [Google Scholar] [CrossRef]
- Li, Z.; He, J.; Yu, T.; Chen, Y.; Huang, W.-F.; Huang, J.; Zhao, Y.; Nie, H.; Su, S. Transcriptional and physiological responses of hypopharyngeal glands in honeybees (Apis mellifera L.) infected by Nosema ceranae. Apidologie 2019, 50, 51–62. [Google Scholar] [CrossRef]
- Gage, S.L.; Kramer, C.; Calle, S.; Carroll, M.; Heien, M.; DeGrandi-Hoffman, G. Nosema ceranae parasitism impacts olfactory learning and memory and neurochemistry in honey bees (Apis mellifera). J. Exp. Biol. 2018, 221, jeb161489. [Google Scholar] [CrossRef]
- Dussaubat, C.; Maisonnasse, A.; Crauser, D.; Beslay, D.; Costagliola, G.; Soubeyrand, S.; Kretzchmar, A.; Le Conte, Y. Flight behavior and pheromone changes associated to Nosema ceranae infection of honey bee workers (Apis mellifera) in field conditions. J. Invertebr. Pathol. 2013, 113, 42–51. [Google Scholar] [CrossRef] [PubMed]
- Higes, M.; Martín-Hernández, R.; Garrido-Bailon, E.; González-Porto, A.V.; García-Palencia, P.; Meana, A.; del Nozal, M.J.; Mayo, R.; Bernal, J.L. Honeybee colony collapse due to Nosema ceranae in professional apiaries. Environ. Microbiol. Rep. 2009, 1, 110–113. [Google Scholar] [CrossRef]
- Botías, C.; Martín-Hernández, R.; Barrios, L.; Meana, A.; Higes, M. Nosema spp. infection and its negative effects on honey bees (Apis mellifera iberiensis) at the colony level. Vet. Res. 2013, 44, 14. [Google Scholar] [CrossRef]
- Burgher-MacLellan, K.L.; Williams, G.R.; Shutler, D.; MacKenzie, K.; Rogers, R.E.L. Optimization of duplex real-time PCR with melting-curve analysis for detecting the microsporidian parasites Nosema apis and Nosema ceranae in Apis mellifera. Can. Entomol. 2010, 142, 271–283. [Google Scholar] [CrossRef]
- Luis, A.R.; García, C.A.Y.; Invernizzi, C.; Branchiccela, B.; Piñeiro, A.M.P.; Morfi, A.P.; Zunino, P.; Antúnez, K. Nosema ceranae and RNA viruses in honey bee populations of Cuba. J. Apic. Res. 2020, 59, 468–471. [Google Scholar] [CrossRef]
- Rangel, J.; Gonzalez, A.; Stoner, M.; Hatter, A.; Traver, B.E. Genetic diversity and prevalence of Varroa destructor, Nosema apis, and N. ceranae in managed honey bee (Apis mellifera) colonies in the Caribbean island of Dominica, West Indies. J. Apic. Res. 2018, 57, 541–550. [Google Scholar] [CrossRef]
- Hall, R.J.; Pragert, H.; Phiri, B.J.; Fan, Q.H.; Li, X.; Parnell, A.; Stanislawek, W.L.; McDonald, C.M.; Ha, H.J.; McDonald, W.; et al. Apicultural practice and disease prevalence in Apis mellifera, New Zealand: A longitudinal study. J. Apic. Res. 2021, 60, 644–658. [Google Scholar] [CrossRef]
- Malfroy, S.F.; Roberts, J.M.K.; Perrone, S.; Maynard, G.; Chapman, N. A pest and disease survey of the isolated Norfolk Island honey bee (Apis mellifera) population. J. Apic. Res. 2016, 55, 202–211. [Google Scholar] [CrossRef]
- Budge, G.E.; Pietravalle, S.; Brown, M.; Laurenson, L.; Jones, B.; Tomkies, V.; Delaplane, K.S. Pathogens as predictors of honey bee colony strength in England and Wales. PLoS ONE 2015, 10, e0133228. [Google Scholar] [CrossRef]
- Bollan, K.A.; Hothersall, J.D.; Moffat, C.; Durkacz, J.; Saranzewa, N.; Wright, G.A.; Raine, N.E.; Highet, F.; Connolly, C.N. The microsporidian parasites Nosema ceranae and Nosema apis are widespread in honeybee (Apis mellifera) colonies across Scotland. Parasitol. Res. 2013, 112, 751–759. [Google Scholar] [CrossRef]
- Fürst, M.A.; McMahon, D.P.; Osborne, J.L.; Paxton, R.J.; Brown, M.J.F. Disease associations between honeybees and bumblebees as a threat to wild pollinators. Nature 2014, 506, 364–366. [Google Scholar] [CrossRef] [PubMed]
- Cilia, G.; Sagona, S.; Giusti, M.; dos Santos, P.E.J.; Nanetti, A.; Felicioli, A. Nosema ceranae infection in honeybee samples from Tuscanian Archipelago (Central Italy) investigated by two qPCR methods. Saudi J. Biol. Sci. 2019, 26, 1553–1556. [Google Scholar] [CrossRef] [PubMed]
- Muñoz, I.; Cepero, A.; Pinto, M.A.; Martín-Hernández, R.; Higes, M.; De la Rúa, P. Presence of Nosema ceranae associated with honeybee queen introductions. Infect. Genet. Evol. 2014, 23, 161–168. [Google Scholar] [CrossRef]
- Canto, E. Arquivo dos Açores—Volume 1; University of Toronto: Ponta Delgada, Portugal, 1878; Volume 1, pp. 232–238. [Google Scholar]
- Ferreira, H.; Henriques, D.; Neves, C.J.; Machado, C.A.S.; Azeved, J.C.; Francoy, T.M.; Pinto, M.A. Historical and contemporaneous human-mediated processes left a strong genetic signature on honey bee populations from the Macaronesian archipelago of the Azores. Apidologie 2020, 51, 316–328. [Google Scholar] [CrossRef]
- Direção Regional da Agricultura (DRA). Anexo I—PSA 2016- Situação Epidemiológica (2008-2015); Secretaria Regional da Agricultura e Ambiente: Angra do Heroísmo, Portugal, 2016.
- Direção Regional da Agricultura (DRA). Programa Sanitário Apícola—Região Autónoma dos Açores; Secretaria Regional da Agricultura e Ambiente: Angra do Heroísmo, Portugal, 2022.
- Direção Regional da Agricultura (DRA). Anexo I—PSA 2021—Situação Epidemiológica (2016–2021); Secretaria Regional da Agricultura e Ambiente: Angra Heroísmo, Portugal, 2021.
- Grupe, A.C.; Quandt, C.A. A growing pandemic: A review of Nosema parasites in globally distributed domesticated and native bees. PLoS Pathog. 2020, 16, e1008580. [Google Scholar] [CrossRef]
- Higes, M.; Martín-Hernández, R.; Meana, A. Nosema ceranae in Europe: An emergent type C nosemosis. Apidologie 2010, 41, 375–392. [Google Scholar] [CrossRef]
- Meana, A.; Llorens-Picher, M.; Euba, A.; Bernal, J.L.; Bernal, J.; García-Chao, M.; Dagnac, T.; Castro-Hermida, J.A.; González-Porto, A.V.; Higes, M.; et al. Risk factors associated with honey bee colony loss in apiaries in Galicia, NW Spain. Span. J. Agric. Res. 2017, 15, e0501. [Google Scholar] [CrossRef]
- Le Conte, Y.; Ellis, M.; Ritter, W. Varroa mites and honey bee health: Can Varroa explain part of the colony losses? Apidologie 2010, 41, 353–363. [Google Scholar] [CrossRef]
- Giacobino, A.; Pacini, A.; Molineri, A.; Rodríguez, G.; Crisanti, P.; Bulacio Cagnolo, N.; Merke, J.; Orellano, E.; Bertozzi, E.; Pietronave, H.; et al. Potential associations between the mite Varroa destructor and other stressors in honeybee colonies (Apis mellifera L.) in temperate and subtropical climate from Argentina. Prev. Vet. Med. 2018, 159, 143–152. [Google Scholar] [CrossRef]
- Little, C.M.; Shutler, D.; Williams, G.R. Associations among Nosema spp. fungi, Varroa destructor mites, and chemical treatments in honey bees, Apis mellifera. J. Apic. Res. 2016, 54, 378–385. [Google Scholar] [CrossRef]
- Michalczyk, M.; Sokol, R.; Bancerz-Kisiel, A. Coexistence between selected pathogens in honey bee workers. J. Apic. Res. 2021, 61, 346–351. [Google Scholar] [CrossRef]
- Pacini, A.; Giacobino, A.; Molineri, A.; Bulacio Cagnolo, N.; Aignasse, A.; Zago, L.; Mira, A.; Izaguirre, M.; Schnittger, L.; Merke, J.; et al. Risk factors associated with the abundance of Nosema spp. in apiaries located in temperate and subtropical conditions after honey harvest. J. Apic. Res. 2016, 55, 342–350. [Google Scholar] [CrossRef]
- Mariani, F.; Maggi, M.; Porrini, M.; Fuselli, S.; Caraballo, G.; Brasesco, C.; Barrios, C.; Principal, J.; Martin, E. Parasitic interactions between Nosema spp. and Varroa destructor in Apis mellifera colonies. Zootec. Trop. 2012, 30, 081–090. [Google Scholar]
- Martín-Hernández, R.; Botías, C.; Bailón, E.G.; Martínez-Salvador, A.; Prieto, L.; Meana, A.; Higes, M. Microsporidia infecting Apis mellifera: Coexistence or competition. Is Nosema ceranae replacing Nosema apis? Environ. Microbiol. 2012, 14, 2127–2138. [Google Scholar] [CrossRef]
- Evans, J.D. Beepath: An ordered quantitative-PCR array for exploring honey bee immunity and disease. J. Invertebr. Pathol. 2006, 93, 135–139. [Google Scholar] [CrossRef] [PubMed]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2020; Available online: https://www.R-project.org/ (accessed on 1 April 2022).
- Botías, C.; Martín-Hernández, R.; Meana, A.; Higes, M. Critical aspects of the Nosema spp. diagnostic sampling in honey bee (Apis mellifera L.) colonies. Parasitol. Res. 2012, 110, 2557–2561. [Google Scholar] [CrossRef]
- Martín-Hernández, R.; Bartolomé, C.; Chejanovsky, N.; Le Conte, Y.; Dalmon, A.; Dussaubat, C.; García-Palencia, P.; Meana, A.; Pinto, M.A.; Soroker, V.; et al. Nosema ceranae in Apis mellifera: A 12 years postdetection perspective. Environ. Microbiol. 2018, 20, 1302–1329. [Google Scholar] [CrossRef]
- Csaki, T.; Heltai, M.; Markolt, F.; Kovacs, B.; Bekesi, L.; Ladanyi, M.; Pentek-Zakar, E.; Meana, A.; Botías, C.; Martín-Hernández, R.; et al. Permanent prevalence of Nosema ceranae in honey bees (Apis mellifera) in Hungary. Acta Vet. Hung. 2015, 63, 358–369. [Google Scholar] [CrossRef][Green Version]
- Shumkova, R.; Georgieva, A.; Radoslavov, G.; Sirakova, D.; Dzhebir, G.; Neov, B.; Bouga, M.; Hristov, P. The first report of the prevalence of Nosema ceranae in Bulgaria. PeerJ 2018, 6, 11. [Google Scholar] [CrossRef]
- Matthijs, S.; de Waele, V.; Vandenberge, V.; Verhoeven, B.; Evers, J.; Brunain, M.; Saegerman, C.; de Winter, P.J.J.; Roels, S.; de Graaf, D.C.; et al. Nationwide screening for bee viruses and parasites in belgian honey bees. Viruses 2020, 12, 890. [Google Scholar] [CrossRef]
- Porrini, C.; Mutinelli, F.; Bortolotti, L.; Granato, A.; Laurenson, L.; Roberts, K.; Gallina, A.; Silvester, N.; Medrzycki, P.; Renzi, T.; et al. The status of honey bee health in italy: Results from the nationwide bee monitoring network. PLoS ONE 2016, 11, e0155411. [Google Scholar] [CrossRef] [PubMed]
- Botías, C.; Martín-Hernández, R.; Garrido-Bailon, E.; Gonzalez-Porto, A.; Martinez-Salvador, A.; De La Rúa, P.; Meana, A.; Higes, M. The growing prevalence of Nosema ceranae in honey bees in Spain, an emerging problem for the last decade. Res. Vet. Sci. 2012, 93, 150–155. [Google Scholar] [CrossRef] [PubMed]
- FNAP. Investigação no PAN 2011–2013. Available online: http://fnap.pt/projectos/projectos-de-investigacao-pan-2011-2013/ (accessed on 13 May 2022).
- Paxton, R.J.; Klee, J.; Korpela, S.; Fries, I. Nosema ceranae has infected Apis mellifera in Europe since at least 1998 and may be more virulent than Nosema apis. Apidologie 2007, 38, 558–565. [Google Scholar] [CrossRef]
- MacInnis, C.I.; Keddie, B.A.; Pernal, S.F. Nosema ceranae (Microspora: Nosematidae): A sweet surprise? investigating the viability and infectivity of N. ceranae spores maintained in honey and on beeswax. J. Econ. Entomol. 2020, 113, 2069–2078. [Google Scholar] [CrossRef]
- Teixeira, É.W.; Guimarães-Cestaro, L.; Alves, M.L.T.M.F.; Martins, M.F.; Luz, C.F.P.d.; Serrão, J.E. Spores of Paenibacillus larvae, Ascosphaera apis, Nosema ceranae and Nosema apis in bee products supervised by the Brazilian Federal Inspection Service. Rev. Bras. Entomol. 2018, 62, 188–194. [Google Scholar] [CrossRef]
- Marín-García, P.J.; Peyre, Y.; Ahuir-Baraja, A.E.; Garijo, M.M.; Llobat, L. The role of Nosema ceranae (Microsporidia: Nosematidae) in honey bee colony losses and current insights on treatment. Vet. Sci. 2022, 9, 12. [Google Scholar] [CrossRef] [PubMed]
- Gregory, P.G.; Evans, J.D.; Rinderer, T.; De Guzman, L. Conditional immune-gene suppression of honeybees parasitized by Varroa mites. J. Insect Sci. 2005, 5, 1–5. [Google Scholar] [CrossRef]
- Yang, X.; Cox-Foster, D.L. Impact of an ectoparasite on the immunity and pathology of an invertebrate: Evidence for host immunosuppression and viral amplification. Proc. Natl. Acad. Sci. USA 2005, 102, 7470–7475. [Google Scholar] [CrossRef]
- Reyes-Quintana, M.; Espinosa-Montaño, L.G.; Prieto-Merlos, D.; Koleoglu, G.; Petukhova, T.; Correa-Benítez, A.; Guzman-Novoa, E. Impact of Varroa destructor and deformed wing virus on emergence, cellular immunity, wing integrity and survivorship of Africanized honey bees in Mexico. J. Invertebr. Pathol. 2019, 164, 43–48. [Google Scholar] [CrossRef]
- Graystock, P.; Goulson, D.; Hughes, W.O.H. Parasites in bloom: Flowers aid dispersal and transmission of pollinator parasites within and between bee species. Proc. R. Soc. B-Biol. Sci. 2015, 282, 7. [Google Scholar] [CrossRef]
- Pettis, J.S.; vanEngelsdorp, D.; Johnson, J.; Dively, G. Pesticide exposure in honey bees results in increased levels of the gut pathogen Nosema. Naturwissenschaften 2012, 99, 153–158. [Google Scholar] [CrossRef] [PubMed]
- Castelli, L.; Branchiccela, B.; Garrido, M.; Invernizzi, C.; Porrini, M.; Romero, H.; Santos, E.; Zunino, P.; Antunez, K. Impact of nutritional stress on honeybee gut microbiota, immunity, and Nosema ceranae infection. Microb. Ecol. 2020, 80, 908–919. [Google Scholar] [CrossRef] [PubMed]
- Invernizzi, C.; Santos, E.; García, E.; Daners, G.; Di Landro, R.; Saadoun, A.; Cabrera, C. Sanitary and nutritional characterization of honeybee colonies in Eucalyptus grandis plantations. Arch. De Zootec. 2011, 60, 1303–1314. [Google Scholar] [CrossRef]
- Gisder, S.; Hedtke, K.; Mockel, N.; Frielitz, M.C.; Linde, A.; Genersch, E. Five-year cohort study of Nosema spp. in Germany: Does climate shape virulence and assertiveness of Nosema ceranae? Appl. Environ. Microbiol. 2010, 76, 3032–3038. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.W.; Chung, W.P.; Wang, C.H.; Softer, L.F.; Huang, W.F. Nosema ceranae infection intensity highly correlates with temperature. J. Invertebr. Pathol. 2012, 111, 264–267. [Google Scholar] [CrossRef]
- Martín-Hernández, R.; Meana, A.; García-Palencia, P.; Marín, P.; Botías, C.; Garrido-Bailón, E.; Barrios, L.; Higes, M. Effect of temperature on the biotic potential of honeybee microsporidia. Appl. Environ. Microbiol. 2009, 75, 2554–2557. [Google Scholar] [CrossRef]
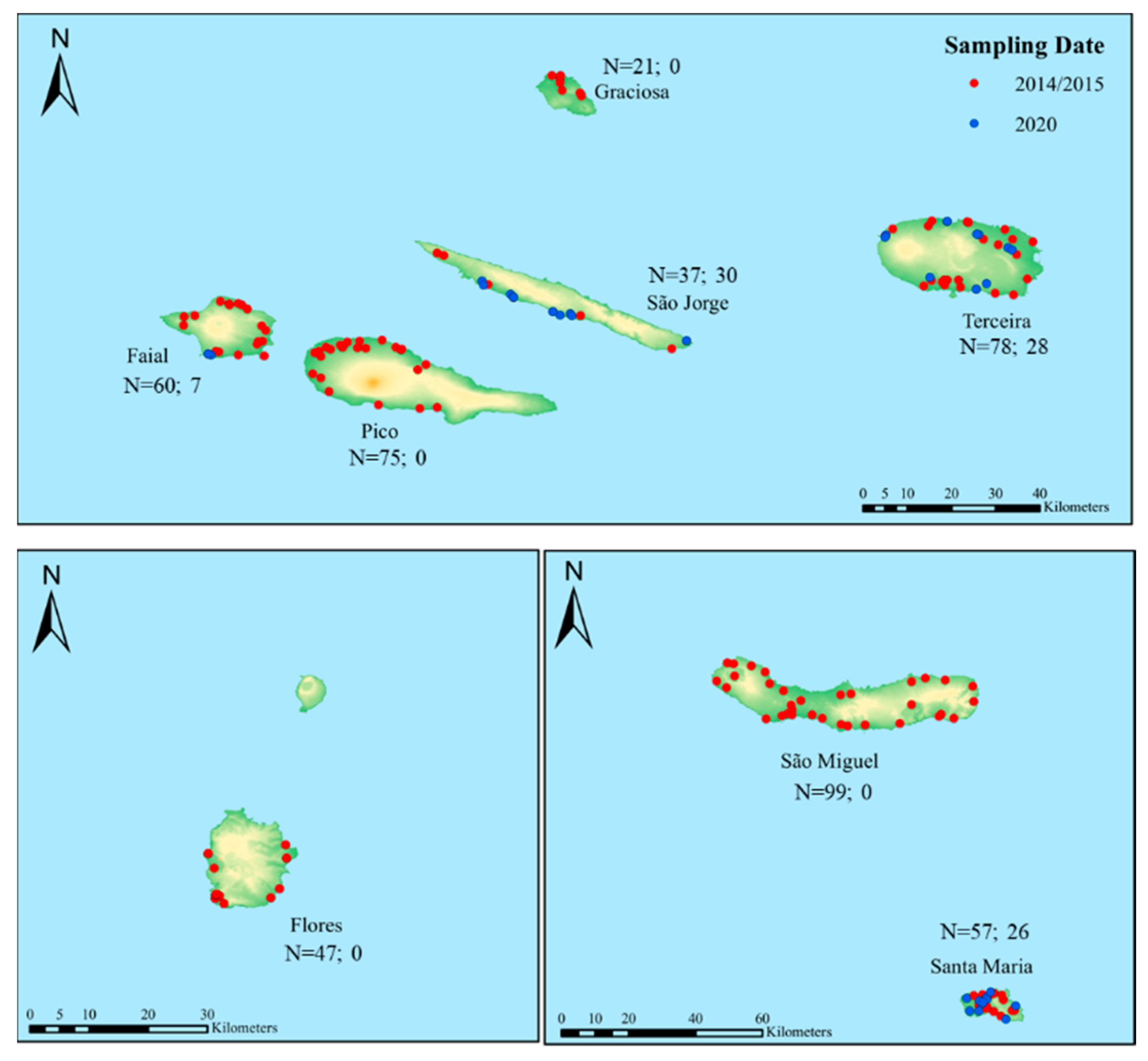
| Island | 2014/2015 | 2020 | ||||||
|---|---|---|---|---|---|---|---|---|
| Colonies | Apiaries | Temperature (°C) * | Rainfall (mm) * | Colonies | Apiaries | Temperature (°C) ** | Rainfall (mm) ** | |
| Santa Maria | 57 | 19 | 22.4/23.1 | 54.7/30.9 | 26 | 12 | 21.4/23.4 | 15.0/36.7 |
| São Miguel | 99 | 33 | 21.6/22.5 | 49.7/58.3 | - | - | 20.8/22.8 | 7.1/19.5 |
| Graciosa | 21 | 7 | 22.0/22.5 | 64.3/91.6 | - | - | *** | *** |
| Terceira | 78 | 26 | 22.5/23.2 | 52.8/35.8 | 28 | 10 | 21.6/23.1 | 9.1/33.8 |
| São Jorge | 37 | 13 | 21.5/21.7 | 67.1/45.3 | 30 | 10 | 21.2/22.8 | 18.0/33.8 |
| Faial | 60 | 20 | 22.1/22.8 | 117.5/63.3 | 7 | 2 | 21.6/22.8 | 16.5/87.0 |
| Pico | 75 | 25 | 22.6/23.3 | 127.6/43.1 | - | - | 22.5/23.4 | 11.8/86.5 |
| Flores | 47 | 13 | 22.3/22.8 | 116.6/165.2 | - | - | 22.3/23.0 | 12.2/131.3 |
| Total | 474 | 156 | 91 | 34 | ||||
| Island | N. ceranae | N. apis | Co-Infection | ||
|---|---|---|---|---|---|
| 2014–2015 | 2020 | 2014–2015 | 2014–2015 | ||
| Multiplex PCR | qPCR | qPCR | Multiplex PCR | Multiplex PCR | |
| Santa Maria | nd (0/57) | nd (0/45) | nd (0/26) | nd (0/57) | nd (0/57) |
| São Miguel | 4.0% (4/99) | 1.1% (1/91) | - | 5.1% (5/99) | 4.0% (4/99) |
| Graciosa | 14.3% (3/21) | 5.6% (1/18) | - | 4.8% (1/21) | nd (0/21) |
| Terceira | 10.3% (8/78) | 1.4% (1/74) | 57.1% (16/28) | 1.3% (1/78) | 1.3% (1/78) |
| São Jorge | 2.7% (1/37) | nd (0/13) | 50.0% (15/30) | 2.7% (1/37) | 2.7% (1/37) |
| Faial | 3.3% (2/60) | nd (0/54) | nd (0/7) | nd (0/60) | nd (0/60) |
| Pico | 50.7% (38/75) | 43.7% (31/71) | - | nd (0/75) | nd (0/75) |
| Flores | nd (0/47) | nd (0/37) | - | 2.1% (1/47) | nd (0/47) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lopes, A.R.; Martín-Hernández, R.; Higes, M.; Segura, S.K.; Henriques, D.; Pinto, M.A. Colonisation Patterns of Nosema ceranae in the Azores Archipelago. Vet. Sci. 2022, 9, 320. https://doi.org/10.3390/vetsci9070320
Lopes AR, Martín-Hernández R, Higes M, Segura SK, Henriques D, Pinto MA. Colonisation Patterns of Nosema ceranae in the Azores Archipelago. Veterinary Sciences. 2022; 9(7):320. https://doi.org/10.3390/vetsci9070320
Chicago/Turabian StyleLopes, Ana Rita, Raquel Martín-Hernández, Mariano Higes, Sara Kafafi Segura, Dora Henriques, and Maria Alice Pinto. 2022. "Colonisation Patterns of Nosema ceranae in the Azores Archipelago" Veterinary Sciences 9, no. 7: 320. https://doi.org/10.3390/vetsci9070320
APA StyleLopes, A. R., Martín-Hernández, R., Higes, M., Segura, S. K., Henriques, D., & Pinto, M. A. (2022). Colonisation Patterns of Nosema ceranae in the Azores Archipelago. Veterinary Sciences, 9(7), 320. https://doi.org/10.3390/vetsci9070320










