Molecular Detection of Malpighamoeba mellificae in Honey Bees
Abstract
:1. Introduction
2. Materials and Methods
2.1. Tested Sample Material
2.2. Sample Disintegration and Nucleic Acid Extraction
2.3. RT-qPCR Assay for Malpighamoeba mellificae
2.4. Sequence Analysis of Amplified Partial Malpighamoeba 18S rRNA Sequences
3. Results
3.1. Sequence Diversity of Malpighamoeba mellificae
3.2. Diagnostic RT-qPCR for Malpighamoeba
3.3. Specificity of MAD-18S Assay
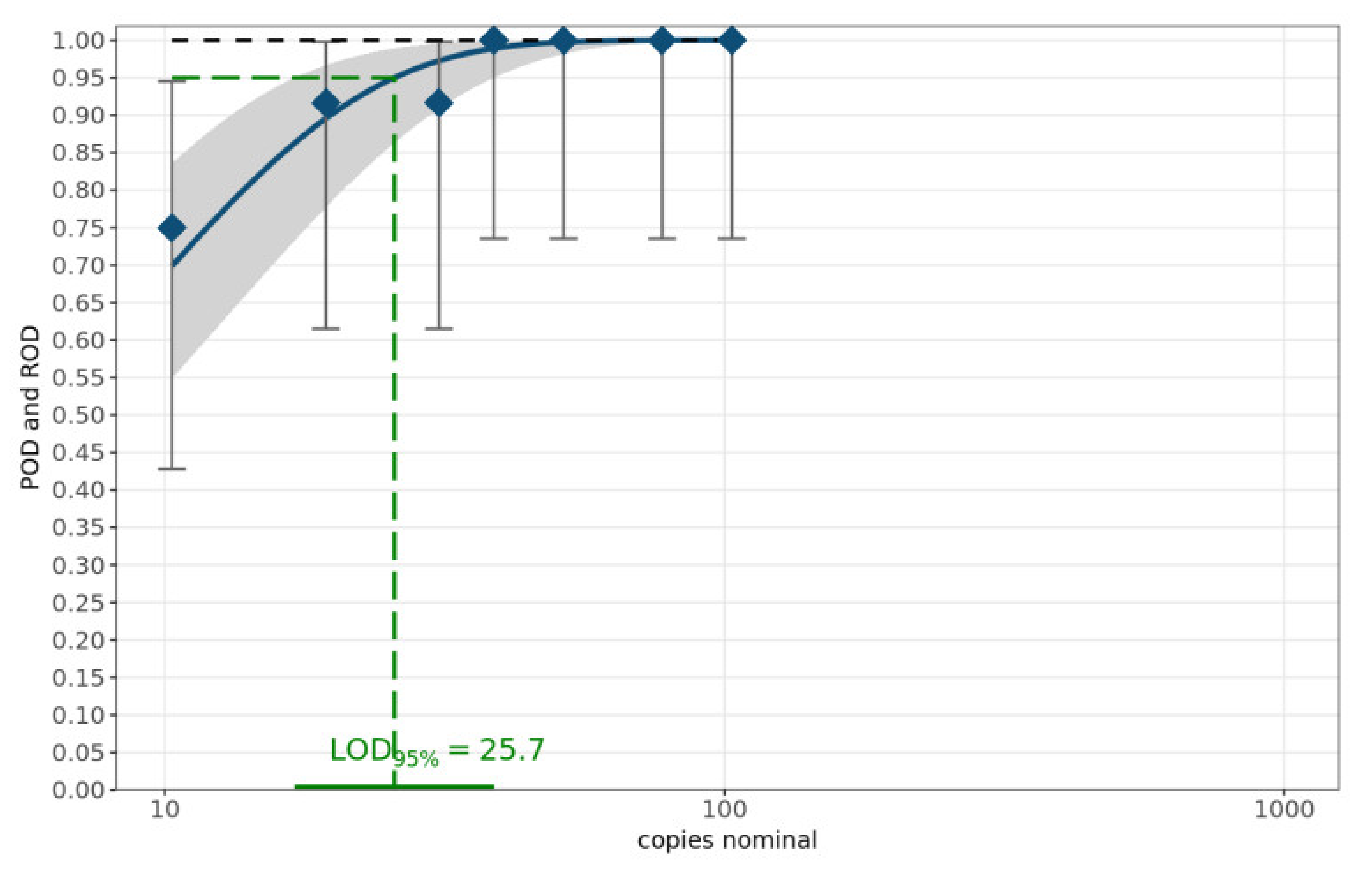
3.4. Sensitivity of MAD-18S Assay
3.5. Investigation of Different Sample Matrices
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Dade, H.A. Anatomy and Dissection of the Honeybee; International Bee Research Association: Cardiff, UK, 1994. [Google Scholar]
- Dow, J.A.; Davies, S.A. The Malpighian tubule: Rapid insights from post-genomic biology. J. Insect Physiol. 2006, 52, 365–378. [Google Scholar] [CrossRef] [PubMed]
- Dow, J.A. Insights into the Malpighian tubule from functional genomics. J. Exp. Biol. 2009, 212, 435–445. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Snodgrass, R.E. Anatomy of the Honey Bee; Comstock Publishing Associates: Ithaca, NY, USA, 1956. [Google Scholar]
- Stell, I. Understanding Bee Anatomy: A Full Colour Guide; Catford Press: London, UK, 2012. [Google Scholar]
- Prell, H. The amoeba-disease of adult bees: A little-noticed spring-time disease. Bee World 1926, 8, 10–13. [Google Scholar] [CrossRef]
- Maaßen, A. Über Bienenkrankheiten. Mitteil Kais Biol. Reichsanst. 1916, 16, 51–58. [Google Scholar]
- Liu, T.P. Scanning electron-microscope observations on the pathological changes of malpighian tubules in the worker honeybee, Apis mellifera, infected by M. mellificae. J. Invertebr. Pathol. 1985, 46, 125–132. [Google Scholar] [CrossRef]
- Bailey, L. The measurement of interrelationships of infections with Nosema apis and Malpighamoeba mellificae of honey-bee populations. J. Invertebr. Pathol. 1968, 12, 175–179. [Google Scholar] [CrossRef]
- Giordani, G. Amoeba disease of the honey bee, Apis mellifera Linnaeus, and an attempt at its chemical control. J. Insect Pathol. 1959, 1, 245–269. [Google Scholar]
- Schuster, F.L. Phylum Rhizopoda. In Handbook of Protoctista; Margulis, L., Corliss, J.O., Melkonian, M., Chapman, D.J., Eds.; Jones and Bartlett: Boston, MA, USA, 1990; pp. 3–18. [Google Scholar]
- Lange, C.E.; Lord, J.C. Protistan Entomopathogens. In Insect Pathology, 2nd ed.; Vega, F.E., Kaya, H.K., Eds.; Elsevier: Amsterdam, The Netherlands, 2012; pp. 368–372. [Google Scholar] [CrossRef]
- Plischuk, S.; Lange, C.E. Detección de Malpighamoeba mellificae (Protista: Amoebozoa) en Apis mellifera (Hymenoptera: Apidae) de Argentina. Rev. Soc. Entomol. Argent. 2010, 69, 299–303. [Google Scholar]
- Wylezich, C.; Höper, D. Meta-Ribosomalomics: RNA sequencing is an unbiased method for parasite detection of different sample types. Front. Microbiol. 2021, 12, 614553. [Google Scholar] [CrossRef] [PubMed]
- Hoffmann, B.; Depner, K.; Schirrmeier, H.; Beer, M. A universal heterologous internal control system for duplex real-time RT-PCR assays used in a detection system for pestiviruses. J. Virol. Methods 2006, 136, 200–209. [Google Scholar] [CrossRef] [PubMed]
- Wylezich, C.; Jürgens, K. Protist diversity in suboxic and sulfidic waters of the Black Sea. Environ. Microbiol. 2011, 13, 2939–2956. [Google Scholar] [CrossRef] [PubMed]
- Diez, B.; Pedros-Alio, C.; Marsh, T.L.; Massana, R. Application of denaturing gradient gel electrophoresis (DGGE) to study the diversity of marine picoeukaryotic assemblages and comparison of DGGE with other molecular techniques. Appl. Environ. Microbiol. 2001, 67, 2942–2951. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wylezich, C.; Meisterfeld, R.; Meisterfeld, S.; Schlegel, M. Phylogenetic analyses of small subunit ribosomal RNA coding regions reveal a monophyletic lineage of euglyphid testate amoebae (Order Euglyphida). J. Eukaryot. Microbiol. 2002, 49, 108–118. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K.; Standley, D.M. MAFFT Multiple Sequence Alignment Software Version 7: Improvements in Performance and Usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bundesamt für Verbraucherschutz und Lebensmittelsicherheit (BVL). Leitlinien zur Einzellabor-Validierung Qualitativer Real-time PCR Methoden. Available online: https://www.bvl.bund.de/SharedDocs/Downloads/07_Untersuchungen/Leitlinien%20zur%20Einzellabor%20Validierung.pdf?__blob=publicationFile&v=5 (accessed on 24 January 2022).
- Web Service. Validation of Qualitative PCR Methods within a Single Laboratory. Available online: https://quodata.de/content/validation-qualitative-pcr-methods-single-laboratory (accessed on 24 January 2022).
- Morgenthaler, O. Von der “Schwindsucht“ der Bienen. Schweiz. Bienen-Ztg. 1925, 48, 279–283. [Google Scholar]
- Bulger, J.W. Malpighamoeba (Prell) in the adult honeybee found in the United States. J. Econ. Entomol. 1928, 21, 376–379. [Google Scholar] [CrossRef]
- Morison, J.D. Amoebic disease of bees in Great Britain. Bee World 1931, 12, 56. [Google Scholar]
- Boncristiani, H.; Ellis, J.D.; Bustamante, T.; Graham, J.; Jack, C.; Kimmel, C.B.; Mortensen, A.; Schmehl, D.R. World Honey Bee Health: The Global Distribution of Western Honey Bee (Apis mellifera L.) Pests and Pathogens. Bee World 2021, 98, 2–6. [Google Scholar] [CrossRef]
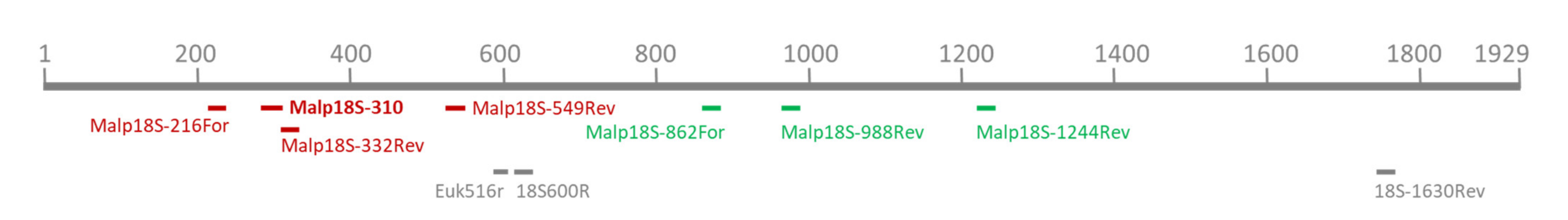
| Name | Sequence (5′–3′) | Reference |
|---|---|---|
| Malp18S-216For | TATACAGATTGTGTAAAAGCG | This study |
| Malp18S-332Rev | TTAGCCTCTATCTAACCTACC | This study |
| Malp18S-549Rev | AAAGCATATCTCGGCATAACCG | This study |
| Malp18S-862For | GGGATTAGATGTATTGGTTGGC | This study |
| Malp18S-988Rev | AATCATCTTCGATCCTTATCCC | This study |
| Malp18S-1244Rev | TCCTACCTTGGTAAAATTTCCC | This study |
| Malp18S-310-FAM | Fam-TACAAGAGGATCTGCCCTATCAACTAT-Tamra | This study |
| 18S-1630Rev | CGACGGGCGGTGTGTACAA | [16] |
| Euk516r | ACCAGACTTGCCCTCC | [17] |
| 18S600R | GCTATTGGAGCTGGAATTACCG | [18] |
| Sample Name | Sample Material | Infection Status | Cq Malpighamoeba | Cq EGFP |
|---|---|---|---|---|
| c1-2020-Mt1 | Malpighian tubules (n = 1) | Verified | 8.5 (0.50) | 27.4 (0.46) |
| c1-2020-Mt2 | Malpighian tubules (n = 1) | Verified | 12.8 (0.11) | 26.2 (0.10) |
| c1-2020-Mt3 | Malpighian tubules (n = 1) | Verified | 9.5 (0.21) | 27.8 (0.10) |
| c1-2020-Mt4 | Malpighian tubules (n = 1) | Verified | 9.4 (0.55) | 26.2 (0.24) |
| c1-2020-ap1 | Abdomen pool (n = 20) | Not verified | 10.8 (0.33) | 29.2 (0.18) |
| c1-2020-ap2 | Abdomen pool (n = 20) | Not verified | 12.3 (0.42) | 27.1 (0.24) |
| c1-2020-ap3 | Abdomen pool (n = 20) | Not verified | 11.1 (0.30) | 27.7 (0.12) |
| c1-2020-ap4 | Abdomen pool (n = 20) | Not verified | 8.9 (0.07) | 27.6 (0.52) |
| c2-2019-Mt | Malpighian tubules (n = 1) | Verified | 18.6 (0.20) | 25.1 (0.06) |
| c2-2019-ap | Abdomen pool (n = 20) | Not verified | 25.1 (0.82) | 26.6 (0.22) |
| c3-2021-Mt | Malpighian tubules (n = 1) | Verified | 16.7 (0.13) | 23.7 (0.20) |
| c3-2021-ap | Abdomen pool (n = 20) | Not verified | 17.9 (0.14) | 24.3 (0.05) |
| c4-2017-Mt | Malpighian tubules (n = 1) | Verified | 14.1 (0.74) | 25.9 (0.63) |
| c4-2017-sup | Supernatant of Mt | Verified | 19.8 (0.03) | 27.1 (0.12) |
| c5-2021-Mt | Malpighian tubules (n = 1) | Verified | 11.9 (0.19) | 24.1 (0.09) |
| c5-2021-ap | Abdomen pool (n = 20) | Not verified | 18.1 (0.26) | 24.6 (0.14) |
| c6-2019-i1 | Intestine (n = 1) | Verified | 24.1 (0.24) | 26.2 (0.12) |
| c6-2019-Mt1 | Malpighian tubules (n = 1) | Verified | 24.6 (0.32) | 26.0 (0.24) |
| c6-2019-i2 | Intestine (n = 1) | Verified | 20.7 (0.30) | 25.0 (0.06) |
| c6-2019-Mt2 | Malpighian tubules (n = 1) | Verified | 24.4 (0.39) | 25.8 (0.05) |
| c6-2019-Mt3 | Malpighian tubules (n = 1) | Verified | 32.1 (0.80) | 26.1 (0.37) |
| c6-2019-sup3 | Supernatant of Mt | Verified | 28.6 (0.36) | 25.9 (0.04) |
| c7-2021-Mt | Malpighian tubules (n = 1) | Not infected | No Cq | 24.1 (0.15) |
| c8-2017-Mt | Malpighian tubules (n = 1) | Verified | 12.7 (0.25) | 22.8 (0.14) |
| c8-2017-sup | Supernatant of Mt | Verified | 17.2 (0.39) | 25.5 (0.14) |
| c9-2020-ap | Abdomen pool (n = 20) | Unknown | 23.1 (0.56) | 29.1 (0.34) |
| c10-2020-ap | Abdomen pool (n = 20) | Unknown | 19.1 (0.53) | 28.8 (0.16) |
| c11-2021-ap | Abdomen pool (n = 20) | Unknown | 18.1 (0.23) | 24.1 (0.15) |
| c12- 2021-ip | Intestine pool (n = 20) | Unknown | 26.0 (0.28) | 22.0 (0.07) |
| c13-2021-f | Feces (fecal spots) | Unknown | 25.2 (0.80) | 22.0 (0.12) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Schäfer, M.O.; Horenk, J.; Wylezich, C. Molecular Detection of Malpighamoeba mellificae in Honey Bees. Vet. Sci. 2022, 9, 148. https://doi.org/10.3390/vetsci9030148
Schäfer MO, Horenk J, Wylezich C. Molecular Detection of Malpighamoeba mellificae in Honey Bees. Veterinary Sciences. 2022; 9(3):148. https://doi.org/10.3390/vetsci9030148
Chicago/Turabian StyleSchäfer, Marc O., Juliane Horenk, and Claudia Wylezich. 2022. "Molecular Detection of Malpighamoeba mellificae in Honey Bees" Veterinary Sciences 9, no. 3: 148. https://doi.org/10.3390/vetsci9030148
APA StyleSchäfer, M. O., Horenk, J., & Wylezich, C. (2022). Molecular Detection of Malpighamoeba mellificae in Honey Bees. Veterinary Sciences, 9(3), 148. https://doi.org/10.3390/vetsci9030148





