Using Blood Transcriptome Analysis to Determine the Changes in Immunity and Metabolism of Giant Pandas with Age
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collection
2.2. RNA Acquisition in Blood Samples
2.3. Library Preparation and Sequencing
- 1.
- mRNA Isolation
- 2.
- mRNA Fragment
- 3.
- First Strand cDNA Synthesis
- 4.
- Second Strand cDNA Synthesis
- 5.
- End repair and Add ‘A’
- 6.
- Adaptor Ligation
- 7.
- PCR
- 8.
- Library Quality Control
- 9.
- Circularization
- 10.
- Sequencing
2.4. Quality Control
- (1)
- Remove reads containing joints (joint contamination);
- (2)
- Remove reads with unknown base N content greater than 5%;
- (3)
- Removal of low-quality reads (low-quality reads were defined as those with a mass value of fewer than 15 bases that accounted for more than 20% of the total number of bases in the reads).
2.5. Gene Enrichment Analysis
3. Results
3.1. Transcriptome Sequencing and Assembly
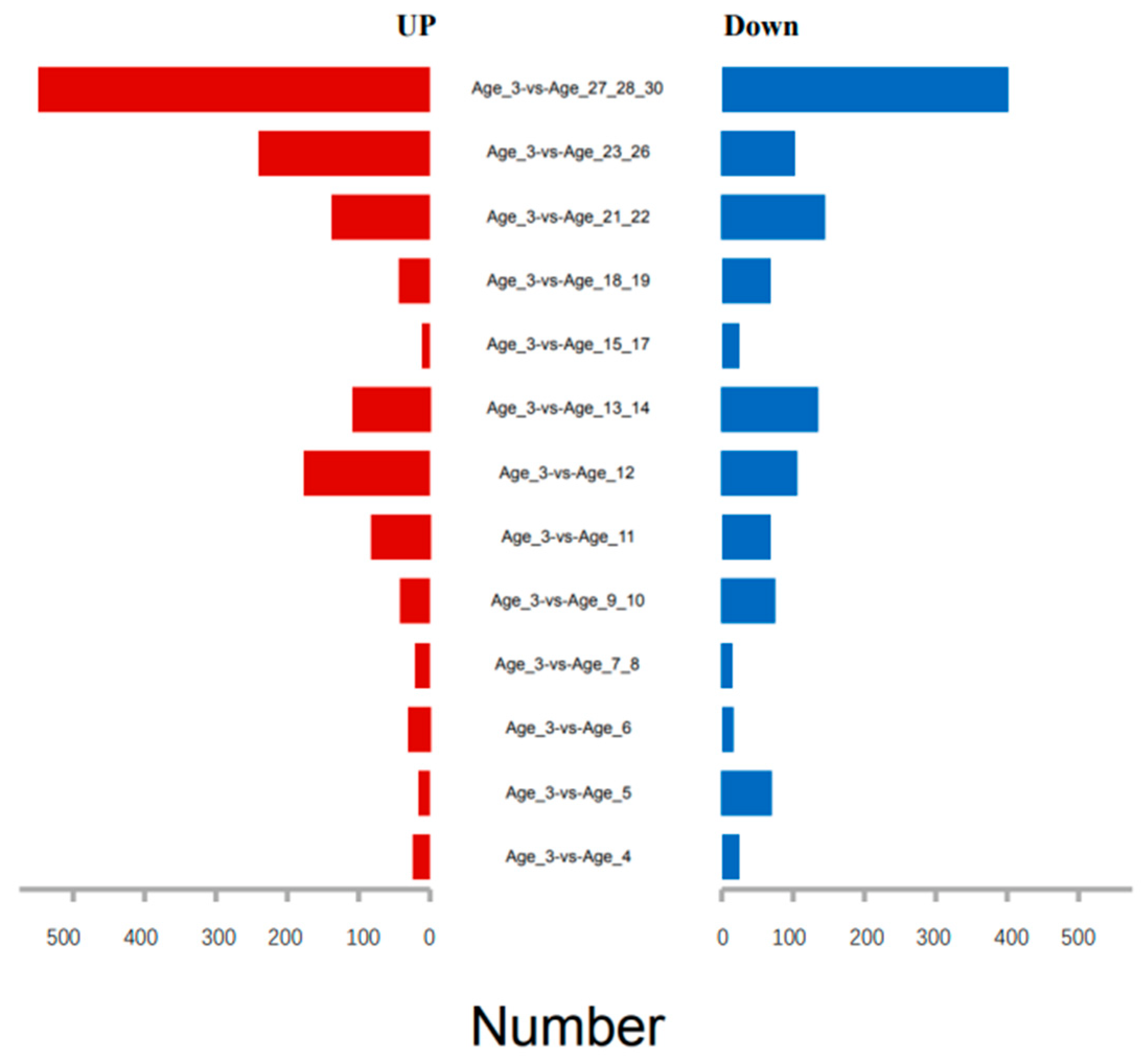
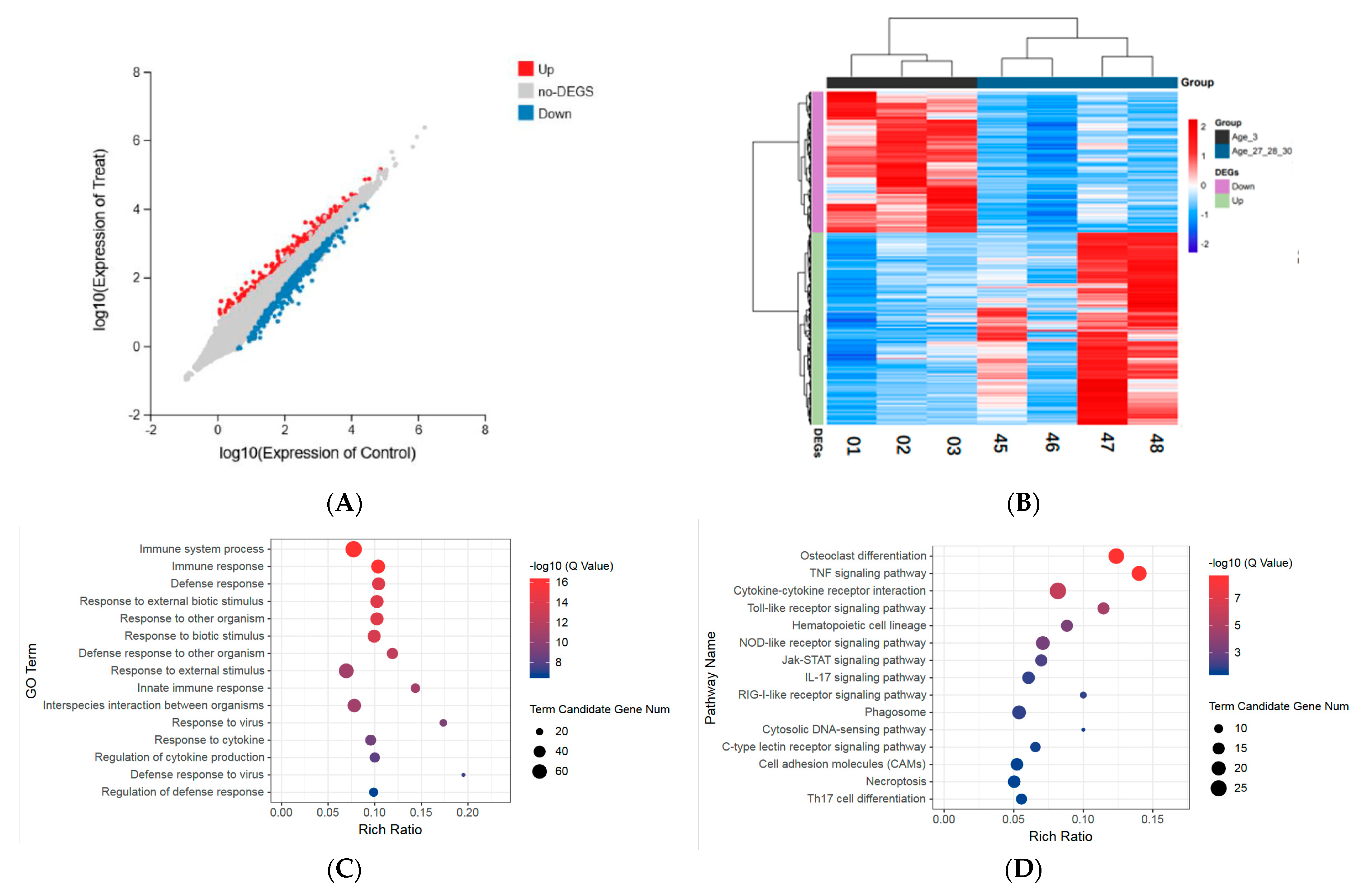
3.2. Identification of Age-Related Differentially Expressed Genes
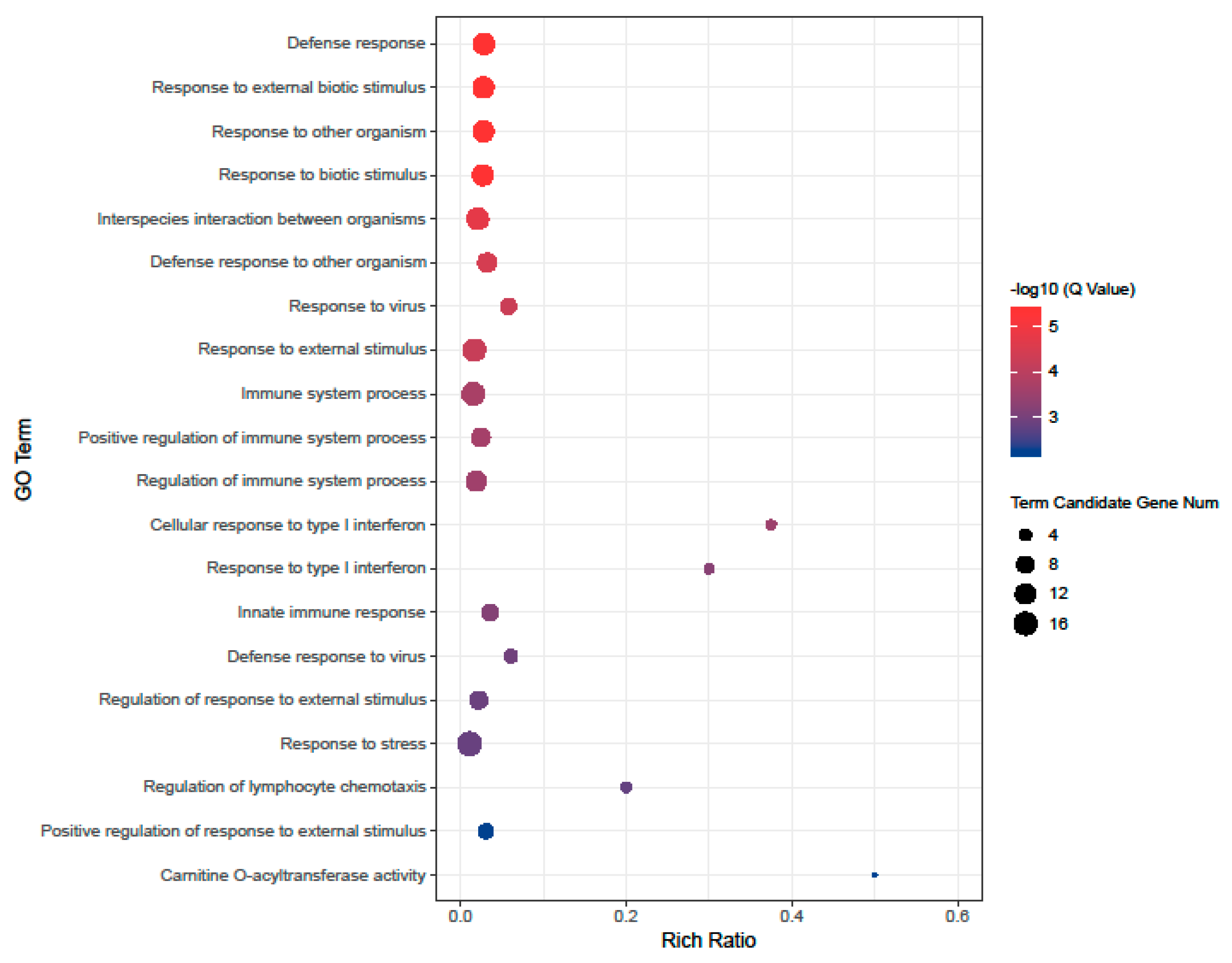
3.3. Gene Ontology Enrichment Analysis of Differentially Expressed Genes
3.4. Pathway Enrichment Analysis of Differentially Expressed Genes
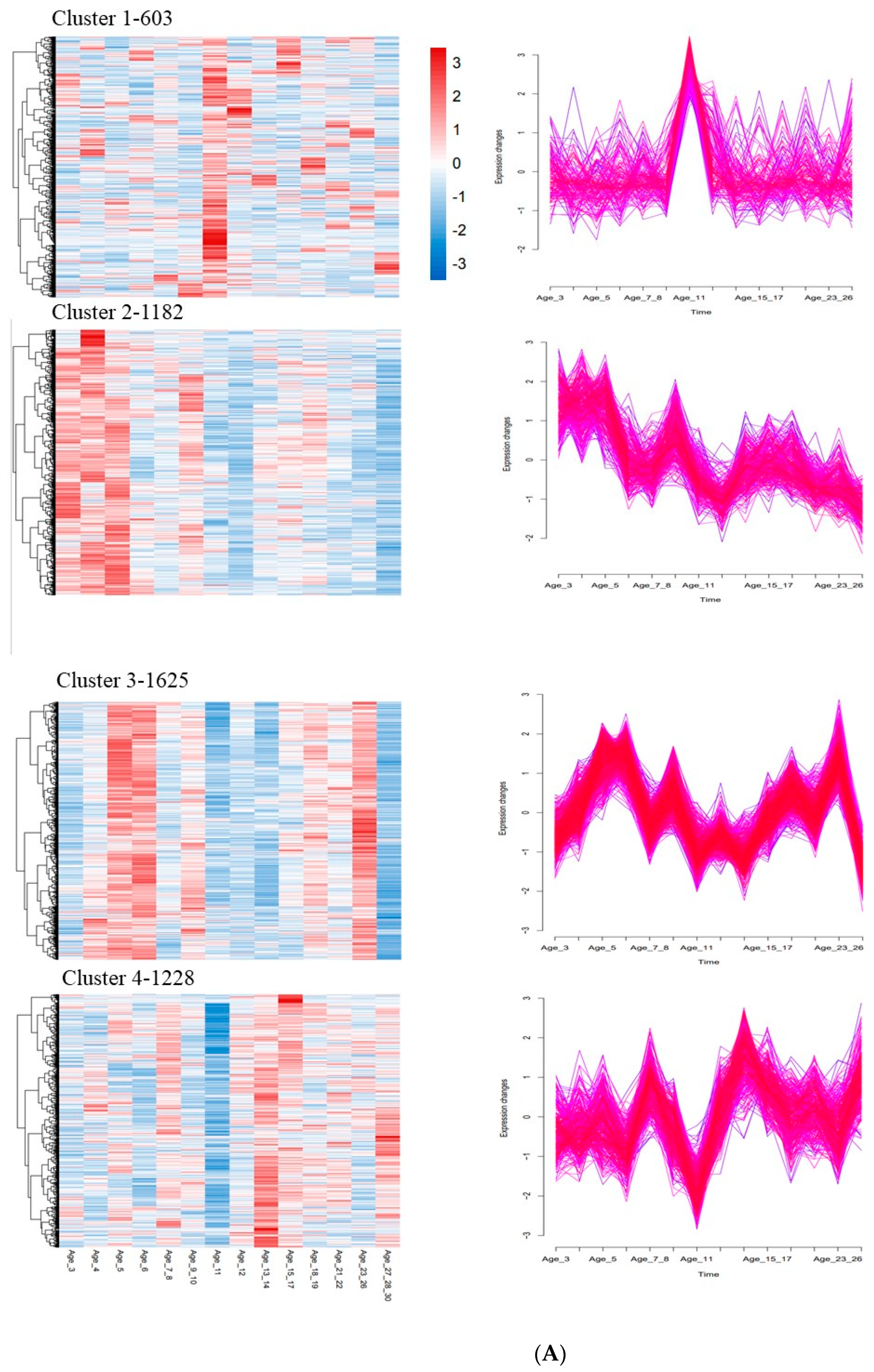
3.5. Time Series Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Huang, J.; Li, Y.-Z.; Du, L.-M.; Yang, B.; Shen, F.-J.; Zhang, H.-M.; Zhang, Z.-H.; Zhang, X.-Y.; Yue, B.-S. Genome-wide survey and analysis of microsatellites in giant panda (Ailuropoda melanoleuca), with a focus on the applications of a novel microsatellite marker system. BMC Genom. 2015, 16, 61. [Google Scholar] [CrossRef] [PubMed]
- Wei, F.; Hu, Y.; Zhu, L.; Bruford, M.W.; Zhan, X.; Zhang, L. Black and white and read all over: The past, present and future of giant panda genetics. Mol. Ecol. 2012, 21, 5660–5674. [Google Scholar] [CrossRef] [PubMed]
- Wei, F.; Wang, X.; Wu, Q. The giant panda gut microbiome. Trends Microbiol. 2015, 23, 450–452. [Google Scholar] [CrossRef] [PubMed]
- Nie, Y.; Speakman, J.R.; Wu, Q.; Zhang, C.; Hu, Y.; Xia, M.; Yan, L.; Hambly, C.; Wang, L.; Wei, W.; et al. Exceptionally low daily energy expenditure in the bamboo-eating giant panda. Science 2015, 349, 171–174. [Google Scholar] [CrossRef] [PubMed]
- Li, R.; Fan, W.; Tian, G.; Zhu, H.; He, L.; Cai, J.; Huang, Q.; Cai, Q.; Li, B.; Bai, Y.; et al. The sequence and de novo assembly of the giant panda genome. Nature 2010, 463, 311–317. [Google Scholar] [CrossRef]
- Zhao, S.; Zheng, P.; Dong, S.; Zhan, X.; Wu, Q.; Guo, X.; Hu, Y.; He, W.; Zhang, S.; Fan, W.; et al. Whole-genome sequencing of giant pandas provides insights into demographic history and local adaptation. Nat. Genet. 2012, 45, 67–71. [Google Scholar] [CrossRef]
- Wei, F.; Hu, Y.; Yan, L.; Nie, Y.; Wu, Q.; Zhang, Z. Giant Pandas Are Not an Evolutionary cul-de-sac: Evidence from Multidisciplinary Research. Mol. Biol. Evol. 2014, 32, 4–12. [Google Scholar] [CrossRef]
- Xie, Y.; Zhou, X.; Zhang, Z.; Wang, C.; Sun, Y.; Liu, T.; Gu, X.; Wang, T.; Peng, X.; Yang, G. Absence of genetic structure in Baylisascaris schroederi populations, a giant panda parasite, determined by mitochondrial sequencing. Parasit Vectors 2014, 7, 606. [Google Scholar] [CrossRef]
- Guo, L.; Yang, S.-L.; Wang, C.-D.; Hou, R.; Chen, S.-J.; Yang, X.-N.; Liu, J.; Pan, H.-B.; Hao, Z.-X.; Zhang, M.-L.; et al. Phylogenetic analysis of the haemagglutinin gene of canine distemper virus strains detected from giant panda and raccoon dogs in China. Virol. J. 2013, 10, 109. [Google Scholar] [CrossRef]
- Feng, N.; Yu, Y.; Wang, T.; Wilker, P.; Wang, J.; Li, Y.; Sun, Z.; Gao, Y.; Xia, X. Fatal canine distemper virus infection of giant pandas in China. Sci. Rep. 2016, 6, 27518. [Google Scholar] [CrossRef]
- Aw, D.; Silva, A.B.; Palmer, D.B. Immunosenescence: Emerging challenges for an ageing population. Immunology 2007, 120, 435–446. [Google Scholar] [CrossRef]
- Sebastiani, P.; Solovieff, N.; Dewan, A.T.; Walsh, K.M.; Puca, A.; Hartley, S.W.; Melista, E.; Andersen, S.; Dworkis, D.A.; Wilk, J.B.; et al. Genetic signatures of exceptional longevity in humans. PLoS ONE 2012, 7, e29848. [Google Scholar] [CrossRef]
- Peters, M.J.; Joehanes, R.; Pilling, L.C.; Schurmann, C.; Conneely, K.N.; Powell, J.; Reinmaa, E.; Sutphin, G.L.; Zhernakova, A.; Schramm, K. The transcriptional landscape of age in human peripheral blood. Nat. Commun. 2015, 6, 1–14. [Google Scholar] [CrossRef]
- Maue, A.C.; Yager, E.J.; Swain, S.L.; Woodland, D.L.; Blackman, M.A.; Haynes, L. T-cell immunosenescence: Lessons learned from mouse models of aging. Trends Immunol. 2009, 30, 301–305. [Google Scholar] [CrossRef]
- Noreen, E.; Bourgeon, S.; Bech, C. Growing old with the immune system: A study of immunosenescence in the zebra finch (Taeniopygia guttata). J. Comp. Physiol. B 2011, 181, 649–656. [Google Scholar] [CrossRef]
- de Magalhães, J.P.; Curado, J.; Church, G.M. Meta-analysis of age-related gene expression profiles identifies common signatures of aging. Bioinformatics 2009, 25, 875–881. [Google Scholar] [CrossRef]
- Kamata, H.; Ohba, S.; Kiba, H.; Tsumagari, S.; Sato, T.; Kanayama, K.; Pan, I.-J.; Watanabe, T.; Zhang, Z.-H.; Hou, R. Seroprevalence of susceptibility to morbillivirus infection on giant panda in captivity. Chin. J. Vet. Sci. 2008, 11, 1167–1170. [Google Scholar]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Charruau, P.; Johnston, R.A.; Stahler, D.R.; Lea, A.; Snyder-Mackler, N.; Smith, D.W.; vonHoldt, B.M.; Cole, S.W.; Tung, J.; Wayne, R.K. Pervasive Effects of Aging on Gene Expression in Wild Wolves. Mol. Biol. Evol. 2016, 33, 1967–1978. [Google Scholar] [CrossRef]
- Grandvaux, N.; Servant, M.J.; Tenoever, B.; Sen, G.C.; Balachandran, S.; Barber, G.N.; Lin, R.; Hiscott, J. Transcriptional profiling of interferon regulatory factor 3 target genes: Direct involvement in the regulation of interferon-stimulated genes. J. Virol. 2002, 76, 5532–5539. [Google Scholar] [CrossRef]
- Glass, W.G.; Rosenberg, H.F.; Murphy, P.M. Chemokine regulation of inflammation during acute viral infection. Curr. Opin. Allergy Clin. Immunol. 2003, 3, 467–473. [Google Scholar] [CrossRef] [PubMed]
- Tedla, N.; Palladinetti, P.; Kelly, M.; Kumar, R.K.; DiGirolamo, N.; Chattophadhay, U.; Cooke, B.; Truskett, P.; Dwyer, J.; Wakefield, D. Chemokines and T lymphocyte recruitment to lymph nodes in HIV infection. Am. J. Pathol. 1996, 148, 1367. [Google Scholar] [PubMed]
- Dumbrepatil, A.B.; Ghosh, S.; Zegalia, K.A.; Malec, P.A.; Hoff, J.D.; Kennedy, R.T.; Marsh, E.N.G. Viperin interacts with the kinase IRAK1 and the E3 ubiquitin ligase TRAF6, coupling innate immune signaling to antiviral ribonucleotide synthesis. J. Biol. Chem. 2019, 294, 6888–6898. [Google Scholar] [CrossRef] [PubMed]
- Tang, M.; Dong, X.; Xiao, L.; Tan, Z.; Luo, X.; Yang, L.; Li, W.; Shi, F.; Li, Y.; Zhao, L.; et al. CPT1A-mediated fatty acid oxidation promotes cell proliferation via nucleoside metabolism in nasopharyngeal carcinoma. Cell Death Dis. 2022, 13, 331. [Google Scholar] [CrossRef]
- Frenk, S.; Houseley, J. Gene expression hallmarks of cellular ageing. Biogerontology 2018, 19, 547–566. [Google Scholar] [CrossRef]
- Yao, Y.; Cai, X.; Ren, F.; Ye, Y.; Wang, F.; Zheng, C.; Qian, Y.; Zhang, M. The Macrophage-Osteoclast Axis in Osteoimmunity and Osteo-Related Diseases. Front. Immunol. 2021, 12, 664871. [Google Scholar] [CrossRef]
- Zuehlke, A.D.; Beebe, K.; Neckers, L.; Prince, T. Regulation and function of the human HSP90AA1 gene. Gene 2015, 570, 8–16. [Google Scholar] [CrossRef]
- Dong, B.; Zhou, Q.; Zhao, J.; Zhou, A.; Harty, R.N.; Bose, S.; Banerjee, A.; Slee, R.; Guenther, J.; Williams, B.R.G.; et al. Phospholipid scramblase 1 potentiates the antiviral activity of interferon. J. Virol. 2004, 78, 8983–8993. [Google Scholar] [CrossRef]
- Kusano, S.; Ikeda, M. Interaction of phospholipid scramblase 1 with the Epstein-Barr virus protein BZLF1 represses BZLF1-mediated lytic gene transcription. J. Biol. Chem. 2019, 294, 15104–15116. [Google Scholar] [CrossRef]
- Luo, W.; Zhang, J.; Liang, L.; Wang, G.; Li, Q.; Zhu, P.; Zhou, Y.; Li, J.; Zhao, Y.; Sun, N.; et al. Phospholipid scramblase 1 interacts with influenza A virus NP, impairing its nuclear import and thereby suppressing virus replication. PLoS Pathog 2018, 14, e1006851. [Google Scholar] [CrossRef]
- Yuan, Y.; Tian, C.; Gong, Q.; Shang, L.; Zhang, Y.; Jin, C.; He, F.; Wang, J. Interactome Map Reveals Phospholipid Scramblase 1 as a Novel Regulator of Hepatitis B Virus X Protein. J. Proteome Res. 2014, 14, 154–163. [Google Scholar] [CrossRef]
- Kalia, N.; Singh, J.; Kaur, M. The role of dectin-1 in health and disease. Immunobiology 2021, 226, 152071. [Google Scholar] [CrossRef]
- Katagiri, T.; Yamazaki, S.; Fukui, Y.; Aoki, K.; Yagita, H.; Nishina, T.; Mikami, T.; Katagiri, S.; Shiraishi, A.; Kimura, S.; et al. JunB plays a crucial role in development of regulatory T cells by promoting IL-2 signaling. Mucosal Immunol. 2019, 12, 1104–1117. [Google Scholar] [CrossRef]
- Grajales-Reyes, G.E.; Iwata, A.; Albring, J.; Wu, X.; Tussiwand, R.; Kc, W.; Kretzer, N.M.; Briseño, C.G.; Durai, V.; Bagadia, P.; et al. Batf3 maintains autoactivation of Irf8 for commitment of a CD8α(+) conventional DC clonogenic progenitor. Nat. Immunol. 2015, 16, 708–717. [Google Scholar] [CrossRef]
- Liu, X.; Zhan, Z.; Li, D.; Xu, L.; Ma, F.; Zhang, P.; Yao, H.; Cao, X. Intracellular MHC class II molecules promote TLR-triggered innate immune responses by maintaining activation of the kinase Btk. Nat. Immunol. 2011, 12, 416–424. [Google Scholar] [CrossRef]
- Pathak, M.; Lal, G. The Regulatory Function of CCR9(+) Dendritic Cells in Inflammation and Autoimmunity. Front. Immunol. 2020, 11, 6326. [Google Scholar] [CrossRef]
- Tu, Z.; Xiao, R.; Xiong, J.; Tembo, K.M.; Deng, X.; Xiong, M.; Liu, P.; Wang, M.; Zhang, Q. CCR9 in cancer: Oncogenic role and therapeutic targeting. J. Hematol. Oncol. 2016, 9, 10. [Google Scholar] [CrossRef]
- Song, J. Adenosine triphosphate energy-independently controls protein homeostasis with unique structure and diverse mechanisms. Protein. Sci. 2021, 30, 1277–1293. [Google Scholar] [CrossRef]
- Huang, X.; Ouyang, Q.; Ran, M.; Zeng, B.; Deng, L.; Hu, S.; Yang, M.; Li, G.; Deng, T.; He, M.; et al. The immune and metabolic changes with age in giant panda blood by combined transcriptome and DNA methylation analysis. Aging 2020, 12, 21777–21797. [Google Scholar] [CrossRef]
- Du, L.; Liu, Q.; Shen, F.; Fan, Z.; Hou, R.; Yue, B.; Zhang, X. Transcriptome analysis reveals immune-related gene expression changes with age in giant panda (Ailuropoda melanoleuca) blood. Aging 2019, 11, 249–262. [Google Scholar] [CrossRef]


| Sample | Age | Gender | Group | Total Raw Reads (M) | Total Clean Reads (M) | Total Clean Bases (Gb) | Clean Reads Q30 (%) | Clean Reads Ratio (%) | Total Mapping (%) |
|---|---|---|---|---|---|---|---|---|---|
| 01 | 3 | Male | Age_3 | 43.82 | 42.97 | 6.45 | 93.11 | 98.05 | 92.69 |
| 02 | 3 | Female | Age_3 | 43.82 | 42.91 | 6.44 | 92.94 | 97.92 | 91.08 |
| 03 | 3 | Female | Age_3 | 43.82 | 43.09 | 6.46 | 93.55 | 98.34 | 92.13 |
| 04 | 4 | Male | Age_4 | 43.82 | 42.96 | 6.44 | 93.63 | 98.04 | 91.93 |
| 05 | 4 | Female | Age_4 | 43.82 | 42.86 | 6.43 | 93.73 | 97.8 | 91.91 |
| 06 | 4 | Male | Age_4 | 43.82 | 42.86 | 6.43 | 93.56 | 97.8 | 91.62 |
| 07 | 5 | Female | Age_5 | 43.82 | 42.87 | 6.43 | 92.84 | 97.83 | 90.83 |
| 08 | 5 | Female | Age_5 | 43.82 | 43.09 | 6.46 | 92.78 | 98.32 | 89.88 |
| 09 | 5 | Female | Age_5 | 45.57 | 43.58 | 6.54 | 89.68 | 95.63 | 86.82 |
| 10 | 6 | Female | Age_6 | 43.82 | 42.89 | 6.43 | 92.75 | 97.88 | 91.05 |
| 11 | 6 | Female | Age_6 | 43.82 | 42.99 | 6.45 | 92.58 | 98.11 | 91.71 |
| 12 | 6 | Male | Age_6 | 43.82 | 42.44 | 6.37 | 89.63 | 96.86 | 85.84 |
| 13 | 6 | Female | Age_6 | 43.82 | 42.11 | 6.32 | 89.45 | 96.1 | 86.55 |
| 14 | 7 | Male | Age_7_8 | 43.82 | 42.62 | 6.39 | 92.59 | 97.25 | 91.56 |
| 15 | 7 | Male | Age_7_8 | 43.82 | 42.6 | 6.39 | 93.09 | 97.21 | 91.08 |
| 16 | 8 | Male | Age_7_8 | 43.82 | 42.91 | 6.44 | 92.89 | 97.91 | 91.24 |
| 17 | 8 | Male | Age_7_8 | 43.82 | 42.95 | 6.44 | 92.87 | 98.01 | 91.26 |
| 18 | 8 | Female | Age_7_8 | 43.82 | 42.89 | 6.43 | 93.33 | 97.87 | 92.38 |
| 19 | 9 | Female | Age_9_10 | 43.82 | 43.11 | 6.47 | 92.5 | 98.39 | 91.93 |
| 20 | 10 | Female | Age_9_10 | 43.82 | 43.04 | 6.46 | 93.48 | 98.22 | 92.35 |
| 21 | 10 | Female | Age_9_10 | 43.82 | 42.63 | 6.39 | 93.5 | 97.28 | 92.16 |
| 22 | 11 | Female | Age_11 | 43.82 | 42.91 | 6.44 | 93.3 | 97.93 | 90.86 |
| 23 | 11 | Female | Age_11 | 43.82 | 43.07 | 6.46 | 93.81 | 98.28 | 91.9 |
| 24 | 11 | Female | Age_11 | 43.82 | 43.11 | 6.47 | 92.94 | 98.37 | 92.51 |
| 25 | 12 | Male | Age_12 | 43.82 | 42.72 | 6.41 | 93.11 | 97.48 | 89.86 |
| 26 | 12 | Male | Age_12 | 43.82 | 43.04 | 6.46 | 93.39 | 98.23 | 91.8 |
| 27 | 12 | Female | Age_12 | 43.82 | 43.07 | 6.46 | 93.16 | 98.29 | 91.63 |
| 28 | 13 | Female | Age_13_14 | 43.82 | 42.15 | 6.32 | 93.54 | 96.19 | 90.35 |
| 29 | 13 | Male | Age_13_14 | 43.82 | 42.99 | 6.45 | 93.34 | 98.1 | 91.47 |
| 30 | 14 | Female | Age_13_14 | 43.82 | 43.32 | 6.5 | 92.79 | 98.87 | 91.19 |
| 31 | 14 | Male | Age_13_14 | 43.82 | 43.26 | 6.49 | 93.33 | 98.71 | 91.69 |
| 32 | 15 | Female | Age_15_17 | 43.82 | 43.09 | 6.46 | 93.03 | 98.32 | 92.25 |
| 33 | 15 | Male | Age_15_17 | 43.82 | 43.14 | 6.47 | 93.19 | 98.45 | 91.73 |
| 34 | 17 | Female | Age_15_17 | 43.82 | 42.99 | 6.45 | 93.69 | 98.11 | 92.44 |
| 35 | 18 | Female | Age_18_19 | 43.82 | 43.06 | 6.46 | 93.24 | 98.26 | 92.34 |
| 36 | 18 | Female | Age_18_19 | 43.82 | 43.11 | 6.47 | 93.2 | 98.38 | 91.62 |
| 37 | 19 | Female | Age_18_19 | 43.82 | 43.1 | 6.47 | 93.39 | 98.36 | 91.25 |
| 38 | 19 | Male | Age_18_19 | 43.82 | 43 | 6.45 | 93.34 | 98.14 | 92.29 |
| 39 | 21 | Female | Age_21_22 | 43.82 | 42.89 | 6.43 | 93.11 | 97.87 | 91.46 |
| 40 | 22 | Female | Age_21_22 | 43.82 | 43.12 | 6.47 | 93.13 | 98.4 | 91.53 |
| 41 | 22 | Female | Age_21_22 | 43.82 | 42.91 | 6.44 | 93.16 | 97.93 | 91.88 |
| 42 | 23 | Male | Age_23_26 | 43.82 | 43.01 | 6.45 | 92.85 | 98.14 | 90.03 |
| 43 | 26 | Female | Age_23_26 | 43.82 | 42.65 | 6.4 | 92.97 | 97.33 | 91.51 |
| 44 | 23 | Female | Age_23_26 | 45.57 | 43.56 | 6.53 | 89.28 | 95.58 | 85.69 |
| 45 | 27 | Female | Age_27_28_30 | 43.82 | 43.03 | 6.45 | 92.73 | 98.18 | 90.75 |
| 46 | 28 | Male | Age_27_28_30 | 43.82 | 42.97 | 6.45 | 93.31 | 98.06 | 90.83 |
| 47 | 30 | Female | Age_27_28_30 | 43.82 | 42.93 | 6.44 | 93.32 | 97.97 | 91.54 |
| 48 | 30 | Female | Age_27_28_30 | 43.82 | 42.72 | 6.41 | 92.99 | 97.48 | 90.06 |
| Gene ID | Gene symbol | log2FC | Q value |
|---|---|---|---|
| 100463753 | CCL5 | 1.197677414 | 0.006639364 |
| 100465129 | TLR2 | 0.672103431 | 0.045445131 |
| 100465831 | LOC100465831 | 1.393709166 | 0.032208904 |
| 100466958 | TMCO6 | 0.625425843 | 0.044165086 |
| 100467144 | TLR8 | 0.772216276 | 0.028726888 |
| 100467227 | SLPI | 1.084231605 | 0.040962187 |
| 100468112 | OAS2 | 1.901277089 | 0.00136348 |
| 100469387 | ISG15 | 2.542014757 | 0.009571346 |
| 100469482 | NOD2 | 1.392587422 | 0.003516809 |
| 100472167 | STAT2 | 1.657003098 | 0.001891366 |
| 100474026 | STAT1 | 1.162051454 | 0.040464308 |
| 100475582 | LOC100475582 | 0.769064873 | 0.044459423 |
| 100475654 | TLR5 | 1.265140332 | 0.007245626 |
| 100475782 | LOC100475782 | 3.167993087 | 6.07 × 10−7 |
| 100476031 | TRIM8 | 1.217417982 | 0.00289373 |
| 100477088 | MX1 | 2.412068631 | 0.018689447 |
| 100477434 | LOC100477434 | 0.813886584 | 0.005457552 |
| 100477995 | RAB20 | 2.011557998 | 6.51 × 10−5 |
| 100481969 | IRF1 | 1.233846742 | 0.000175493 |
| 100482061 | SLC11A1 | 1.498952435 | 2.74 × 10−5 |
| 100483325 | LOC100483325 | 2.131324274 | 0.01898041 |
| 105238896 | LOC105238896 | 1.108664282 | 0.041700258 |
| 109488883 | LOC109488883 | 1.392799023 | 0.000629463 |
| BGI_novel_G000507 | BGI_novel_G000507 | 1.074982321 | 0.043726236 |
| BGI_novel_G000508 | BGI_novel_G000508 | 0.802736093 | 0.013117263 |
| BGI_novel_G000816 | BGI_novel_G000816 | 1.80240837 | 6.94 × 10−5 |
| BGI_novel_G000830 | BGI_novel_G000830 | 2.064624645 | 0.000209942 |
| BGI_novel_G000834 | BGI_novel_G000834 | 2.921364095 | 3.50 × 10−12 |
| Gene ID | Gene symbol | log2FC | Q value |
|---|---|---|---|
| 100463670 | SYNCRIP | −0.657043606 | 0.022428329 |
| 100463973 | SATB1 | −1.087420722 | 0.001571307 |
| 100464341 | ILF2 | −0.599904586 | 0.03021565 |
| 100464393 | LOC100464393 | −1.100916666 | 0.000807226 |
| 100465982 | LEF1 | −1.200422244 | 0.000864714 |
| 100467178 | BACH2 | −1.407743785 | 0.015084227 |
| 100467628 | LTB | −0.84441179 | 0.004005136 |
| 100469077 | LAX1 | −0.942370345 | 0.044459423 |
| 100469792 | RHOH | −0.883131268 | 0.036375059 |
| 100471131 | CD79B | −1.964581188 | 9.70 × 10−7 |
| 100471141 | CD19 | −1.433097254 | 0.00042723 |
| 100471593 | BCL11B | −1.084197888 | 0.020106663 |
| 100471626 | CD79A | −1.163510008 | 0.006749405 |
| 100472031 | LOC100472031 | −0.865494399 | 0.010478855 |
| 100475265 | WNT10B | −1.55344294 | 0.028829421 |
| 100475502 | CD7 | −0.815269322 | 0.00519229 |
| 100476508 | MS4A1 | −1.032444512 | 0.001300313 |
| 100476528 | TCF7 | −0.861882414 | 0.032397565 |
| 100476544 | KLHL6 | −1.037565296 | 0.00887367 |
| 100477968 | STON2 | −1.409667727 | 0.024177379 |
| 100478290 | LOC100478290 | −1.161973596 | 0.01488409 |
| 100478982 | IMPDH2 | −0.777026522 | 0.008127539 |
| 100479355 | BLK | −1.452355267 | 0.008778106 |
| 100479879 | FCRL1 | −1.998071764 | 3.56 × 10−5 |
| 100480638 | ATM | −1.481610202 | 6.80 × 10−8 |
| 100482026 | SOX4 | −2.850078573 | 3.40 × 10−10 |
| 100482462 | CD3E | −0.758174358 | 0.038685191 |
| 100482465 | CR2 | −1.415727356 | 0.003516809 |
| 105240103 | LOC105240103 | −2.821011447 | 2.31 × 10−8 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, S.; Li, C.; Yan, W.; Jin, S.; Wang, K.; Wang, C.; Gong, H.; Wu, H.; Fu, X.; Deng, L.; et al. Using Blood Transcriptome Analysis to Determine the Changes in Immunity and Metabolism of Giant Pandas with Age. Vet. Sci. 2022, 9, 667. https://doi.org/10.3390/vetsci9120667
Liu S, Li C, Yan W, Jin S, Wang K, Wang C, Gong H, Wu H, Fu X, Deng L, et al. Using Blood Transcriptome Analysis to Determine the Changes in Immunity and Metabolism of Giant Pandas with Age. Veterinary Sciences. 2022; 9(12):667. https://doi.org/10.3390/vetsci9120667
Chicago/Turabian StyleLiu, Song, Caiwu Li, Wenjun Yan, Senlong Jin, Kailu Wang, Chengdong Wang, Huiling Gong, Honglin Wu, Xue Fu, Linhua Deng, and et al. 2022. "Using Blood Transcriptome Analysis to Determine the Changes in Immunity and Metabolism of Giant Pandas with Age" Veterinary Sciences 9, no. 12: 667. https://doi.org/10.3390/vetsci9120667
APA StyleLiu, S., Li, C., Yan, W., Jin, S., Wang, K., Wang, C., Gong, H., Wu, H., Fu, X., Deng, L., Lei, C., He, M., Wang, H., Cheng, Y., Wang, Q., Lin, S., Huang, Y., Li, D., & Yang, X. (2022). Using Blood Transcriptome Analysis to Determine the Changes in Immunity and Metabolism of Giant Pandas with Age. Veterinary Sciences, 9(12), 667. https://doi.org/10.3390/vetsci9120667








