Replicative Deformed Wing Virus Found in the Head of Adults from Symptomatic Commercial Bumblebee (Bombus terrestris) Colonies
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collection
2.2. Extraction of Total RNA
2.3. qRT-PCR Assays to Detect and Quantify the Deforming Wing Virus (DWV)
2.4. Strand-Specific RT-PCR
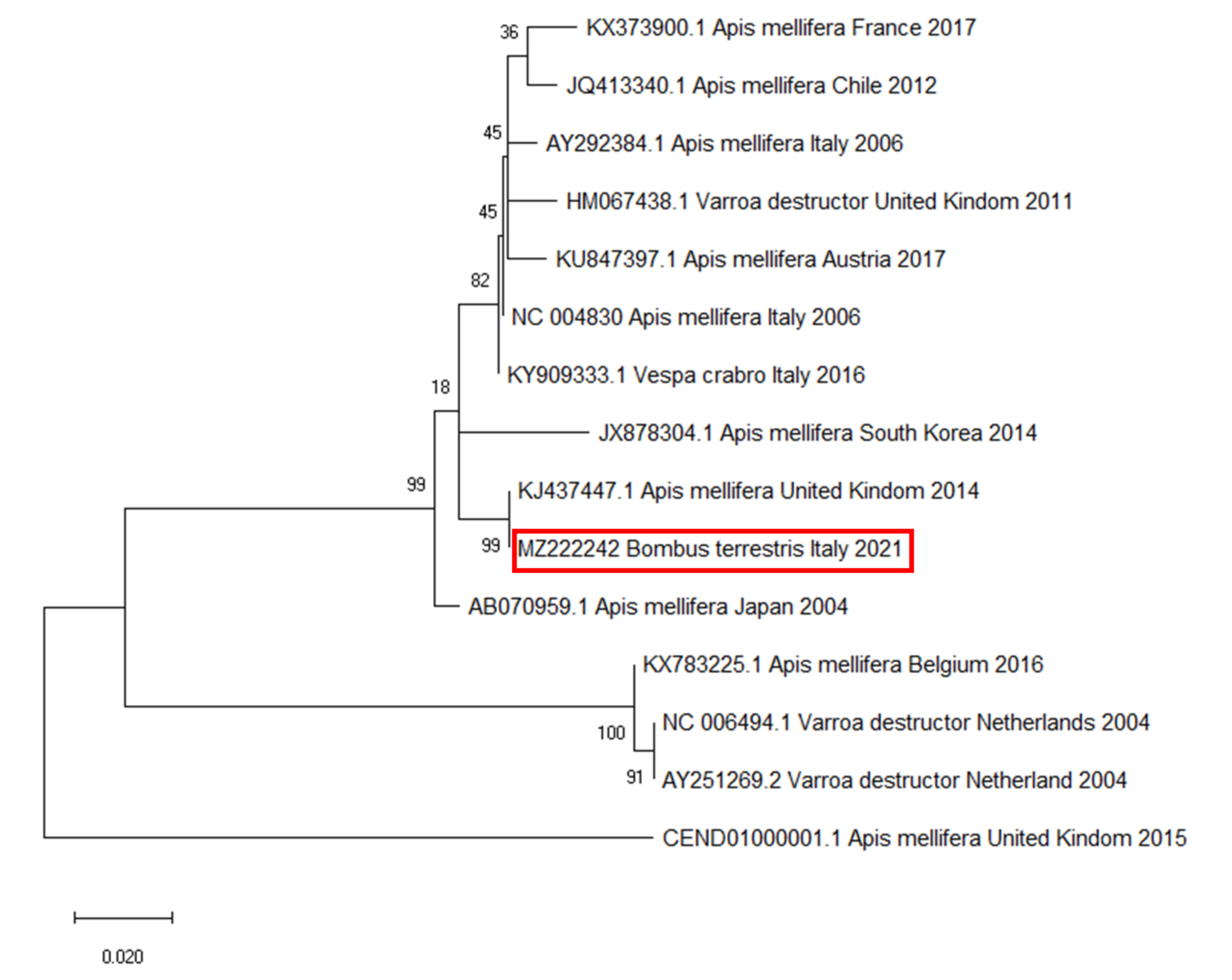
2.5. Phylogenetic Analysis
2.6. Statistical Analysis
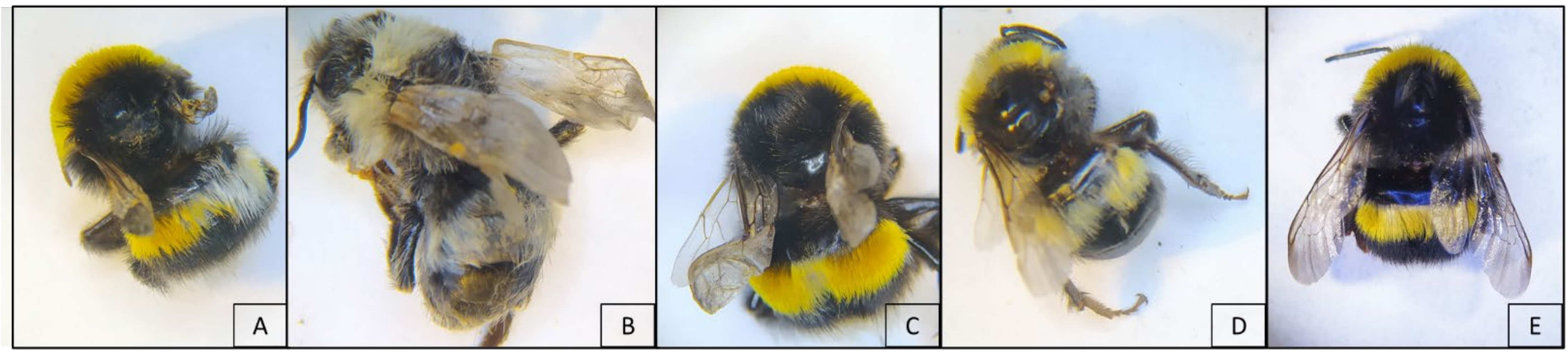
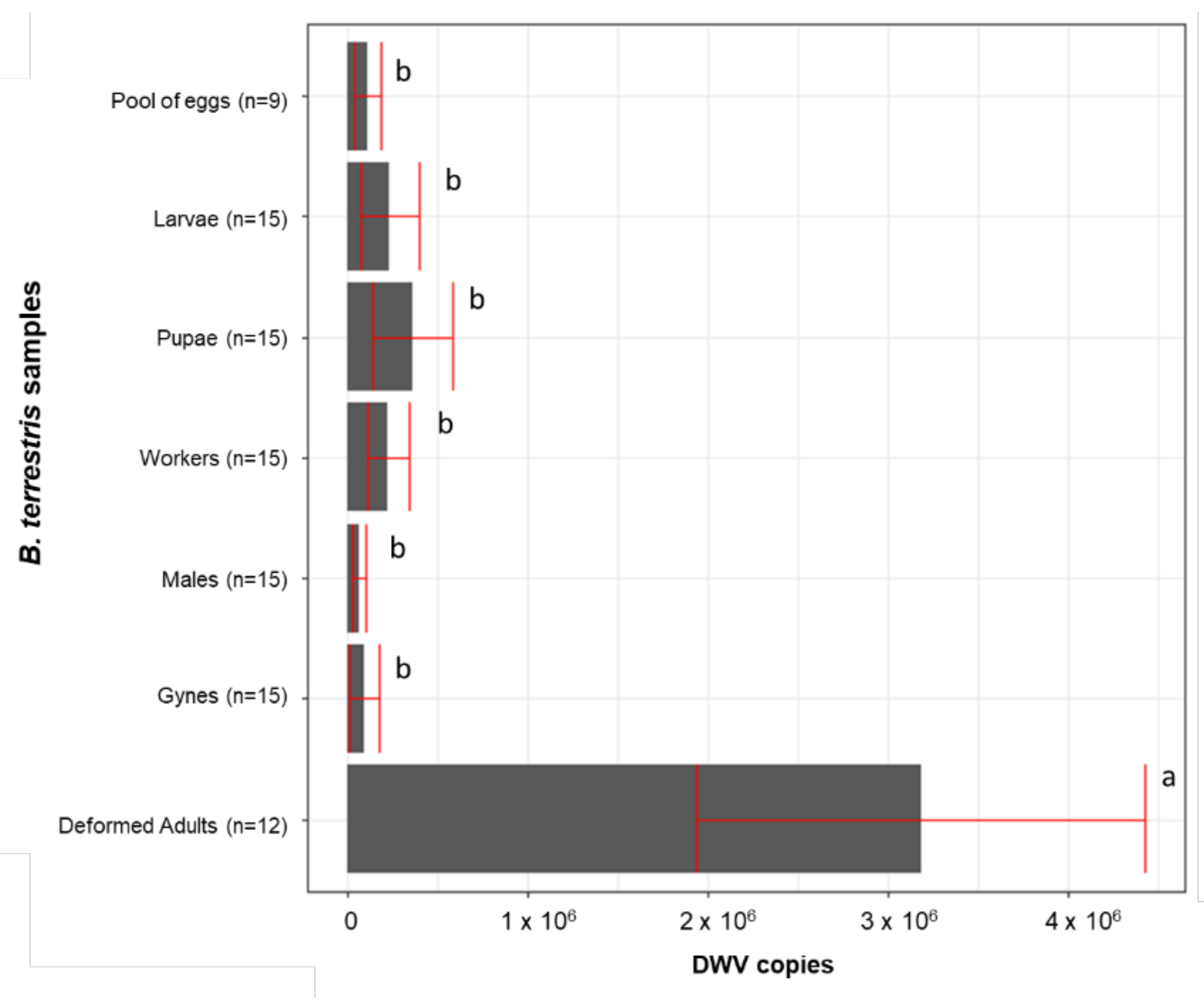
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- De Miranda, J.R.; Genersch, E. Deformed wing virus. J. Invertebr. Pathol. 2010, 103, S48–S61. [Google Scholar] [CrossRef]
- Genersch, E.; Aubert, M. Emerging and re-emerging viruses of the honey bee (Apis mellifera L.). Vet. Res. 2010, 41, 54. [Google Scholar] [CrossRef]
- McMenamin, A.J.; Genersch, E. Honey bee colony losses and associated viruses. Curr. Opin. Insect Sci. 2015, 8, 121–129. [Google Scholar] [CrossRef]
- Steinhauer, N.; Kulhanek, K.; Antúnez, K.; Human, H.; Chantawannakul, P.; Chauzat, M.P.; van Engelsdorp, D. Drivers of colony losses. Curr. Opin. Insect Sci. 2018, 26, 142–148. [Google Scholar] [CrossRef] [PubMed]
- Buendía, M.; Martín-Hernández, R.; Ornosa, C.; Barrios, L.; Bartolomé, C.; Higes, M. Epidemiological study of honeybee pathogens in Europe: The results of Castilla-La Mancha (Spain). Span. J. Agric. Res. 2018, 16, e0502. [Google Scholar] [CrossRef]
- Martin, S.J.; Highfield, A.C.; Brettell, L.; Villalobos, E.M.; Budge, G.E.; Powell, M.; Nikaido, S.; Schroeder, D.C. Global honey bee viral landscape altered by a parasitic mite. Science 2012, 336, 1304–1306. [Google Scholar] [CrossRef] [PubMed]
- Mordecai, G.J.; Wilfert, L.; Martin, S.J.; Jones, I.M.; Schroeder, D.C. Diversity in a honey bee pathogen: First report of a third master variant of the Deformed Wing Virus quasispecies. ISME J. 2016, 10, 1264–1273. [Google Scholar] [CrossRef]
- McMahon, D.P.; Natsopoulou, M.E.; Doublet, V.; Fürst, M.; Weging, S.; Brown, M.J.F.; Gogol-Döring, A.; Paxton, R.J. Elevated virulence of an emerging viral genotype as a driver of honeybee loss. Proc. Biol. Sci. 2016, 283, 20160811. [Google Scholar] [CrossRef] [PubMed]
- Ryabov, E.V.; Wood, G.R.; Fannon, J.M.; Moore, J.D.; Bull, J.C.; Chandler, D.; Mead, A.; Burroughs, N.; Evans, D.J. A virulent strain of deformed Wing Virus (DWV) of honeybees (Apis mellifera) prevails after varroa destructor-mediated, or in vitro, transmission. PLoS Pathog. 2014, 10, e1004230. [Google Scholar] [CrossRef]
- Yue, C.; Schroder, M.; Gisder, S.; Genersch, E. Vertical-transmission routes for deformed wing virus of honeybees (Apis mellifera). J. Gen. Virol. 2007, 88, 2329–2336. [Google Scholar] [CrossRef]
- Ball, B.V.; Allen, M.F. The prevalence of pathogens in honey bee (Apis mellifera) colonies infested with the parasitic mite Varroa jacobsoni. Ann. Appl. Biol. 1988, 113, 237–244. [Google Scholar] [CrossRef]
- Nordström, S. Distribution of deformed wing virus within honey bee (Apis mellifera) brood cells infested with the ectoparasitic mite Varroa destructor. Exp. Appl. Acarol. 2003, 29, 293–302. [Google Scholar] [CrossRef] [PubMed]
- Shen, M.; Cui, L.; Ostiguy, N.; Cox-Foster, D. Intricate transmission routes and interactions between picorna-like viruses (Kashmir bee virus and sacbrood virus) with the honeybee host and the parasitic varroa mite. J. Gen. Virol. 2005, 86, 2281–2289. [Google Scholar] [CrossRef] [PubMed]
- Lanzi, G.; de Miranda, J.R.; Boniotti, M.B.; Cameron, C.E.; Lavazza, A.; Capucci, L.; Camazine, S.M.; Rossi, C. Molecular and biological characterization of deformed wing virus of honeybees (Apis mellifera L.). J. Virol. 2006, 80, 4998–5009. [Google Scholar] [CrossRef]
- Gisder, S.; Aumeier, P.; Genersch, E. Deformed wing virus: Replication and viral load in mites (Varroa destructor). J. Gen. Virol. 2009, 90, 463–467. [Google Scholar] [CrossRef] [PubMed]
- Mazzei, M.; Carrozza, M.L.; Luisi, E.; Forzan, M.; Giusti, M.; Sagona, S.; Tolari, F.; Felicioli, A. Infectivity of DWV associated to flower pollen: Experimental evidence of a horizontal transmission route. PLoS ONE 2014, 9, e113448. [Google Scholar] [CrossRef]
- Mockel, N.; Gisder, S.; Genersch, E. Horizontal transmission of deformed wing virus: Pathological consequences in adult bees (Apis mellifera) depend on the transmission route. J. Gen. Virol. 2011, 92, 370–377. [Google Scholar] [CrossRef]
- Chen, Y.; Evans, J.; Feldlaufer, M. Horizontal and vertical transmission of viruses in the honey bee, Apis mellifera. J. Invertebr. Pathol. 2006, 92, 152–159. [Google Scholar] [CrossRef]
- Mazzei, M.; Forzan, M.; Cilia, G.; Sagona, S.; Bortolotti, L.; Felicioli, A. First detection of replicative deformed wing virus (DWV) in Vespa velutina nigrithorax. Bull. Insectol. 2018, 71, 211–216. [Google Scholar]
- Forzan, M.; Sagona, S.; Mazzei, M.; Felicioli, A. Detection of deformed wing virus in Vespa crabro. Bull. Insectol. 2017, 70, 261–265. [Google Scholar]
- Sébastien, A.; Lester, P.J.; Hall, R.J.; Wang, J.; Moore, N.E.; Gruber, M.A.M. Invasive ants carry novel viruses in their new range and form reservoirs for a honeybee pathogen. Biol. Lett. 2015, 11, 20150610. [Google Scholar] [CrossRef] [PubMed]
- Gisder, S.; Genersch, E. Viruses of commercialized insect pollinators. J. Invertebr. Pathol. 2017, 147, 51–59. [Google Scholar] [CrossRef] [PubMed]
- Gusachenko, O.N.; Woodford, L.; Balbirnie-Cumming, K.; Ryabov, E.V.; Evans, D.J. Evidence for and against deformed wing virus spillover from honey bees to bumble bees: A reverse genetic analysis. Sci. Rep. 2020, 10, 16847. [Google Scholar] [CrossRef] [PubMed]
- Singh, R.; Levitt, A.L.; Rajotte, E.G.; Holmes, E.C.; Ostiguy, N.; van Engelsdorp, D.; Lipkin, W.I.; dePamphilis, C.W.; Toth, A.L.; Cox-Foster, D.L. RNA viruses in hymenopteran pollinators: Evidence of Inter-Taxa Virus transmission via pollen and potential impact on non-apis hymenopteran species. PLoS ONE 2010, 5, e14357. [Google Scholar] [CrossRef]
- Tehel, A.; Brown, M.J.; Paxton, R.J. Impact of managed honey bee viruses on wild bees. Curr. Opin. Virol. 2016, 19, 16–22. [Google Scholar] [CrossRef] [PubMed]
- Ravoet, J.; de Smet, L.; Meeus, I.; Smagghe, G.; Wenseleers, T.; de Graaf, D.C. Widespread occurrence of honey bee pathogens in solitary bees. J. Invertebr. Pathol. 2014, 122, 55–58. [Google Scholar] [CrossRef]
- Eyer, M.; Chen, Y.P.; Schäfer, M.O.; Pettis, J.; Neumann, P. Small hive beetle, Aethina tumida, as a potential biological vector of honeybee viruses. Apidologie 2009, 40, 419–428. [Google Scholar] [CrossRef]
- Potts, S.G.; Imperatriz-Fonseca, V.; Ngo, H.T.; Aizen, M.A.; Biesmeijer, J.C.; Breeze, T.D.; Dicks, L.V.; Garibaldi, L.A.; Hill, R.; Settele, J.; et al. Safeguarding pollinators and their values to human well-being. Nature 2016, 540, 220–229. [Google Scholar] [CrossRef] [PubMed]
- Velthuis, H.H.W.; van Doorn, A. A century of advances in bumblebee domestication and the economic and environmental aspects of its commercialization for pollination. Apidologie 2006, 37, 421–451. [Google Scholar] [CrossRef]
- Lecocq, T.; Rasmont, P.; Harpke, A.; Schweiger, O. Improving international trade regulation by considering intraspecific variation for invasion risk assessment of commercially traded species: The Bombus terrestris case. Conserv. Lett. 2016, 9, 281–289. [Google Scholar] [CrossRef]
- Colla, S.R.; Otterstatter, M.C.; Gegear, R.J.; Thomson, J.D. Plight of the bumble bee: Pathogen spillover from commercial to wild populations. Biol. Conserv. 2006, 129, 461–467. [Google Scholar] [CrossRef]
- Imhoof, B.; Schmid-Hempel, P. Colony success of the bumble bee, Bombus terrestris, in relation to infections by two protozoan parasites, Crithidia bombi and Nosema bombi. Insectes Soc. 1999, 46, 233–238. [Google Scholar] [CrossRef]
- Folly, A.J.; Koch, H.; Stevenson, P.C.; Brown, M.J.F. Larvae act as a transient transmission hub for the prevalent bumblebee parasite Crithidia bombi. J. Invertebr. Pathol. 2017, 148, 81–85. [Google Scholar] [CrossRef] [PubMed]
- Shykoff, J.A.; Schmid-Hempel, P. Incidence and effects of four parasites in natural populations of bumble bees in Switzerland. Apidologie 1991, 22, 117–125. [Google Scholar] [CrossRef]
- Otterstatter, M.C.; Whidden, T.L. Patterns of parasitism by tracheal mites (Locustacarus buchneri) in natural bumble bee populations. Apidologie 2004, 35, 351–357. [Google Scholar] [CrossRef][Green Version]
- McMenamin, A.J.; Flenniken, M.L. Recently identified bee viruses and their impact on bee pollinators. Curr. Opin. Insect Sci. 2018, 26, 120–129. [Google Scholar] [CrossRef]
- Meeus, I.; de Miranda, J.R.; de Graaf, D.C.; Wäckers, F.; Smagghe, G. Effect of oral infection with Kashmir bee virus and Israeli acute paralysis virus on bumblebee (Bombus terrestris) reproductive success. J. Invertebr. Pathol. 2014, 121, 64–69. [Google Scholar] [CrossRef] [PubMed]
- Pascall, D.J.; Tinsley, M.C.; Clark, B.L.; Obbard, D.J.; Wilfert, L. Virus prevalence and genetic diversity across a wild bumblebee community. Front. Microbiol. 2021, 12. [Google Scholar] [CrossRef]
- Wang, H.; Meeus, I.; Piot, N.; Smagghe, G. Systemic Israeli acute paralysis virus (IAPV) infection in bumblebees (Bombus terrestris) through feeding and injection. J. Invertebr. Pathol. 2018, 151, 158–164. [Google Scholar] [CrossRef]
- Fürst, M.A.; McMahon, D.P.; Osborne, J.L.; Paxton, R.J.; Brown, M.J.F.F. Disease associations between honeybees and bumblebees as a threat to wild pollinators. Nature 2014, 506, 364–366. [Google Scholar] [CrossRef]
- Genersch, E.; Yue, C.; Fries, I.; de Miranda, J.R. Detection of Deformed wing virus, a honey bee viral pathogen, in bumble bees (Bombus terrestris and Bombus pascuorum) with wing deformities. J. Invertebr. Pathol. 2006, 91, 61–63. [Google Scholar] [CrossRef]
- Radzevičiūtė, R.; Theodorou, P.; Husemann, M.; Japoshvili, G.; Kirkitadze, G.; Zhusupbaeva, A.; Paxton, R.J. Replication of honey bee-associated RNA viruses across multiple bee species in apple orchards of Georgia, Germany and Kyrgyzstan. J. Invertebr. Pathol. 2017, 146, 14–23. [Google Scholar] [CrossRef] [PubMed]
- Alger, S.A.; Burnham, P.A.; Boncristiani, H.F.; Brody, A.K. RNA virus spillover from managed honeybees (Apis mellifera) to wild bumblebees (Bombus spp.). PLoS ONE 2019, 14, e0217822. [Google Scholar] [CrossRef] [PubMed]
- Manley, R.; Temperton, B.; Doyle, T.; Gates, D.; Hedges, S.; Boots, M.; Wilfert, L. Knock-on community impacts of a novel vector: Spillover of emerging DWV-B from Varroa-infested honeybees to wild bumblebees. Ecol. Lett. 2019, 22, 1306–1315. [Google Scholar] [CrossRef]
- de Sousa Pereira, K.; Meeus, I.; Smagghe, G. Honey bee-collected pollen is a potential source of Ascosphaera apis infection in managed bumble bees. Sci. Rep. 2019, 9, 1–9. [Google Scholar] [CrossRef]
- Bogo, G.; de Manincor, N.; Fisogni, A.; Galloni, M.; Bortolotti, L. Effects of queen mating status, pre-diapause weight and pupae’s sex on colony initiation in small-scale rearing of Bombus terrestris. Apidologie 2017, 48, 845–854. [Google Scholar] [CrossRef]
- Cilia, G.; Cabbri, R.; Maiorana, G.; Cardaio, I.; Dall’Olio, R.; Nanetti, A. A novel TaqMan ® assay for Nosema ceranae quantification in honey bee, based on the protein coding gene Hsp70. Eur. J. Protistol. 2018, 63, 44–50. [Google Scholar] [CrossRef]
- Cilia, G.; Sagona, S.; Giusti, M.; dos Santos, P.E.J.; Nanetti, A.; Felicioli, A. Nosema ceranae infection in honeybee samples from Tuscanian Archipelago (Central Italy) investigated by two qPCR methods. Saudi J. Biol. Sci. 2019, 26, 1553–1556. [Google Scholar] [CrossRef]
- Chen, Y.P.; Higgins, J.A.; Feldlaufer, M.F. Quantitative real-time reverse transcription-PCR analysis of deformed wing virus infection in the honeybee (Apis mellifera L.). Appl. Environ. Microbiol. 2005, 71, 436–441. [Google Scholar] [CrossRef]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Dalmon, A.; Diévart, V.; Thomasson, M.; Fouque, R.; Vaissière, B.E.; Guilbaud, L.; le Conte, Y.; Henry, M. Possible spillover of pathogens between bee communities foraging on the same floral resource. Insects 2021, 12, 122. [Google Scholar] [CrossRef]
- Brenton-Rule, E.C.; Dobelmann, J.; Baty, J.W.; Brown, R.L.; Dvorak, L.; Grangier, J.; Masciocchi, M.; McGrannachan, C.; Shortall, C.R.; Schmack, J.; et al. The origins of global invasions of the German wasp (Vespula germanica) and its infection with four honey bee viruses. Biol. Invasions 2018, 20, 3445–3460. [Google Scholar] [CrossRef]
- Huwiler, M.; Papach, A.; Cristina, E.; Yañez, O.; Williams, G.R.; Neumann, P. Deformed wings of small hive beetle independent of virus infections and mites. J. Invertebr. Pathol. 2020, 172, 107365. [Google Scholar] [CrossRef] [PubMed]
- Nanetti, A.; Ellis, J.D.; Cardaio, I.; Cilia, G. Detection of Lotmaria passim, Crithidia mellificae and replicative forms of deformed Wing Virus and Kashmir Bee Virus in the Small Hive Beetle (Aethina tumida). Pathogens 2021, 10, 372. [Google Scholar] [CrossRef] [PubMed]
- McMahon, D.P.; Fürst, M.A.; Caspar, J.; Theodorou, P.; Brown, M.J.F.; Paxton, R.J. A sting in the spit: Widespread cross-infection of multiple RNA viruses across wild and managed bees. J. Anim. Ecol. 2015, 84, 615–624. [Google Scholar] [CrossRef]
- Meeus, I.; Brown, M.J.F.; de Graaf, D.C.; Smagghe, G. Effects of invasive parasites on bumble bee declines. Conserv. Biol. 2011, 25, 662–671. [Google Scholar] [CrossRef]
- Jabal-Uriel, C.; Martín-Hernández, R.; Ornosa, C.; Higes, M.; Berriatua, E.; de la Rua, P. First data on the prevalence and distribution of pathogens in bumblebees (Bombus terrestris and Bombus pascuorum) from Spain. Span. J. Agric. Res. 2017, 15, e05SC01. [Google Scholar] [CrossRef]
- Arismendi, N.; Riveros, G.; Zapata, N.; Smagghe, G.; González, C.; Vargas, M. Occurrence of bee viruses and pathogens associated with emerging infectious diseases in native and non-native bumble bees in southern Chile. Biol. Invasions 2021, 1–15. [Google Scholar] [CrossRef]
- Evison, S.E.F.; Roberts, K.E.; Laurenson, L.; Pietravalle, S.; Hui, J.; Biesmeijer, J.C.; Smith, J.E.; Budge, G.; Hughes, W.O.H. Pervasiveness of parasites in pollinators. PLoS ONE 2012, 7, e30641. [Google Scholar] [CrossRef] [PubMed]
- Sachman-Ruiz, B.; Narváez-Padilla, V.; Reynaud, E. Commercial Bombus impatiens as reservoirs of emerging infectious diseases in central Mexico. Biol. Invasions 2015, 17, 2043–2053. [Google Scholar] [CrossRef]
- Levitt, A.L.; Singh, R.; Cox-Foster, D.L.; Rajotte, E.; Hoover, K.; Ostiguy, N.; Holmes, E.C. Cross-species transmission of honey bee viruses in associated arthropods. Virus Res. 2013, 176, 232–240. [Google Scholar] [CrossRef]
- Li, J.; Peng, W.; Wu, J.; Strange, J.P.; Boncristiani, H.; Chen, Y. Cross-species infection of deformed wing virus poses a new threat to pollinator conservation. J. Econ. Entomol. 2011, 104, 732–739. [Google Scholar] [CrossRef] [PubMed]
- Gamboa, V.; Ravoet, J.; Brunain, M.; Smagghe, G.; Meeus, I.; Figueroa, J.; Riaño, D.; de Graaf, D.C. Bee pathogens found in Bombus atratus from Colombia: A case study. J. Invertebr. Pathol. 2015, 129, 36–39. [Google Scholar] [CrossRef]
- Reynaldi, F.; Sguazza, G.; Albicoro, F.; Pecoraro, M.; Galosi, C. First molecular detection of co-infection of honey bee viruses in asymptomatic Bombus atratus in South America. Braz. J. Biol. 2013, 73, 797–800. [Google Scholar] [CrossRef]
- Graystock, P.; Goulson, D.; Hughes, W.O.H. The relationship between managed bees and the prevalence of parasites in bumblebees. PeerJ 2014, 2, e522. [Google Scholar] [CrossRef] [PubMed]
- Graystock, P.; Blane, E.J.; McFrederick, Q.S.; Goulson, D.; Hughes, W.O.H. Do managed bees drive parasite spread and emergence in wild bees? Int. J. Parasitol. Parasites Wildl. 2016, 5, 64–75. [Google Scholar] [CrossRef] [PubMed]
- Ribeiro, M.F.; Duchateau, M.J.; Velthuis, H.H.W. Comparison of the effects of two kinds of commercially available pollen on colony development and queen production in the bumble bee Bombus terrestris L (Hymenoptera, Apidae). Apidologie 1996, 27, 133–144. [Google Scholar] [CrossRef]
- Yañez, O.; Piot, N.; Dalmon, A.; de Miranda, J.R.; Chantawannakul, P.; Panziera, D.; Amiri, E.; Smagghe, G.; Schroeder, D.; Chejanovsky, N. Bee viruses: Routes of infection in hymenoptera. Front. Microbiol. 2020, 11, 943. [Google Scholar] [CrossRef]
- Gurel, F.; Gosterit, A. Effects of different stimulation methods on colony initiation and development of Bombus terrestris L. (Hymenoptera: Apidae) queens. Appl. Entomol. Zool. 2008, 43, 113–117. [Google Scholar] [CrossRef][Green Version]
- Dalmon, A.; Peruzzi, M.; le Conte, Y.; Alaux, C.; Pioz, M. Temperature-driven changes in viral loads in the honey bee Apis mellifera. J. Invertebr. Pathol. 2019, 160, 87–94. [Google Scholar] [CrossRef] [PubMed]
- Graystock, P.; Yates, K.; Evison, S.E.F.; Darvill, B.; Goulson, D.; Hughes, W.O.H. The Trojan hives: Pollinator pathogens, imported and distributed in bumblebee colonies. J. Appl. Ecol. 2013, 50, 1207–1215. [Google Scholar] [CrossRef]
- Meeus, I.; Mosallanejad, H.; Niu, J.; de Graaf, D.C.; Wäckers, F.; Smagghe, G. Gamma irradiation of pollen and eradication of Israeli acute paralysis virus. J. Invertebr. Pathol. 2014, 121, 74–77. [Google Scholar] [CrossRef] [PubMed]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cilia, G.; Zavatta, L.; Ranalli, R.; Nanetti, A.; Bortolotti, L. Replicative Deformed Wing Virus Found in the Head of Adults from Symptomatic Commercial Bumblebee (Bombus terrestris) Colonies. Vet. Sci. 2021, 8, 117. https://doi.org/10.3390/vetsci8070117
Cilia G, Zavatta L, Ranalli R, Nanetti A, Bortolotti L. Replicative Deformed Wing Virus Found in the Head of Adults from Symptomatic Commercial Bumblebee (Bombus terrestris) Colonies. Veterinary Sciences. 2021; 8(7):117. https://doi.org/10.3390/vetsci8070117
Chicago/Turabian StyleCilia, Giovanni, Laura Zavatta, Rosa Ranalli, Antonio Nanetti, and Laura Bortolotti. 2021. "Replicative Deformed Wing Virus Found in the Head of Adults from Symptomatic Commercial Bumblebee (Bombus terrestris) Colonies" Veterinary Sciences 8, no. 7: 117. https://doi.org/10.3390/vetsci8070117
APA StyleCilia, G., Zavatta, L., Ranalli, R., Nanetti, A., & Bortolotti, L. (2021). Replicative Deformed Wing Virus Found in the Head of Adults from Symptomatic Commercial Bumblebee (Bombus terrestris) Colonies. Veterinary Sciences, 8(7), 117. https://doi.org/10.3390/vetsci8070117






