Abstract
Environmental DNA (eDNA) analysis is related to screening genetic material of various organisms in environmental samples. Honey represents a natural source of exogenous DNA, which allows for the detection of different honey bee pathogens and parasites. In the present study, we extracted DNA from 20 honey samples from different regions in Bulgaria and tested for the presence of DNA of the ectoparasitic mite Varroa destructor, as well as Nosema apis and Nosema ceranae. Only Nosema ceranae was detected, showing up in 30% of all samples, which confirms the widespread prevalence of this pathogen. All positive samples were found in plain regions of the country, while this pathogen was not detected in mountainous parts. None of the samples gave positive amplifications for the Nosema apis and Varroa mite. The obtained results from this study confirm previous observations that eDNA contained in honey is a potent source for effective biomonitoring of actual diseases in the honey bee.
1. Introduction
The Western honey bee (Apis mellifera Linnaeus, 1758) is a species with economic, agricultural, and environmental significance. Honey bees are known as the most effective pollinators because they pollinate various crops, fruits, and vegetables [1,2], thus supporting the existence of a large number of animal and plant species, including humans [3,4].
Honey bees are threatened by various pests and pathogens, some of which inflict heavy annual losses in the beekeeping sector. Among these threats, the ectoparasitic mite Varroa destructor, the fungal pathogens Nosema apis and Nosema ceranae, often acting simultaneously with some honey bee-associated viruses, are suspected to contribute considerably to honey bee colony losses [5,6,7,8]. A large number of methods and techniques have been developed for the identification of various honey bee pathogens [9,10,11]. One of the most widely used methods for the detection of a particular pathogen is the amplification of coding and/or noncoding DNA regions, followed by a sequence analysis of the amplified DNA fragment [12,13,14].
Most often forager bees and, less frequently, house bees, drones, or brood are used as a source of biological material for the identification of a particular pathogen. Alternatively, a large number of honey bee pathogens may be detected in honey bee products—usually, honey, propolis, and bee wax [14,15,16,17,18].
The term environmental DNA (eDNA) is most commonly used for the genetic material isolated directly from environmental samples (soil, sediment, water, etc.) without any obvious signs of biological material from external source eDNA [19]. This approach is a credible strategy for correct identification of cryptic species or juvenile life stages, elusive, or invasive organisms, including many pathogens that might be difficult to sample and/or identify [20,21]. Environmental DNA has been obtained from ancient and modern animals and plants samples, thus identifying not only single organisms but whole communities as well. This technique has recently been implemented successfully in the beekeeping sector. For example, the analysis of eDNA in pollen and some bee products (wax, propolis, and honey) has been used to detect the level of environmental contaminants and pollutants and to identify their origin [22,23]. Additionally, some bee products and, in particular, the analysis of eDNA in honey (i.e., derived from pollen) can be used for the evaluation of honey bee foraging behavior and the identification of the botanical composition of pollen [24,25]. Moreover, since plant-sucking insects produce honeydew, which honey bees regularly collect, these Hemiptera species can be identified on the basis of DNA traces in honey [26]. The detection of fungal and bacterial DNA in honey can be used for forensic and food safety analyses [27]. The following approaches have been adopted in the analysis of DNA contained in honey: (i) use of specific PCR primers to identify a particular species; (ii) use of universal primers for barcoding—the mitochondrial genes that are the most commonly used in bacteria are the small subunit of 16S DNA gene and cytochrome c oxidase subunit I gene (cox1) [17,28,29]; (iii) use of metabarcoding in combination with next generation sequencing (NGS) to allow determining the taxonomic composition of pooled honey eDNA [30,31]. As for the identification of single pathogens in honey, this approach is a non-invasive and relatively fast way to assess the health status of bee colonies as well as to identify some diseases without clinical symptoms [28,29].
Considering the above, we have conducted, for the first time in Bulgaria, a purposeful study on the detection of some of the most widely distributed pathogens of honey bees by using honey of different botanical and geographical origin as a source of eDNA. We have decided to use this relatively new approach due to the fact that it represents an alternative, yet effective, strategy for establishing the epidemiological status and monitoring of various honey bee diseases.
2. Materials and Methods
2.1. Honey Samples
A total of 20 honey samples, originating from four different regions in Bulgaria (Northeastern, Western, Central and South Bulgaria) referred to as KV, ST, SF, and SM, respectively, were produced and collected in the summer of 2020 (Figure 1). In each geographical location, five honey samples were taken from single hives in each apiary. There was no bias concerning the obtained honey samples. The first two regions comprise generally flat plains, while the last two regions are situated in the mountainous parts of the country (Vitosha and the Rhodope Mountains, respectively). The honey samples were collected from hives within the same apiary. According to all producers, the apiaries had no visible clinical symptoms of any diseases.
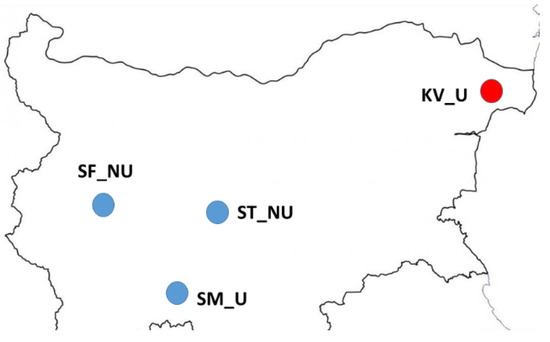
Figure 1.
Map showing the locations of 20 honey samples in Bulgaria. Sample names indicate the region in Bulgaria (SF, Sofia City Province; SM, Smolyan; ST, Strelcha; KV, Kavarna). Habitat indicator (U, urban area; NU, non-urban area). Blue circles—polyfloral honey; Red circle—monofloral honey.
According to the Bulgarian Food Safety Agency (BFSA), the results of an epizootiological survey in different areas of the country in 2020 showed that bee colonies are most often affected by varroasis and nosemosis, while in the previous year varroasis predominated as a cause of mortality in bee colonies. As far as the regions that we have studied are concerned, from 1 January to 30 April 2020, a total of 243 samples were tested; 23 of them were positive for varroasis, and 56—for nosemosis. The predominant part of the studied samples showed the presence of V. destructor and N. ceranae in Central and Northeastern Bulgaria. Moreover, in the regions observed in 2020, 37% of the samples showed a mixed invasion of varroasis and nosemosis, 33.3%—only varroasis, and 14.8%—only nosemosis. These data gave us reason to focus our research particularly on the molecular detection of V. destructor and N. ceranae in the honey samples.
2.2. DNA Extraction for Identification of eDNA from V. destructor, N. apis, and N. ceranae in Honey Samples, Using CTAB Protocol with a Spin Column
Honey is mainly composed of carbohydrates, which comprise about 95% of honey’s dry weight. It has been noted that the high concentration of sugars reduces the amount of DNA yields and inhibits PCR reactions [32]. Therefore, as an initial step in the process of DNA isolation, it is necessary to reduce the sugar content. Briefly, for each honey sample, a total of 50 g of honey was divided into four 50 mL conical centrifuge tubes (12.5 g for each tube). Subsequently, 37.5 mL ddH2O were added to each tube and incubated at 40 °C for 30 min to dissolve the honey. After the complete dissolution of the honey, the tubes were centrifuged for 30 min at 5000× g at room temperature, and the supernatant was discarded. The obtained pellet was re-suspended in 5 mL of ddH2O, and the content of the four tubes was pooled together, and then diluted in 50 mL ddH2O. These steps were repeated three times. After the last centrifugation and discard of the supernatant, the pellet was dissolved in 1 mL of an autoclaved CTAB DNA extraction buffer (2% (w/v) cetyltrimethylammonium bromide, containing 100 mM Tris-HCl, pH = 8, 20 mM EDTA, pH = 8, 1.4 M NaCl), transferred in a 1.5 mL tube. Subsequently, 18 μL of RNase A solution (10 mg/mL, Cat. no. EN0531, Thermo Fisher Scientific, Inc. Waltham, MA, USA) were added to each honey sample and incubated for 30 min at 65 °C. CTAB buffer is suitable for DNA isolating because the CTAB is a cationic surfactant, which is why it binds with lipids from the cell membrane and provokes cell lysis. Additionally, in cells with a high concentration of sugars (as with honey) CTAB binds to the polysaccharides when the salt concentration is high, thus removing polysaccharides from the solution. After this incubation step, 30 µL of proteinase K (20 mg/mL) were added in each tube, and all samples were further incubated at 56 °C for 90 min with gentle shaking. After the inactivation of proteinase K for 10 min at 70 °C, the samples were transferred to nucleospin columns to obtain a high yield of DNA. Total DNA was isolated with a GeneMATRIX Tissue Kit (Cat. no. E3550, EURx Ltd., Gdansk, Poland), according to the manufacturer’s instructions. The quantity and quality of the extracted eDNA were checked spectrophotometrically and by 1% agarose gel electrophoresis in 1X TAE buffer staining with SimpliSafe™ (Cat. no. E4600, EURx Ltd., Gdansk, Poland) under UV light. The DNA was stored at −20 °C until PCR amplifications.
2.3. PCR Analysis
PCR amplification was performed using the PCR primer sets presented in Table 1.

Table 1.
Information on PCR primers used in this study to amplify eDNA from the honey samples.
We decided to use two primer sets to detect the presence of V. destructor (for fragments of coxI and cytb genes) in order to increase the possibility of identifying the ectoparasitic mite.
Before proceeding to PCR identification of the presence of some pathogens and parasites, we initially checked for possible amplification of eDNA from honey bees in honey samples. According to Ruttner’s morphometric analysis [37], A. m. macedonica exists as a native honey bee in Bulgaria. This honey bee subspecies belongs to the C evolutionary lineage, which is why we performed amplification of the mitochondrial coxI-coxII intergenic region according to the recommendation by Utzeri et al. [33]
The PCR analysis used one primer set for each reaction (single PCR analysis). The PCR analysis was carried out using a Little Genius thermocycler (BIOER Technology Co., Ltd., Hangzhou, China) in a total volume of 50 μL. The PCR mixture contained 25 µL of NZYTaq 2× Colorless Master Mix (Cat. No. MB04002, Nzytech, Lisboa, Portugal) and 0.4 µM of each specific primer (FOR/REV). The PCR conditions were as follows: initial denaturation at 94 °C for 5 min; 35 cycles (denaturation at 94 °C for 30 s; primer annealing at 56 °C for 30 s; extension at 72 °C for 1 min), and final extension at 72 °C for 10 min. The PCR products were visualized by 1% agarose gel staining with SimpliSafe™ (Cat. no. E4600, EURx Ltd., Gdansk, Poland) under UV light. The fragment size was determined using Gene-Ruler™ 100 bp Ladder Plus (Cat. No. SM0323, Thermo Fisher Scientific Inc., Waltham, MA, USA). When there was no visible amplification or smears on the gel after the first PCR reaction, 5 µL were taken from the reaction mixture and were used as the DNA template for a second PCR reaction. This approach is a standardized procedure to suppress the formation of artificial chimeras during PCR amplification [38]. A second PCR reaction was not initiated when there was visible amplification after the first reaction. The successfully amplified products were purified by a PCR purification kit (Gene Matrix, PCR clean-up kit, EURx, Poland) and sequenced in both directions by a PlateSeq kit (Eurofins Genomics Ebersberg, Gdansk, Germany).
2.4. Sequence and Phylogenetic Analysis
All five obtained DNA sequences were manually edited and aligned in MEGA v7.0.20 [39], using the MUSCLE algorithm [40]. BLASTN (http://www.ncbi.nlm.nih.gov/BLAST/ (accessed on 15 October 2021)) was performed to compare the similarity and validity of the obtained sequences [41]. After the alignment, all five obtained sequences of N. ceranae showed complete homology. Therefore, only one of them was deposited in GenBank—under accession number MG657260.
In order to evaluate any geographic linkage, the obtained sequence of N. ceranae from Bulgaria was aligned with 18 other isolates from different geographical locations, using the reference sequence NW_020169312, a part of ASM98816v1 Nosema ceranae genome assembly [42]. Sequences were analyzed by polymorphic single-nucleotide polymorphism position.
3. Results
Environmental DNA extracted from all honey samples was successfully amplified with respect to the mitochondrial coxI–coxII intergenic region of Apis mellifera, obtaining fragments of the expected size. Thus, DNA was successfully isolated from all samples, and PCR amplification was not inhibited by any contaminants in the isolation process. Therefore, eDNA isolated from all honey samples was suitable for the molecular detection of three pathogens in a single PCR analysis.
When using the two pairs of primers for V. destructor, we found no amplification of the mitochondrial genes coxI and cytb, even after a second PCR. Apparently, the excessive size of the expected fragments in the presence of the highly degraded eDNA does not allow successful amplification. In contrast to the results regarding the ectoparasitic mite, eDNA in the honey samples was successfully amplified with respect to the fungal pathogen N. ceranae. We detected this pathogen only in plain regions—the KV honey samples (20%, 4/20) as well as in the ST samples (10%, 2/20). In the studied mountainous regions (SF and SM), the presence of N. ceranae was not established.
The results from the sequence analysis of the 16S rDNA from Bulgaria and 18 other sequences available in GenBank (https://www.ncbi.nlm.nih.gov/genbank/ (accessed on 25 October 2021) revealed that the newly obtained sequence from Bulgaria is practically identical to the sequences from Morocco, Lebanon, Egypt, Turkey, Argentina, Switzerland, Germany, France, and Japan, when compared with the reference sequence (GenBank Acc. no. NW 020169312) (Table 2). Our findings also show that it is difficult to find any geographical relationship between the different haplotypes. Moreover, the sequence obtained from a single isolate showed different nucleotide polymorphism from those obtained from distinct isolates in the same country (GenBank Acc. no. LC493173, LC510233 and LC510243 from Japan) (Table 2). Conversely, there are isolates with identical haplotypes in samples of very different geographical origin (GenBank Acc. no. LC510243 from Japan and GenBank Acc. no. HM581509 from Mexico).

Table 2.
N. ceranae sequence variation among 19 isolates from different geographical locations. A 218-bp segment of the 16r DNA from 18 different geographical sites is aligned with the reference sequence NW_020169312 part of the ASM98816v1 N. ceranae genome assembly [42]. Variable nucleotides are indicated. Sequence positions above each column are given from the start of the reference sequence.
In contrast to N. ceranae, the other fungal pathogen, N. apis, did not show amplification in any of the tested samples.
4. Discussion
Honey has always been an attractive and relevant subject of study, not only because of its nutritional value and content of micro and macronutrients, but also because of its widespread use as an alternative therapy (apitherapy) in medicine [43,44]. However, there are other biological molecules in the composition of honey, which have been used and studied relatively rarely, so far. Undoubtedly one of them, which has recently aroused considerable research interest in various scientific aspects, is eDNA [45,46].
In this study, we used conventional PCR analyses for molecular detection of some of the most widely distributed honey bee pathogens in honey samples. In this pioneering study for Bulgaria, we tried to use the non-invasive, relatively fast and innovative approach for DNA isolation in honey samples in order to evaluate and monitor bees’ health status. Honey bee DNA is always present in honey and can be used as a positive control even for the presence of traces of eDNA [33]. Therefore, the first step in this study was to detect the presence of bee eDNA and to assess its quality and quantity. This is a necessary condition that allows for the search and amplification of eDNA from other biological objects. After receiving the amplification of the mitochondrial coxI-coxII intergenic region in the subspecies A. m. macedonica in all tested samples, we proceeded with analyses for molecular detection of some bee pathogens.
As anticipated, no successful application was obtained for the ectoparasitic mite V. destructor. Apparently, the size of the expected fragments when using the two pairs of primers for the two mitochondrial genes coxI and cytb (929 bp and 985 bp, respectively) is too large to allow the application of the highly fragmented DNA in honey samples, as confirmed by previous studies [17,33]. The possibility of a negative result due to the absence of V. destructor DNA is extremely low.
It is also worth mentioning that a reason for the unsuccessful identification of the V. destructor might be not only the large size of the expected PCR fragments for coxI and cytb genes, but also the origin of the honey samples provided for analysis. Most of the studies in this direction use mixed honey from several hives in an apiary, or commercial honey, which largely resembles the mixed honey provided by beekeepers for commercial purposes [14,17,33]. This approach significantly increases the possibility of detecting a pathogen in honey samples.
Another limitation associated with the absence of residual mite DNA in honey may be the life cycle of this honey bee parasite. It is well known that the adult mites usually live and feed under the abdominal plates of adult bees primarily on the underside of the metasoma region [47]. At this location, the probability of V. destructor getting into the honey cells is very low.
Another pathogen that we were unable to identify in any of the analyzed samples was N. apis. This is not very surprising, given the reduction in the prevalence of this fungal pathogen and its replacement with N. ceranae in many geographical regions [48,49]. Our results confirm this trend, as N. ceranae was detected in 25% of the analyzed honey samples, only in the plain regions in the country (ST and KV).
Several explanations for these observations are possible. First, processes of increasing urbanization, observed in many countries, create favorable conditions for the spread of various pathogens in honey bees [50,51]. In such sites, contact between humans and animals, domestic and wild, is inevitable, which facilitates the dissemination of various pathogens [52]. The effects of increasing urbanization have been found to be additive rather than interactive, i.e., urbanization imposes changes not only on managed honey bee colonies but on feral populations with respect to their pathogens [53]. What is worrying is the fact that many of the beekeeping practices used and applied have also drastically changed the intensity of the invasion of various pathogens in honey bees [54,55]. In this regard, widespread prevalence of N. ceranae in poorly managed bee colonies has been found [56,57]. Our research has confirmed these observations, as we have found only infestation with N. ceranae in apiaries located in the KV honey samples (20% infestation level) and the ST honey samples (10% infestation level), but not in the mountainous parts in the country (the Vitosha Mountain—SF honey samples and the Rhodope Mountains—SM honey samples). The negative results obtained in the mountainous parts in the country could be due to the failure of amplification of the tested primer pairs, since we have not used primers for N. ceranae identification already tested by other authors [17,57]. Several PCR conditions were tested, with the same negative results for the presence of N. ceranae in SF and SM honey samples. The negative results for N. ceranae in the mountainous parts of the country could be due to for the presence of different strains across samples, having mutations in the primer regions that could affect the amplification efficiency. It will be interesting to further explore this issue.
The predominance of this pathogen only in the plains of the country could also be geographically determined. The mountainous regions are geographically isolated, and the opportunities for movement of people, goods, and livestock there are much more limited than in the plains. Moreover, exposure to insecticides in field conditions is much larger than in mountainous regions, which can increase colony susceptibility to various pathogens [58]. Another reason related to the establishment of N. ceranae only in the plains of the country may be associated with the fact that these regions provided a diet composed of pollen from diverse plants. In contrast, in the mountainous parts of the country the diet most often consists of pollen from a single plant species. As is well-known, the plains provide much greater opportunity for diverse honey vegetation than the mountainous regions (providing low-nutritional quality), which leads to a higher degree of infection [59].
Environmental conditions also affect N. ceranae dissemination and infestation rate, as it has been found that these microsporidia develop favorably in warmer climates [48,60]. In Bulgaria, the development of bee colonies in mountainous areas, e.g., the Vitosha Mountain (SF samples) and the Rhodope Mountains (SM samples), starts usually in May–June, i.e., about two months later than on the plains, which also contributes to the wider spread of N. ceranae in the plains.
The sequence alignment of N. ceranae isolate from Bulgaria along with other sequences available in the GenBank database (http://www.ncbi.nlm.nih.gov/ (accessed on 28 October 2021) has not revealed any differences among them, except for the single nucleotide polymorphism. These haplotypes are not specific in terms of geographical origin, so it is difficult to determine which isolate is specific for a given area, as well as to look for possible dependence regarding the spread of nosemosis worldwide. This is not surprising, given that our phylogenetic analysis of rDNA has not shown significant differences between sequences [61].
The present study on the use of eDNA analysis of honey samples for the molecular identification of some parasites and pathogens in bees has been, to our knowledge, the first research applying this innovative approach in Bulgaria. Although only one pathogen has been detected (N. ceranae), the obtained results (albeit preliminary) provide possibilities for the use of eDNA in monitoring the health status of bee colonies, the epidemiology, and the spread of some of the most common bee diseases.
Author Contributions
Conceptualization, P.H. and G.R.; methodology, D.S. and R.S.; software, B.N.; validation, N.P., R.B. and R.S.; formal analysis, P.H.; investigation, D.S.; resources, P.H.; data curation, G.R.; writing—original draft preparation, P.H.; writing—review and editing, R.B.; visualization, B.N.; supervision, R.S.; project administration, P.H.; funding acquisition, P.H. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Bulgarian National Science Fund of the Ministry of Education and Science, grant number KП-06-H56/11 12.12.2021. The APC was funded by grant number KП-06-H56/11 12.12.2021.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
All data are available upon request.
Acknowledgments
The authors wish to thank Maya Marinova (IBPhBME-BAS, Bulgaria) for the English editing.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Aizen, M.A.; Garibaldi, L.A.; Cunningham, S.A.; Klein, A.M. How much does agriculture depend on pollinators? Lessons from long-term trends in crop production. Ann. Bot. 2009, 103, 1579–1588. [Google Scholar] [CrossRef] [PubMed]
- Klatt, B.K.; Holzschuh, A.; Westphal, C.; Clough, Y.; Smit, I.; Pawelzik, E.; Tscharntke, T. Bee pollination improves crop quality, shelf life and commercial value. Proc. R. Soc. B 2014, 281, 20132440. [Google Scholar] [CrossRef] [PubMed]
- Alvarez-Suarez, J.M.; Tulipani, S.; Romandini, S.; Bertoli, E.; Battino, M. Contribution of Honey in Nutrition and Human Health: A Review. Med. J. Nutr. Metab. 2009, 3, 15–23. [Google Scholar] [CrossRef]
- Magrach, A.; González-Varo, J.P.; Boiffier, M.; Vilà, M.; Bartomeus, I. Honeybee Spillover Reshuffles Pollinator Diets and Affects Plant Reproductive Success. Nat. Ecol. Evol. 2017, 1, 1299–1307. [Google Scholar] [CrossRef]
- Goblirsch, M. Nosema ceranae disease of the honey bee (Apis mellifera). Apidologie 2018, 49, 131–150. [Google Scholar] [CrossRef] [Green Version]
- van Dooremalen, C.; Cornelissen, B.; Poleij-Hok-Ahin, C.; Blacquière, T. Single and interactive effects of Varroa destructor, Nosema spp., and imidacloprid on honey bee colonies (Apis mellifera). Ecosphere 2018, 9, e02378. [Google Scholar] [CrossRef] [Green Version]
- Grozinger, C.M.; Flenniken, M.L. Bee viruses: Ecology, pathogenicity, and impacts. Annu. Rev. Entomol. 2019, 64, 205–226. [Google Scholar] [CrossRef]
- Morfin, N.; Goodwin, P.H.; Guzman-Novoa, E. Interaction of field realistic doses of clothianidin and Varroa destructor parasitism on adult honey bee (Apis mellifera L.) health and neural gene expression, and antagonistic effects on differentially expressed genes. PLoS ONE 2020, 15, e0229030. [Google Scholar] [CrossRef] [Green Version]
- Gisder, S.; Möckel, N.; Linde, A.; Genersch, E. A cell culture model for Nosema ceranae and Nosema apis allows new insights into the life cycle of these important honey bee-pathogenic microsporidia. Environ. Microbiol. 2011, 13, 404–413. [Google Scholar] [CrossRef]
- Lamei, S.; Hu, Y.O.O.; Olofsson, T.C.; Andersson, A.F.; Forsgren, E.; Vásquez, A. Improvement of identification methods for honeybee specific Lactic Acid Bacteria; future approaches. PLoS ONE 2017, 12, e0174614. [Google Scholar] [CrossRef]
- Cameron, T.C.; Wiles, D.; Beddoe, T. Current Status of Loop-Mediated Isothermal Amplification Technologies for the Detection of Honey Bee Pathogens. Front. Vet. Sci. 2021, 8, 659683. [Google Scholar] [CrossRef]
- Garrido-Bailón, E.; Higes, M.; Martínez-Salvador, A.; Antúnez, K.; Botías, C.; Meana, A.; Prieto, L.; Martín-Hernández, R. The prevalence of the honeybee brood pathogens Ascosphaera apis, Paenibacillus larvae and Melissococcus plutonius in Spanish apiaries determined with a new multiplex PCR assay. Microb. Biotechnol. 2013, 6, 731–739. [Google Scholar] [CrossRef]
- Arismendi, N.; Bruna, A.; Zapata, N.; Vargas, M. PCR-specific detection of recently described Lotmaria passim (Trypanosomatidae) in Chilean apiaries. J. Invertebr. Pathol. 2016, 134, 1–5. [Google Scholar] [CrossRef]
- Milićević, V.; Radojičić, S.; Kureljušić, J.; Šekler, M.; Nešić, K.; Veljović, L.; Maksimović Zorić, J.; Radosavljević, V. Molecular detection of black queen cell virus and Kashmir bee virus in honey. AMB Expr. 2018, 8, 128. [Google Scholar] [CrossRef] [PubMed]
- Bakonyi, T.; Derakhshifar, I.; Grabensteiner, E.; Nowotny, N. Development and evaluation of PCR assays for the detection of Paenibacillus larvae in honey samples: Comparison with isolation and biochemical characterization. Appl. Environ. Microbiol. 2003, 69, 1504–1510. [Google Scholar] [CrossRef] [Green Version]
- Giersch, T.; Berg, T.; Galea, F.; Hornitzky, M. Nosemaceranae infects honey bees (Apis mellifera) and contaminates honey in Australia. Apidologie 2009, 40, 117–123. [Google Scholar] [CrossRef] [Green Version]
- Ribani, A.; Utzeri, V.J.; Taurisano, V.; Fontanesi, L. Honey as a source of environmental DNA for the detection and monitoring of honey bee pathogens and parasites. Vet. Sci. 2020, 7, 113. [Google Scholar] [CrossRef]
- Ribani, A.; Utzeri, V.J.; Taurisano, V.; Galuppi, R.; Fontanesi, L. Analysis of honey environmental DNA indicates that the honey bee (Apis mellifera L.) trypanosome parasite Lotmaria passim is widespread in the apiaries of the North of Italy. J. Invertebr. Pathol. 2021, 184, 107628. [Google Scholar] [CrossRef] [PubMed]
- Thomsen, P.F.; Willerslev, E. Environmental DNA–An emerging tool in conservation for monitoring past and present biodiversity. Biol. Conserv. 2015, 183, 4–18. [Google Scholar] [CrossRef]
- Nevers, M.B.; Byappanahalli, M.N.; Morris, C.C.; Shively, D.; Przybyla-Kelly, K.; Spoljaric, A.M.; Dickey, J.; Roseman, E.F. Environmental DNA (eDNA): A tool for quantifying the abundant but elusive round goby (Neogobius melanostomus). PLoS ONE 2018, 13, e0191720. [Google Scholar] [CrossRef] [PubMed]
- Ruppert, K.M.; Kline, R.J.; Rahman, M.S. Past, present, and future perspectives of Environmental DNA (eDNA) metabarcoding: A systematic review in methods, monitoring, and applications of global eDNA. Glob. Ecol. Conserv. 2019, 17, e00547. [Google Scholar] [CrossRef]
- Bargańska, Ż.; Ślebioda, M.; Namieśnik, J. Honey bees and their products: Bioindicators of environmental contamination. Crit. Rev. Environ. Sci. Technol. 2016, 46, 235–248. [Google Scholar] [CrossRef]
- Girotti, S.; Ghini, S.; Ferri, E.; Bolelli, L.; Colombo, R.; Serra, G.; Porrini, C.; Sangiorgi, S. Bioindicators and biomonitoring: Honeybees and hive products as pollution impact assessment tools for the Mediterranean area. Euro-Mediterr. J. Environ. Integr. 2020, 5, 62. [Google Scholar] [CrossRef]
- Hawkins, J.; de Vere, N.; Griffith, A.; Ford, C.R.; Allainguillaume, J.; Hegarty, M.J.; Baillie, L.; Adams-Groom, B. Using DNA metabarcoding to identify the floral composition of honey: A new tool for investigating honey bee foraging preferences. PLoS ONE 2015, 10, e0134735. [Google Scholar] [CrossRef] [Green Version]
- Utzeri, V.J.; Ribani, A.; Schiavo, G.; Bertolini, F.; Bovo, S.; Fontanesi, L. Application of next generation semiconductor based sequencing to detect the botanical composition of monofloral, polyfloral and honeydew honey. Food Control 2018, 86, 342–349. [Google Scholar] [CrossRef]
- Balkanska, R.; Stefanova, K.; Stoikova–Grigorova, R. Main honey botanical components and techniques for identification: A review. J. Apic. Res. 2020, 59, 852–861. [Google Scholar] [CrossRef]
- Olivieri, C.; Marota, I.; Rollo, F.; Luciani, S. Tracking plant, fungal, and bacterial DNA in honey specimens. J. Forensic Sci. 2012, 57, 222–227. [Google Scholar] [CrossRef]
- Forsgren, E.; Laugen, A.T. Prognostic value of using bee and hive debris samples for the detection of American foulbrood disease in honey bee colonies. Apidologie 2014, 45, 10–20. [Google Scholar] [CrossRef] [Green Version]
- Revainera, P.D.; Quintana, S.; de Landa, G.F.; Iza, C.G.; Olivera, E.; Fuentes, G.; Plischuk, S.; Medici, S.; Ruffinengo, S.; Marcangelli, J.; et al. Molecular detection of bee pathogens in honey. J. Insects Food Feed 2020, 6, 467–474. [Google Scholar] [CrossRef]
- Soares, S.; Grazina, L.; Costa, J.; Amaral, J.S.; Oliveira, M.B.P.P.; Mafra, I. Botanical authentication of Lavender (Lavandula spp.) honey by a novel DNA-barcoding approach coupled to high resolution melting analysis. Food Control 2018, 86, 367–373. [Google Scholar] [CrossRef] [Green Version]
- Lucek, K.; Galli, A.; Gurten, S.; Hohmann, N.; Maccagni, A.; Patsiou, T.; Willi, Y. Metabarcoding of honey to assess differences in plant-pollinator interactions between urban and non-urban sites. Apidologie 2019, 50, 317–329. [Google Scholar] [CrossRef]
- Soares, S.; Amaral, J.S.; Oliveira, M.B.P.P.; Mafra, I. Improving DNA isolation from honey for the botanical origin identification. Food Control 2015, 48, 130–136. [Google Scholar] [CrossRef] [Green Version]
- Utzeri, V.J.; Ribani, A.; Fontanesi, L. Authentication of honey based on a DNA method to dierentiate Apis mellifera subspecies: Application to Sicilian honey bee (A. m. siciliana) and Iberian honey bee (A. m. iberiensis) honeys. Food Control 2018, 91, 294–301. [Google Scholar] [CrossRef]
- Solignac, M.; Cornuet, J.M.; Vautrin, D.; Le Conte, Y.; Anderson, D.; Evans, J.; Cros-Arteil, S.; Navajas, M. The invasive Korea and Japan types of Varroa destructor, ectoparasitic mites of the Western honey bee (Apis mellifera), are two partly isolated clones. Proc. R. Soc. Lond. B Biol. Sci. 2005, 272, 411–419. [Google Scholar] [CrossRef] [Green Version]
- Navajas, M.; Anderson, D.L.; de Guzman, L.I.; Huang, Z.Y.; Clement, J.; Zhou, T.; Le Conte, Y. New Asian types of Varroa destructor: A potential new threat for world apiculture. Apidologie 2010, 41, 181–193. [Google Scholar] [CrossRef] [Green Version]
- Martín-Hernández, R.; Meana, A.; Prieto, L.; Salvador, A.M.; Garrido-Bailón, E.; Higes, M. Outcome of colonization of Apis mellifera by Nosema ceranae. Appl. Environ. Microbiol. 2007, 73, 6331–6338. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ruttner, F. Biogeography and Taxonomy of Honey Bees; Springer: Berlin/Heidelberg, Germany, 1988. [Google Scholar]
- Smyth, R.P.; Schlub, T.E.; Grimm, A.; Venturi, V.; Chopra, A.; Mallal, S.; Davenport, M.P.; Mak, J. Reducing chimera formation during PCR amplification to ensure accurate genotyping. Gene 2010, 469, 45–51. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Z.; Schwartz, S.; Wagner, L.; Miller, W. A greedy algorithm for aligning DNA sequences. J. Comput. Biol. 2000, 7, 203–214. [Google Scholar] [CrossRef]
- Pelin, A.; Selman, M.; Aris-Brosou, S.; Farinelli, L.; Corradi, N. Genome analyses suggest the presence of polyploidy and recent human-driven expansions in eight global populations of the honeybee pathogen Nosema ceranae. Environ. Microbiol. 2015, 17, 4443–4458. [Google Scholar] [CrossRef] [PubMed]
- Basa, B.; Belay, W.; Tilahun, A.; Teshale, A. Review on medicinal value of honeybee products: Apitherapy. Adv. Biol. Res. 2016, 10, 236–247. [Google Scholar] [CrossRef]
- Jovetić, M.; Trifković, J.; Stanković, D.; Manojlović, D.; Milojković-Opsenica, D. Mineral content as a tool for the assessment of honey authenticity. J. AOAC Int. 2017, 100, 862–870. [Google Scholar] [CrossRef] [PubMed]
- Langlois, V.S.; Allison, M.J.; Bergman, L.C.; To, T.A.; Helbing, C.C. The need for robust qPCR-based eDNA detection assays in environmental monitoring and species inventories. Environ. DNA 2021, 3, 519–527. [Google Scholar] [CrossRef]
- Beng, K.C.; Corlett, R.T. Applications of environmental DNA (eDNA) in ecology and conservation: Opportunities, challenges and prospects. Biodivers. Conserv. 2020, 29, 2089–2121. [Google Scholar] [CrossRef]
- Ramsey, S.D.; Ochoa, R.; Bauchan, G.; Gulbronson, C.; Mowery, J.D.; Cohen, A.; Lim, D.; Joklik, J.; Cicero, J.M.; Ellis, J.D.; et al. Varroa destructor feeds primarily on honey bee fat body tissue and not hemolymph. Proc. Natl. Acad. Sci. USA 2019, 116, 1792–1801. [Google Scholar] [CrossRef] [Green Version]
- Emsen, B.; Guzman-Novoa, E.; Hamiduzzaman, M.M.; Eccles, L.; Lacey, B.; Ruiz-Pérez, R.A.; Nasr, M. Higher prevalence and levels of Nosema ceranae than Nosema apis infections in Canadian honey bee colonies. Parasitol. Res. 2016, 115, 175–181. [Google Scholar] [CrossRef] [PubMed]
- Pacini, A.; Mira, A.; Molineri, A.; Giacobino, A.; Bulacio Cagnolo, N.; Aignasse, A.; Zago, L.; Izaguirre, M.; Merke, J.; Orellano, E.; et al. Distribution and prevalence of Nosema apis and N. ceranae in temperate and subtropical eco-regions of Argentina. J. Invertebr. Pathol. 2016, 141, 34–37. [Google Scholar] [CrossRef]
- Giraudeau, M.; Mousel, M.; Earl, S.; McGraw, K. Parasites in the city: Degree of urbanization predicts poxvirus and coccidian infections in house finches (Haemorhous mexicanus). PLoS ONE 2014, 9, e86747. [Google Scholar] [CrossRef] [Green Version]
- Leong, M.; Kremen, C.; Roderick, G.K. Pollinator interactions with yellow starthistle (Centaurea solstitialis) across urban, agricultural, and natural landscapes. PLoS ONE 2014, 9, e86357. [Google Scholar] [CrossRef] [PubMed]
- Patz, J.A.; Daszak, P.; Tabor, G.M.; Aguirre, A.A.; Pearl, M.; Epstein, J.; Wolfe, N.D.; Kilpatrick, A.M.; Foufopoulos, J.; Molyneux, D.; et al. Unhealthy landscapes: Policy recommendations on land use change and infectious disease emergence. Environ. Health Perspect. 2004, 112, 1092–1098. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Youngsteadt, E.; Appler, R.H.; López-Uribe, M.M.; Tarpy, D.R.; Frank, S.D. Urbanization increases pathogen pressure on feral and managed honey bees. PLoS ONE 2015, 10, e0142031. [Google Scholar] [CrossRef] [Green Version]
- Decourtye, A.; Alaux, C.; Le Conte, Y.; Henry, M. Toward the protection of bees and pollination under global change: Present and future perspectives in a challenging applied science. Curr. Opin. Insect. Sci. 2019, 35, 123–131. [Google Scholar] [CrossRef] [PubMed]
- Wakgari, M.; Yigezu, G. Honeybee keeping constraints and future prospects. Cogent Food Agric. 2021, 7, 1872192. [Google Scholar] [CrossRef]
- Brosi, B.J.; Delaplane, K.S.; Boots, M.; de Roode, J.C. Ecological and evolutionary approaches to managing honeybee disease. Nat. Ecol. Evol. 2017, 1, 1250–1262. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Steinhauer, N.; Kulhanek, K.; Antúnez, K.; Human, H.; Chantawannakul, P.; Chauzat, M.P. Drivers of colony losses. Curr. Opin. Insect Sci. 2018, 26, 142–148. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Evans, J.D.; Zhou, L.; Boncristiani, H.; Kimura, K.; Xiao, T.; Litkowski, A.M.; Pettis, J.S. Asymmetrical coexistence of Nosema ceranae and Nosema apis in honey bees. J. Invertebr. Pathol. 2009, 101, 204–209. [Google Scholar] [CrossRef] [PubMed]
- Zhang, G.; St. Clair, A.L.; Dolezal, A.; Toth, A.L.; O’Neal, M. Honey bee (Hymenoptera: Apidea) pollen forage in a highly cultivated agroecosystem: Limited diet diversity and its relationship to virus resistance. J. Econ. Entomol. 2020, 113, 1062–1072. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.J.; Smart, M.D.; Anelli, C.M.; Sheppard, W.S. Honey bees (Apis mellifera) reared in brood combs containing high levels of pesticide residues exhibit increased susceptibility to Nosema (Microsporidia) infection. J. Invertebr. Pathol. 2012, 109, 326–329. [Google Scholar] [CrossRef]
- Chen, Y.W.; Chung, W.P.; Wang, C.H.; Solter, L.F.; Huang, W.F. Nosema ceranae infection intensity highly correlates with temperature. J. Invertebr. Pathol. 2012, 111, 264–267. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).