Abstract
The increasing demand for animal-derived foods has led to intensive and large-scale livestock production with the consequent formation of large amounts of manure. Livestock manure is widely used in agricultural practices as soil fertilizer worldwide. However, several antibiotic residues, antibiotic resistance genes (ARGs) and antibiotic-resistant bacteria are frequently detected in manure and manure-amended soils. This review explores the role of manure in the persistence and dissemination of ARGs in the environment, analyzes the procedures used to decrease antimicrobial resistance in manure and the potential impact of manure application in public health. We highlight that manure shows unique features as a hotspot for antimicrobial gene dissemination by horizontal transfer events: richness in nutrients, a high abundance and diversity of bacteria populations and antibiotic residues that may exert a selective pressure on bacteria and trigger gene mobilization; reduction methodologies are able to reduce the concentrations of some, but not all, antimicrobials and microorganisms. Conjugation events are often seen in the manure environment, even after composting. Antibiotic resistance is considered a growing threat to human, animal and environmental health. Therefore, it is crucial to reduce the amount of antimicrobials and the load of antimicrobial resistant bacteria that end up in soil.
1. Introduction
Livestock production is an extremely dynamic activity, highly influenced not only by human population growth, but also by competition for natural resources and by human health and environmental concerns. In fact, the human population in 2050, according to the Department of Economic and Social Affairs of the United Nations, is estimated to be 9.735 billion [1], with the consequent increasing demand for livestock products [2]. The need to increase the potential of livestock production has led to developments in breeding, nutrition and animal health [3]. Therefore, antibiotics are not only used globally with prophylactic and metaphylactic purposes and for the treatment of animal infectious disease, but are also used in subtherapeutic doses to promote animal growth [4,5].
The administration of antibiotics to animals through feed and drinking water, or by other routes such as injection, has become increasingly important in intensive food-animal production [4,6,7]. Despite restrictions on the use of antibiotics in animal feed as growth promoters, some of these compounds are still used illegally on some small farms [6]. In European Union countries, the use of antibiotics as growth promoters was banned in 2006 [8].
The extensive use and misuse of antimicrobial agents in human and veterinary medicine led to the emergence and selection of antimicrobial resistance in bacteria, with potential adverse consequences for human and animal health [9]. Resistant bacteria and their genes can disseminate between humans and animals by the food chain and spread in the environment, which is the reason why antimicrobial resistance must be handled by a holistic approach, a concept designated as One Health [10]. After administration, many antibiotics that are used in food-producing animals are poorly absorbed in the animal gut, resulting in their excretion into the environment, without degradation, in their active metabolite forms [6].
The increasing demand for animal-derived products has led to intensive and large-scale livestock breeding, with the consequent production of a huge amount of livestock manure [11]. Animal manure is directly used as organic fertilizer in the agricultural sector and consists, mostly, of animal feces [12]. This became a common practice in many countries of the world as an alternative to chemical fertilizers for arable soils of low fertility [13,14].
Due to anthropogenic activities, livestock manure may act as a reservoir for antibiotic residues and bacteria carrying different antibiotic resistance genes (ARGs) that confer resistance to many clinically important antibiotics [14,15]. The main focus of the current review will be to explore how manure can work as a hotspot for antimicrobial resistance gene dissemination. Manure gives to the soil a unique environment for the spread of antibiotic resistance genes by horizontal gene transfer (HGT) mechanisms [16,17]. It is rich in nutrients and presents dense and highly diverse bacterial populations and accumulates antibiotic residues and/or metals that act as a selective pressure and may trigger the exchange of bacterial DNA. These elements mix with agricultural soil bacterial communities and irrigation water, reaching microbial communities from different ecosystems, including water streams, and spread to the environment, wildlife and human and other animal communities [18]. Herein, we will discuss the composition of manure, highlighting the most frequent antibiotics and ARGs associated with mobile genetic elements and the HGT mechanisms described so far in manure, to better understand how manure can be a source of antimicrobial resistance spread, the potential impact in public health and the procedures used to reduce this risk and their limitations.
2. Composition of Manure
The intensive production of meat, milk, eggs and other agricultural products for increasing human consumption needs results in the generation of large amounts of manure during animal growth. The mixture of animal feces, urine, feed remains, soil, wash waters and bedding materials to absorb waste such as wood chips, wheat straw, flax straw, sawdust or even peanut and rice hulls, give rise to the product that has been used as organic soil fertilizer on several farms [11]. Manure from different animals have different physical properties, different nutrient contents and specific application rates on the land. This complexity of manure contents makes it more difficult to manage when compared to chemical fertilizers [19].
In addition to the nutrients excreted by animals, including nitrogen, phosphorus, potassium and sulfur, manure also contains heavy metals, such as cadmium, cobalt, copper (Cu), lead, manganese, nickel, selenium and zinc (Zn), which are considered long-term soil contaminants since they are not degraded [19,20,21,22].
Furthermore, the heavy metals present in manure may exert a long-term and continuous selective pressure on ARGs via co-resistance or cross-resistance because the resistance genes for antibiotics and metals are often located together in the same plasmid or other mobile genetic element (MGE) [23]. A strong relationship between MGEs, metals and metal resistance genes and ARGs present in manure has been previously determined [24]. Therefore, heavy metals may also promote the long-term persistence of ARGs during manure management, storage and disposal [25,26,27].
The use of manure as a fertilizer not only provides nutrients and organic material during agricultural practices but also facilitates the spread of beneficial or pathogenic microorganisms in soils [28,29]. In fact, the bacterial communities of manure and soil are highly diverse [30]. The microbial population present in manure is highly diverse [29], and mainly depends on the animal species [31]. Manure is an important source of bacteria, which can cause serious illness both in animals and humans. The major manure-borne pathogens are zoonotic bacteria, such as Salmonella spp., Escherichia coli, Campylobacter spp., Listeria monocytogenes, Yersinia enterocolitica; and protozoa like Cryptosporidium parvum and Giardia lamblia [32]. Other manure-borne pathogens and phages are also present, but they are more unusual [33]. Moreover, Enterococcus, Bacillus and Clostridium species are also found in manure [29]. These bacteria are colonizers of the intestinal tract of animals and are shed with feces. Their potential dissemination into environment and humans firstly depends on the ability to survive in manure after excretion, which can vary from a few days to several months. For instance, Bacillus spp., due to the ability to form spores, can resist unfavorable growth conditions for long periods of time [29]. Moreover, several Salmonella enterica serotypes were shown to persist in soil for at least 21 days after manure application, while they were rarely present before [34]. The application of manure also leads to the spread of these bacteria in the environment, namely in water [18].
Animal feces and manure are important reservoirs of antibiotics and antibiotic-resistant bacteria. Nowadays, ARGs are considered as emerging contaminants, considering the potential risks and their origin, mobility and elimination in the environment [35]. The use of antibiotics during animal growth favors the appearance of antibiotic-resistant bacteria in the gut microbiota, which consequently may be released within feces and be disseminated by manure application [30]. The application of organic fertilizers, as well as inorganic fertilizers to a less extent, has been shown to impact the soil microbial community and it is the major driver of the shaping of the antibiotic resistome [36,37].
Antibiotics and Antimicrobial Resistance Genes in Manure
A strong correlation between the high use of certain antibiotics in livestock (and their residues) and the presence of ARGs and MGEs, such as plasmids, integrative conjugative elements, transposons and integrons, involved in the spread of ARGs via HGT pathways from manure to soil microbes, has been found [38,39,40]. ARGs and MGEs, especially integrons and transposons, in manure have been showed to be closely related, and their increased abundance is related to manure application [24,37]. Manure has a higher impact on the abundance and diversity of ARGs in soil than chemical fertilizers [37].
As most antibiotics have polar functional groups and high solubility in water, many antibiotics used in livestock production are poorly absorbed in the animal gut, resulting in the excretion of 30–90% of the parent compound via feces or urine [5]. Antibiotic metabolites can also be excreted with antimicrobial activity or can return to the initial active compound [41]. One example is the conjugation in the liver of sulfamethazine with sugars after its administration; after its excretion, bacteria are able to degrade the sugars and convert it back to its bioactive form [42]. Therefore, active forms of veterinary antimicrobials are often found in manure and may disseminate in the environment after its application on soil [5]. Yet, the stability and efficacy of antibiotics in the environment depends on their physio-chemical properties, types of soil, climatic conditions and other environmental factors [5,43]. Non-degraded antibiotics in manure and in soil may act as a selective pressure and contribute to the emergence and dissemination of antimicrobial resistance determinants [44].
In general, ARG concentrations in livestock waste is higher than in human waste, and in manured soil, levels of ARGs can be 28,000 times higher compared to un-manured soil [35]. However, antibiotic-resistant bacteria are also abundant in manure from animals with no history of antibiotic treatment, suggesting that the animal gastrointestinal microbiota harbor intrinsically antibiotic-resistant bacteria [14].
One hundred and nine ARGs, associated with resistance to antibiotics extensively used in livestock farming and representing the main classes applied in human and veterinary medicine, have been detected in fresh chicken, pig and bovine manure collected from 12 large-scale Chinese farms [24,45]. ARG abundance and content may differ depending on the species [24,35,45,46]. For instance, in the previously mentioned Chinese farms, a higher diversity and abundance of ARGs was seen in chicken manure, followed by pig manure and finally by bovine manure [24,47], while dairy manure collected from two Canadian farms had a slightly higher variety of ARGs [46]. Common to both studies is the observation that the species is not the only influencing factor of ARG distribution, as this varied from farm to farm and with the sampling year. Dairy manure slurry also had higher levels of ARGs than a mix of dry stack equine, bovine and ovine manure [48]. In addition to the antibiotic administration patterns [49], feed administration, exposure to heavy metals, animal age, time and manure nitrogen content may all contribute to the ARG content in manure [24,46,48]. The major concern about the presence of ARGs in manure is the possibility of their transfer to the soil bacteria via HGT mechanisms, which can promote the spread of antimicrobial resistance among different microbial communities [30,37,48]. In this context, it is important to understand the differences in the content of antibiotic-resistant bacteria and ARGs in soils in response to manure application. This response mainly depends on the characteristics of applied manures [50]. For instance, swine manure application was shown to have a higher impact on the soil ARG frequency than dairy manure [46]. Different ARG compositions were found between pig and chicken feces. For example, sulfonamide-resistant Escherichia coli from different host species had significantly different distributions of the resistance determinant sul2 gene, which was found less frequently in swine samples than in other animals’ samples [51]. After a one-time manure application, persistence times in soil vary among different ARGs [48]. However, ARGs are stable in the microbial community of a soil that regularly receives manure applications [36,37,52].
Diverse classes of antibiotics are used in food-animal production, which depends on the animal, the objective of the given antibiotic (therapeutic, prophylaxis, growth promotion) and country policy rules.
Tetracyclines and sulfonamides are two classes of antibiotics often used in veterinary medicine due to their broad-spectrum activity, relatively low toxicity and low price [53,54]. After administration, most tetracyclines are excreted as active compounds through feces and urine, which are spread in the environment by manure fertilization practices [55,56]. These compounds have a high affinity for soil organic matter [57]. Additionally, animals’ gut microbiota show a very high prevalence of tetracycline resistance [30,54].
The tet gene family, involved in the active efflux of tetracycline compounds, ribosomal protection or the enzymatic modification of antimicrobial agents [58], has been reported as one of the most frequently detected ARGs in animal manure [21,25]. After manure application, tet gene levels in the soil are increased significantly and manure application amended with tetracyclines promotes the accumulation of tet genes and modifies the soil bacterial composition [54,55,59,60]. These genes are often embedded in plasmids (isolated from manure and animal bacteria) [61,62] and inserted in transposons, which facilitates their dissemination [54]. For example, the genes tetW, tetO and tetQ are very common in pig and cattle manure, suggesting that they are stably maintained in the animals’ gut microbiota [63].
Sulfonamides have been widely used in clinical and veterinary medicine to treat bacterial and protozoal infections [64]. They inhibit the dihydropteroate synthase (DHPS), inhibiting the folic acid biosynthesis pathway, which is necessary for the synthesis of DNA and RNA precursors, leading to the non-replication of bacterial DNA and the non-synthesis of bacterial proteins [65]. To date, three sul genes have been identified [66]: sul1 is frequently identified in class 1 integrons in slurry and soil environments, sul2 was first identified on a broad host range Escherichia coli plasmid and has been found on small nonconjugative resistance plasmids of the IncQ family, while the sul3 gene has been identified in isolates from different sources that also carry a class 1 integron [64,65,67]. Nowadays, sul genes occur in a wide range of bacterial species, because they are often located on transposable elements of self-transferable or mobilizable broad host range plasmids [68,69].
In contrast to tetracyclines, sulfonamide compounds do not sorb strongly to the soil, and have been detected in surface water, groundwater and soil pore water [70]. Nevertheless, these antibiotics and their resistance genes have been found in soil and manure [21,52,64,68,71,72]. Mutations in the chromosomal DHPS gene that codes for the inhibition of folic acid biosynthesis (folP) or the acquisition of an alternative DHPS gene (sul) are responsible for sulfonamide resistance [64,65]. The sul genes occur in a wide range of bacterial species because they are frequently located on mobilizable plasmids and in mobile genetic elements, such as class 1 integrons and transposons [64,65,67,68,69]. These elements carry multiple antibiotic resistance genes that are co-selected by sulphonamides [64,69,72]. The presence of sul genes in pig farms and cattle waste lagoons seems to be associated with the use of sulfonamide [30,73]. Sulfadiazine was found in turkey and chicken manure in significant amounts [74], suggesting that poultry manure could be a source of sul genes. Several studies reported sulfonamide resistance genes as the most detected ARGs in manured soils. In general, the sul1 gene is the most frequently detected ARG in manures, manured soil samples, agricultural and non-agricultural soils, wastewater and surface water in feedlots, evidencing their extensive ability to spread [75,76].
β-lactams are one of the most widely used groups of antibiotics, including in veterinary medicine [77]. β-lactam residues, β-lactam-resistant bacteria and β-lactam-resistant determinants have been found in dairy cattle manure because β-lactam antibiotics are often used to treat mastitis [14,15,40,78,79]. Pig manure can also work as a reservoir for the transferable amoxicillin antibiotic resistance genes blaTEM. These are often embedded in IncN plasmids [40]. IncN plasmids are also associated with blaCTX-M genes, coding for extended-spectrum β-lactamase (ESBL)—which have been found in bacteria from pigs, farmers and farm environments, such as manure [80]. These findings may suggest that IncN plasmids play a key role in β-lactamase gene dissemination in manure and soil bacteria. Beyond their insertion in mobilizable plasmids, ESBL genes are located in insertion sequences, like ISEcp1 and ISCR1, which facilitate their spread [81].
Macrolide antibiotics, such as erythromycin and tylosin, are often administered together with lincosamides and streptogramins in livestock production [82,83]. Tylosin is not completely metabolized in the gut and up to three quarters of the antibiotic can be excreted in the urine and feces [84]; tylosin residues were reported in swine manure [83,84]. Macrolide-resistant bacteria, carrying erythromycin ribosome methylation (erm) genes and/or macrolide efflux (mef) genes, are also excreted in feces [83,84]. Moreover, the binding site for erythromycin overlaps binding sites for other macrolides, lincosamides and streptograminB (MLSB) antibiotics, leading to cross-resistance in the MLSB antibiotic family due to emr encoded genes [83]. Therefore, the use of tylosin increases the resistance to MLSB of animal gut microbiota [85]. Specifically, its use at sub-therapeutic concentrations has been correlated with increasing Enterococcus spp. macrolide resistance [86]. Several studies report erm genes in manured soil [82,87,88,89,90]. Swine manure seems to have a higher content of erm genes than bovine manure samples [85], and a high diversity. Various erm genes have been found in swine waste lagoons: ermA, ermB, ermC, ermF, ermG, ermT, ermQ and ermX, with ermB and ermF being the most prevalent. Remarkably, the ermB gene was never detected in non-manured soils or before the first spring manure amendment, which is strongly suggestive of the introduction of this gene into soil via manure application [89]. The erm(B) gene has often been found to be associated with the presence of tetM, probably because these genes are frequently integrated together on the Tn916-Tn1545 family of conjugative transposons [91].
Colistin is an old antibiotic widely used in veterinary medicine for the prevention and treatment of gastrointestinal infections caused by Enterobacteriaceae [92,93]. Colistin is poorly absorbed through the pig gastrointestinal tract and is subsequently excreted in feces. It not only contaminates the soil but may also contribute to the development and spread of colistin resistance by exerting pressure selection on animal gut microbiota [94]. Recently, the plasmid-mediated and mobilizable colistin resistance genes, mcr, have been identified from bacteria worldwide [92,93]. Ten different mcr genes (mcr-1 to mcr-10) and variants have been described [95], even though the mcr-1 gene is the most reported worldwide, especially in animal samples. This may suggest that the colistin mobilizable resistance gene reached humans via mcr from animal bacteria [96]. The presence of colistin in manure correlates with the finding of the mcr-1 gene both in pig and poultry manure samples [96,97]. Additionally, this gene has been detected in bovine and horse manure [98]. Livestock manure seems to be an important reservoir of mcr-encoding plasmids, mostly carried by colistin-resistant Escherichia coli on diverse plasmid replicon types: IncX4, IncI1, IncFII, IncFIB, IncX1 and IncQ1 [97,99,100].
Quinolones are also used in intensive animal production and might be found in manure. The most frequent resistance is encoded by mutations in the topoisomerase genes, involved in DNA replication, which are chromosomal. Yet, there are plasmid resistances genes, such as in the qnr family, that may confer resistance to quinolones when mutations in the chromosome are already present [101]. They can be transferred to a recipient cell, but they might not give clinical resistance to the host cell.
3. ARGs Spread by HGT Mechanisms
HGT is the process of the transfer of genetic information between organisms that are not directly related, and includes the spread of antibiotic resistance genes among bacteria [102]. There are three main mechanisms of HGT in bacteria: conjugation, natural transformation and transduction, which involve plasmids, naked DNA and bacteriophages, respectively. Moreover, MGEs, such as integrons, insertion sequences and transposons, also play a synergistic role in resistance dissemination [16]. Conjugation is considered the most common mechanism for HGT between different bacteria and occurs when a donor bacterium directly transfer genes to another recipient cell, while in transduction and natural transformation, there is no direct contact between the donor and recipient cells [103].
HGT can promote rather large-scale changes in a bacterial genome, allowing for the evolution of genes and bacteria, which in the ultimate instance may result in the emergence of resistant bacteria [104].
Fertilized soils may be hotspots for HGT occurrence once all necessary elements are present, including ARGs provided by manure, the natural soil resistome, bacterial cells vehiculated by manure that may also carry ARGs, soil microbiota and antibiotic residues from manure and irrigation water that create a selective pressure in bacteria. The presence of antibiotic residues, even at subinhibitory concentrations, can stimulate HGT events and ARG transfer [105]. Dong et al. showed that extracellular ARGs, such as those detected in manure, maintain their transforming ability [106]. Animal manure represents a major source of antibiotics and ARGs in the environment [107], creating a favorable environment for HGT events [37,48]. Organic fertilization has been demonstrated to accelerate ARG dissemination in soil by HGT events, as compared to inorganic fertilization [36]. Furthermore, manure is often stored before being applied to soil, allowing for HGT and resistance selection during storage. The application of a mathematical model concluded that it is crucial to know HGT rates, antibiotic influx and the length of storage time in order to prevent the spread of resistance; for instance, reducing antibiotic inflow only controls resistance if the gene transfer rate is low [108].
The co-occurrence of heavy metals and antibiotics in manure may increase the HGT pathways of ARGs and promote an increased emergence of antibiotic-resistant bacteria [23]. Many studies show not only a positive correlation between the presence of many ARGs and Cu levels in soil, but also the ability of Cu to promote the transfer of ARGs via conjugation [109]. The antimicrobial properties of Cu may explain its ability to promote conjugative transfer not only within bacterial genera but also across different bacterial genera. The overproduction of reactive oxygen species, elevated SOS response and enhanced cell membrane permeability might be the underlying mechanisms in the increase of conjugative activity [103]. In particular, sul1 and sul2 genes are strictly associated with levels of Cu, Zn and mercury. Other specific ARGs, such as tetM, blaCTX-M and blaOXA genes, are more closely associated with chromium; tetW with nickel; and tetM with nickel, iron and lead levels [21,110]. Finally, quaternary ammonium compounds used as common disinfectants in pig farms may also enhance the co-selection of ARGs [111].
The rapid evolution, proliferation and spread of antibiotic resistance is especially promoted by plasmid mobilization via conjugation [112]. Conjugation requires physical cell-to-cell contact, with the donor being responsible for the formation of the sexual (F) pilus. Due to this contact, the bacterial environment and cell density influence the outcome frequency of conjugation events [105]. In general, plasmids are composed of the core genes required for essential functions, such as replication, transfer and maintenance, and accessory functions, encoding antibiotic or heavy metal resistance, catabolic functions and virulence or pathogenicity determinants; they often have a mosaic structure [40]. There are several worldwide reports of plasmids harboring extended-spectrum beta-lactamase (ESBL) genes, ampC genes, quinolone resistance genes (qnr), mcr genes and carbapenemase genes from environmental samples, including manure [112]. These plasmid-borne genes are mainly located on broad host range IncP-1, IncQ, IncN, IncW and IncF replicon-type plasmids, which are important vectors for the dissemination of ARGs between distantly related genera and species [30,105,112,113]. Salmonella plasmids were shown to persist for long periods in a North Carolina commercial swine farm environment after land manure application; 14 strains belonging to six serotypes were able to transfer ARGs embedded in plasmids to Escherichia coli JM109 by conjugation, and sequencing analysis revealed that plasmid-mediated ARG transfer occurred among the different Salmonella serotypes [112]. This result highlights that conjugal transfer occurs in the field. Plasmid pSN1216-29 has been isolated from cow manure [114] and was demonstrated to be able to conjugate with a broad range of bacterial genera extracted from soil and cow manure [115]; despite the fact that this plasmid does not carry accessory genes, one transposition event can lead to antimicrobial resistance acquisition, followed by horizontal dissemination.
The traF gene, involved in the assembly of the F pilus in Escherichia coli, has been detected at a high rate in multidrug-resistant Escherichia spp. from livestock manure, suggesting the possible occurrence of high levels of conjugation with the consequent spread of ARGs [23].
As mentioned above, the use of wastewater and manure for the irrigation and fertilization of soil may be the main uses responsible for the vast dissemination of the mcr-1 gene. Recently, two Escherichia coli harboring mcr-1 were isolated from horse (n = 1) and bovine (n = 1) manure, and the plasmid carrying mcr-1 isolated from the horse manure was successfully transferred by conjugation [116]. The sul1 and sul2 genes from pig slurry and manured agricultural soil samples were transferred from a variety of Gram-negative bacterial species to Escherichia coli K-12 CV601 and Pseudomonas putida UWC1 recipient cells by conjugation at different rates, indicating their presence on different mobile elements [64]. Livestock manure has a high prevalence of class 1 integrons carrying different ARGs as gene cassettes. The association between integrons and the sul1 gene may contribute to the dissemination of sulfonamide resistance in arable soils [117]. The presence of an antibiotic and/or its residues in manure increases the activity of integrases and transposases and consequently increases the excision/integration of gene cassettes in integrons [30].
A wide dissemination of blaCTX-M-1 has been reported in Escherichia coli in pig farming, including animals, manure and farmworkers. Despite the high diversity of the isolates, blaCTX-M-1 was associated with similar conjugative and broad host range IncN plasmids, suggesting their horizontal mobilization between the different isolates [80].
Plasmids belonging to different incompatibility types and carrying a variety of resistance genes, including blaTEM-1, sul1, sul2 and sul3, have been isolated from piggery manure, with most of them being able to transfer by conjugation, highlighting the potential for the transfer of resistance genes in manure [40].
Lateral erm gene transfer in a manure environment has also been reported between Gram-negative and Gram-positive bacteria. The molecular analysis of ermB genes from human, swine and poultry Enterococcus faecium isolates suggests that the HGT of antibiotic resistance genes plays a greater role in the dissemination of resistant determinants than the direct transmission of resistant strains [118]. The soil-borne equine and zoonotic pathogen Rhodococcus equi has been shown to be able to conjugate with other environmental Actinobacteria that share the habitat, leading to macrolide resistance due to the dissemination of erm(46) embedded in plasmid pRErm46; conjugation between two Rhodococcus equi strains also occurred in equine manure at environmental temperatures (22 and 30 °C) [119].
LowGC type plasmids are a group of conjugative plasmids that are abundant in pig manure and manured soils [62,120]. The accessory regions, which encode ARGs and mobile element-derived genes, have highly variable composition. However, after conjugative transfer, this plasmid structure is genetically unstable, which may have contributed to the significant diversity of this type of plasmid [62,121]. LowGC plasmids are closely involved with the horizontal dissemination of sul genes in manure and soil and have a long-term persistence in manured soils [62,120]. These plasmids can spread to a wide range of bacterial species, and sul2 gene can integrate into the chromosome through an intracellular mechanism mediated by insertion sequences located in the plasmid [122]. Jechalke et al. demonstrated that the effect of swine manure containing an antibiotic, in this particular case sulfadiazine, on the horizontal dissemination of sul genes embedded into LowGC plasmids also depends on the soil traits (bulk soil or rhizosphere) and the developmental stage of the plant species [120]. LowGC plasmids may also be involved in the dissemination into the environment and soil of tetY genes [121]. Acinetobacter species were identified as natural hosts for LowGC plasmids in manure and soil. This finding is particularly relevant since Acinetobacter spp. comprise both environmental and nosocomial species and the spread of ARGs located on LowGC plasmids is facilitated between closely related bacteria [111,121].
The conjugative Tn916-like transposon, associated with tetracycline resistance, has been identified in two isolates belonging to the Bacillus cereus group isolated from pig manure, and transfer to Enterococcus faecalis JH2-2 by conjugation was demonstrated [123].
The transfer of plasmid-mediated tetracycline and gentamicin resistance from indigenous soil bacteria to Pseudomonas putida KT2442 by conjugation has been shown to occur immediately after the amendment of soil with cattle manure, while the transfer rate after 29 weeks was insignificant when compared with unfertilized soil; the application of other fertilizers, such as sewage sludge and municipal solid waste compost also had similar effects [17]. This result highlights that the duration of persistence of ARGs in soil is not crucial, and the increase in ARG abundance due to manure application, even if for a short period of time, is enough to promote the horizontal dissemination of antimicrobial resistance.
A conjugative plasmid carrying a multidrug efflux pump encoding gene, oqxAB, especially associated with olaquindox and previously extensively used as a growth promoter in pigs, and chloramphenicol resistance have been detected in Escherichia coli isolated from swine manure [124]. This plasmid has since been shown to be transferred to Escherichia coli N43 by conjugation [125].
Transduction is the process of the transfer of genetic material between bacteria through the action of a virus, named a bacteriophage or simply a phage. Phages have been determined to be the most abundant biological entities on Earth and are a driving force in bacterial diversity and evolution [126], since they are able to regulate the dynamic of the bacterial population as well as facilitate HGT events. It is estimated that phage-like elements have been involved in the acquisition of 20% of the bacterial genome [127]. Agricultural soils are rich in bacteria and phages, in a ratio 1:10 to 1:100 [128], and manure slurry has been shown to have a higher prevalence of phages than other environmental sources [129]. Environmental phages harbor ARGs and genes conferring resistance to aminoglycosides, β-lactams and sulfonamides have been detected in fertilized soils [130]. There are studies that demonstrate the transmission of virulence genes by transduction in soil [18] or of resistance genes through a phage isolated from sewage effluent [131]; however, there are no studies exploring the involvement of transduction in the transfer of resistance genes within agricultural soil, despite the high potential for the occurrence of this HGT mechanism in this environment [18].
Natural transformation is characterized by the acquisition of naked DNA from the environment by competent recipient cells. Extracellular DNA has been shown to maintain its transformability over long periods of time [132], and there are several naturally competent bacterial species that inhabit soil, such as Acinetobacter baylyi, Bacillus subtilis and Pseudomonas stutzeri, or are part of the animal digestive tract, like Campylobacter spp. [133]. Several studies demonstrated that DNA adsorbed to different particles of the soil is protected against nuclease activity, which allows it to persist unchanged and available for uptake by competent cells [105,133]. Calcium is essential to the natural transformation of some species, and its presence in some natural environments has been shown to induce natural competence. Swine manure has a high calcium content, which suggests that natural transformation can occur in this environment [105], though there are as yet no studies demonstrating this.
A fourth HGT mechanism, transformation mediated by membrane vesicles (MVs), has been proposed [134]. MVs are released from all living cells, are biologically active and contain different components, such as genetic material, including chromosomal and plasmid DNA, as well as different types of RNA. These vesicles can mediate the transfer of ARGs between bacteria and can persist over time in natural environments [134,135]. Environmental and external stimuli induce the release of MVs; for instance, the exposure to subinhibitory antibiotics of different classes may induce MV release and HGT mediated by MVs [134]. Although there are no studies focused on the presence of MVs in manure and their potential involvement in HGT events, manure and manured soils can be a privileged platform for HGT mediated by MVs due to the presence of antimicrobials in manure and the stress response triggered by the land application of manure in soil resident bacterial communities [135].
The acquisition of antibiotic resistance genes and MGEs through HGT pathways often has a biological cost for the host cell in terms of reduced growth, competition and/or infectivity [136,137]. This metabolic burden often reduces the competitiveness of host bacteria in the absence of antibiotic pressure [30]. However this fitness reduction can be ameliorated within a few generations after plasmid acquisition through the evolution of compensatory mechanisms, which may restore fitness without compromising resistance to antibiotics [30,136,137]. Due to the long-term persistence of antibiotic pressure, organic farming practices may have favored the maintenance of acquired genes by HGT pathways in host bacteria of soil and/or manure and, consequently, this may have led to antibiotic-resistant bacteria emergence and persistence in the manure environment [30].
4. Manure Treatment
The control and elimination of antimicrobial resistance in agricultural fields are urgently needed [138]. The sewage treatment, composting or anaerobic digestion process of manure are some of the techniques that are used to reduce the concentrations of antimicrobials, ARGs and microorganisms that are introduced in soil through manure fertilization practices [139].
Composting is a spontaneous and biological process of aerobic digestion that involves the mineralization and humification of organic matter [12,140]. This bio-oxidative process involves environmental microorganisms, which break down organic materials, resulting in the formation of a stable final product, known as compost [12,140,141]. Composting is a suitable option for manure management, which implies the reduction of volume of the animal wastes and the elimination or reduction of pathogenic microorganisms, with economic, environmental and public health advantages. The composting procedure requires controlled conditions. Parameters such as bulk density, porosity, particle size, nutrient content, carbon/nitrogen ratio, temperature, pH, moisture and oxygen supply determine the optimal conditions of microbial development and organic matter degradation [12].
The elimination of antibiotic residues seems to be effective during this process [12]. For example, chlortetracycline and sulfadiazine residues were completely removed from antibiotic-spiked swine manure within 21 and 3 days, respectively, while 17–31% of spiked ciprofloxacin remained in the composting mass. Similar results were observed by Esperón et al. in antibiotic-spiked poultry manure, with a 90% decrease in the concentration of ciprofloxacin and doxycycline after composting for 3 weeks [141]. In addition, β-lactams have a very sensitive ring structure which is easily cleaved by the phosphate, ammonia and hydroxyl ions present in the composting environment, resulting in a degradation product with no antimicrobial activity [142]. Unfortunately, the elimination of ARGs may not be as straightforward.
The abundance of ARGs in livestock manure during composting is highly variable, essentially due to three main factors: the reproduction or death of intestinal microorganisms carrying ARGs in animal manure; the content of antibiotic residues and heavy metals that may exert selective pressure on ARG-carrying bacteria; and, finally, the possibility of the horizontal spread of ARGs [143]. Several studies have investigated changes in ARG and MGE content during composting, but the conclusions are inconsistent. For instance, the abundance of tetC, tetG, tetW, tetX, sul2, drfA1, drfA7, ermB, ermF, ermQ, ermX and intI1 genes after chicken manure composting with the addition of bamboo charcoal for 26 days was found to decrease significantly, while sul1 increased [144]; the composting of poultry manure for 10 weeks also led to a significant reduction of tetA, tetB tetK, tetM, tetQ, tetS, tetW, ermB, qnrS and blaTEM genes and an increased abundance of sul1, sul2, tetY and aadA genes [141]. The absolute abundances of intI1 and intI2, genes coding for integron integrases, and different erm, sul and tet ARGs were reduced by up to 45% after the composting of pig manure with cotton stalks, with good aeration, for 40 days [143]. Thermophilic composting was shown to be an effective method for the elimination of the mcr-1 gene in livestock manure, which was completely undetectable after 22 days of composting at a high temperature (44–65 °C) [98]. Furthermore, blaTEM, sul3 and ermB gene abundance decreased significantly during the composting of chicken manure in comparison to simple storage for 6 weeks [145]. In contrast, the abundance of tetG, ermF, and tetA did not change during composting, while there was an increase in tetW and tetO [146]. Different MGEs were also detected in most of the bovine, chicken and pig manure composts [24].
Obviously, composting conditions play a key role in the success of ARG elimination in manure. Temperature is pointed out as a critical factor for ARG reduction during composting because high temperatures may kill most of the bacterial species [47]. However, if thermophilic microorganisms are the hosts of ARGs, their abundance may increase during the thermophilic stage of composting [143].
The inconsistent results obtained by these studies call for the improvement of this process by adjusting the composting parameters and by adding adsorbents/surfactants or decomposition agents. The addition of natural zeolite during composting may be able to reduce some ARGs and accelerate the removal of pathogenic bacteria in compost, minimizing environmental risks [147,148]. This effect could be due to the porous structure and ability to reduce selective pressure and ARG co-selection by heavy metals, which implies decreased rates of microbiological contact and then HGT through the reduction of conjugation [147]. Similar effects are observed with the addition of biochar during chicken and swine manure composting due to its ability to increase the temperature, thereby prolonging the thermophilic phase. Moreover, biochar decreased copper and zinc levels with a consequent reduction of co-selection pressure by heavy metals [144,149]. However, the efficiency and the extension of the removal of ARGs is dependent on the type of biochar and manure [149]. Superabsorbent polymers have also been shown to be useful in the reduction of ARGs and MGEs in swine manure compost [150]. The application of an iron-based material and phosphate-dissolving inoculant during composting appears to be a promising method for the efficient removal of ARGs. On the other hand, the addition of red mud hindered the removal of tetracycline resistance genes and affected the shaping of bacterial communities during composting [151].
Anaerobic digestion is another widely used technique for manure treatment, associated with the reduction of organic matter pollution, microbial pathogens and veterinary antibiotic residues and the production of methane-rich biogas, a renewable energy source [152]. However, there are no consistent results and no key conclusions concerning the fate of ARGs during the anaerobic digestion process [153]. Some studies have pointed to anaerobic digestion as an alternative to reduce ARG content in different animal manures, although their complete removal has rarely been achieved [152,154,155,156,157]. The efficient removal of ARGs seems to be temperature dependent, since in thermophilic conditions (55 °C) ARG removal rates are higher than in mesophilic conditions (35 °C) [152]. For instance, tetM, tetQ, gyrA and sul1 levels were reduced with high efficiency during the anaerobic digestion of dairy manure with a high temperature treatment, while the increase in Bacteroidetes was responsible for the increases in tetC, tetM, tetQ, tetX, and sul1 under moderate and mesophilic temperatures. Moreover, the inhibition of HGT at high temperatures may also contribute to the decreased content of ARGs in the thermophilic system [155]. In another study, the relative abundances of intI1, sul1, sul2, tetA, tetO and tetX were evaluated during thermophilic and mesophilic swine manure anaerobic digestion conditions. All these ARGs decreased in abundance at the thermophilic temperature, while only intI1, sul1 and tetO decreased and sul2, tetA and tetX increased at the mesophilic temperature [157].
Furthermore, it is generally considered that anaerobic digestion could reduce the content of the ermB gene in swine manure, but in sewage sludge, the ermB gene generally increased after anaerobic digestion [153]. The substrate matrix types may also influence the efficiency of the ARG removal process due to the different physiochemical proprieties of different animal manures [153,158]. On the other hand, the microbial community needed for the degradation of different animal manures may be distinct and its evolution is, at the same time, conditioned by the presence of ARGs and the selective pressure of antibiotic residues [153].
In general, reduction techniques appear to be a sustainable and environmentally friendly process to reduce the risk of antimicrobial resistance dissemination and its impact on environmental and public health. Yet, conjugation was shown to occur even after 6 weeks of chicken manure composting [145], suggesting that reduction techniques are still not completely able to abolish the horizontal transfer of ARGs, although they can reduce it [159]. Therefore, more studies on composting parameters are needed to improve its efficiency.
5. Impact on Human, Animal and Environmental Health/One Health Perspective
Antimicrobial resistance is recognized as one of the “One Health” challenges that requires close observation and implementation strategies along the potential human–animal–environment bacterial spread chain [139,160].
The introduction of manure containing antibiotics and ARGs into the environment has a significant effect on the spread of resistance in the human community because human microbiota and pathogens may acquire MGEs carrying antibiotic resistance determinants by HGT events, even between distantly related species [30,111]. The environment acts as a bridge to different compartments; animal to manure to soil to water to sediment and, at the same time, it acts as a reservoir of MGEs that may interact and spread to other compartments or to human and animal hosts (Figure 1) [161].
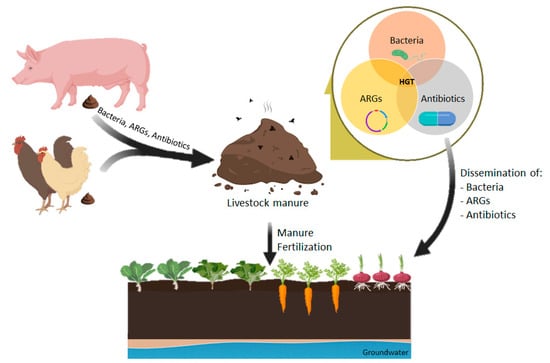
Figure 1.
Dissemination of antimicrobial resistance associated with manure application in agricultural soils.
Resistance determinants can be transferred from animal to animal or animal to human either directly via contact, or indirectly through the food chain, water, sludge-fertilized soils and manure. The human transmission of antibiotic-resistant bacteria and ARGs from agricultural sources is mostly foodborne [139]. Antibiotic-resistant bacteria are often found in vegetables and fruits which are cultivated in animal-manured soil [162,163]. Since these fresh products contaminated by manure or irrigation water are often consumed raw, their intake may result in the ingestion of resistant bacteria which may colonize the human gut or pass through the intestine and pose a threat to public health [163,164]. The microbial communities of fresh produce reflect the soil where it grows. Two studies reported a higher frequency of β-lactam-resistant bacteria in manure-amended soil and raise the possibility that these β-lactamase encoded genes could spread from manure-amended soil bacteria to human pathogens [14,79]. Food-producing animals, and their related food products in all stages of processing, also contain abundant quantities of antibiotics, resistant bacteria and their resistance genes that spread into humans through food chain, increasing the possibility of resistant determinant exchange between human, environmental and animal microbiomes [165]. Furthermore, in countries with poor water treatment and sewage conditions, there is an increased risk of manure-borne resistant bacteria and resistant gene transmission from animals [139].
The widespread use of antibiotics in human clinical settings and the agricultural and livestock industries has exerted a selective pressure for the selection and survival of antibiotic-resistant bacteria [166]. The survival and spread of antibiotic-resistant bacteria and ARGs is a threat and a major concern for human, animal and environmental health due to the disastrous impact on clinical infection outcomes [139,166]. Antimicrobial resistance is one of the major causes responsible for the reduction of antimicrobial therapy effectiveness and the increasing incidence, severity and costs of infections in clinical settings [139].
Moreover, anthropogenic activities have a great impact on the environmental resistome because large amounts of antimicrobials are produced annually and, through diverse pathways, such as manure fertilization, are spread into the environment (Figure 1) [139]. Thus, intensive animal production is often considered the major cause responsible for the increased environmental burden of antibiotics [167,168]. Antibiotics in the soil can also cause adverse effects in soil plants [35,167]. It is crucial to reduce the amounts of antimicrobials that end up in soil, either due to the prudent use of antimicrobials or by composting techniques.
Overall, manure must be seen has a hotspot for the dissemination of antibiotics, metals and antimicrobial resistance genes, with impacts on public and environmental health, and measures must be taken to reduce their impact on society.
Preventive actions to decrease the risk of antimicrobial resistance should keep being implemented [160]. In the livestock sector, the maintenance of good animal health should be encouraged by implementing practices such as decreased animal density in feedlots and improved nutritional programs [35].
Finally, the development and implementation of Good Agricultural Practices [111] may help to limit or even reduce contamination and the dissemination of antibiotics and resistance determinants through the human–animal–environment chain.
6. Conclusions
Manure application has been shown to increase the diversity and abundance of ARGs and MGEs in manure-amended soils. Although ARG persistence in soil may vary from a few days to some months, they can be taken up by bacteria through HGT mechanisms, including soil microbiota and potential animal and human pathogenic bacteria, which may reach humans via the food chain. Different organic fertilizers impact ARG diversity and HGT events differently and this point should be taken into account when choosing a fertilizer. Different composting techniques have been shown to lead to varied reduction levels of specific ARGs, highlighting the importance of knowing local antimicrobial resistance patterns before choosing which composting technique to use. Furthermore, more studies on composting parameters are needed to improve its efficiency.
Author Contributions
Writing—original draft preparation, T.L., S.D. and G.J.D.S.; writing—review and editing, S.D. and G.J.D.S. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Acknowledgments
The Faculty of Pharmacy of the University of Coimbra and Center for Neurosciences and Cell Biology through “Fundação para a Ciência e a Tecnologia, projecto Estratégico: UID/NEU/04539/2013”. Tiago Lima acknowledges Fundação para a Ciência e a Tecnologia (FCT) for his PhD grant (SFRH/BD/132555/2017).
Conflicts of Interest
The authors declare no conflict of interest.
References
- Baker, S.J.; Payne, D.J.; Rappuoli, R.; De Gregorio, E. Technologies to address antimicrobial resistance. Proc. Natl. Acad. Sci. USA 2018, 115, 12887–12895. [Google Scholar] [CrossRef]
- Nabarro, D.; Wannous, C. The potential contribution of Iivestock to food and nutrition security: The application of the One Health approach in livestock policy and practice. Rev. Sci. Tech. 2014, 33, 475–485. [Google Scholar] [CrossRef]
- Thornton, P.K. Livestock production: Recent trends, future prospects. Philos. Trans. R. Soc. Lond. Ser. B Biol. Sci. 2010, 365, 2853–2867. [Google Scholar] [CrossRef]
- Zhou, L.J.; Ying, G.G.; Liu, S.; Zhang, R.Q.; Lai, H.J.; Chen, Z.F.; Pan, C.G. Excretion masses and environmental occurrence of antibiotics in typical swine and dairy cattle farms in China. Sci. Total Environ. 2013, 444, 183–195. [Google Scholar] [CrossRef]
- Sarmah, A.K.; Meyer, M.T.; Boxall, A.B. A global perspective on the use, sales, exposure pathways, occurrence, fate and effects of veterinary antibiotics (VAs) in the environment. Chemosphere 2006, 65, 725–759. [Google Scholar] [CrossRef]
- Liu, C.; Chen, Y.; Li, X.; Zhang, Y.; Ye, J.; Huang, H.; Zhu, C. Temporal effects of repeated application of biogas slurry on soil antibiotic resistance genes and their potential bacterial hosts. Environ. Pollut. 2020, 258, 113652. [Google Scholar] [CrossRef]
- Lekagul, A.; Tangcharoensathien, V.; Yeung, S. Patterns of antibiotic use in global pig production: A systematic review. Vet. Anim. Sci. 2019, 7, 100058. [Google Scholar] [CrossRef]
- Castanon, J.I. History of the use of antibiotic as growth promoters in European poultry feeds. Poult. Sci. 2007, 86, 2466–2471. [Google Scholar] [CrossRef]
- Friedman, N.D.; Temkin, E.; Carmeli, Y. The negative impact of antibiotic resistance. Clin. Microbiol. Infect. 2016, 22, 416–422. [Google Scholar] [CrossRef]
- McEwen, S.A.; Collignon, P.J. Antimicrobial Resistance: A One Health Perspective. Microbiol. Spectr. 2018, 6. [Google Scholar] [CrossRef]
- Shober, A.L.; Maguire, R.O. Manure Management. In Reference Module in Earth Systems and Environmental Sciences; Elsevier: Toronto, ON, Canada, 2014. [Google Scholar] [CrossRef]
- Bernal, M.P.; Alburquerque, J.A.; Moral, R. Composting of animal manures and chemical criteria for compost maturity assessment. A review. Bioresour. Technol. 2009, 100, 5444–5453. [Google Scholar] [CrossRef]
- Zhao, L.; Dong, Y.H.; Wang, H. Residues of veterinary antibiotics in manures from feedlot livestock in eight provinces of China. Sci. Total Environ. 2010, 408, 1069–1075. [Google Scholar] [CrossRef]
- Udikovic-Kolic, N.; Wichmann, F.; Broderick, N.A.; Handelsman, J. Bloom of resident antibiotic-resistant bacteria in soil following manure fertilization. Proc. Natl. Acad. Sci. USA 2014, 111, 15202–15207. [Google Scholar] [CrossRef]
- Lee, J.; Shin, S.G.; Jang, H.M.; Kim, Y.B.; Lee, J.; Kim, Y.M. Characterization of antibiotic resistance genes in representative organic solid wastes: Food waste-recycling wastewater, manure, and sewage sludge. Sci. Total Environ. 2017, 579, 1692–1698. [Google Scholar] [CrossRef]
- Cheng, J.H.; Tang, X.Y.; Cui, J.F. Effect of long-term manure slurry application on the occurrence of antibiotic resistance genes in arable purple soil (entisol). Sci. Total Environ. 2019, 647, 853–861. [Google Scholar] [CrossRef]
- Riber, L.; Poulsen, P.H.; Al-Soud, W.A.; Skov Hansen, L.B.; Bergmark, L.; Brejnrod, A.; Norman, A.; Hansen, L.H.; Magid, J.; Sorensen, S.J. Exploring the immediate and long-term impact on bacterial communities in soil amended with animal and urban organic waste fertilizers using pyrosequencing and screening for horizontal transfer of antibiotic resistance. FEMS Microbiol. Ecol. 2014, 90, 206–224. [Google Scholar] [CrossRef]
- van Overbeek, L.S.; van Doorn, J.; Wichers, J.H.; van Amerongen, A.; van Roermund, H.J.; Willemsen, P.T. The arable ecosystem as battleground for emergence of new human pathogens. Front. Microbiol. 2014, 5, 104. [Google Scholar] [CrossRef]
- Huang, P.; Li, Y.; Sumner, M. Handbook of Soil Sciences: Resource Management and Environmental Impacts, 2nd ed.; Taylor & Francis: Boca Raton, FL, USA, 2011. [Google Scholar]
- Sheppard, S.C.; Sanipelli, B. Trace elements in feed, manure, and manured soils. J. Environ. Qual. 2012, 41, 1846–1856. [Google Scholar] [CrossRef]
- Guo, T.; Lou, C.; Zhai, W.; Tang, X.; Hashmi, M.Z.; Murtaza, R.; Li, Y.; Liu, X.; Xu, J. Increased occurrence of heavy metals, antibiotics and resistance genes in surface soil after long-term application of manure. Sci. Total Environ. 2018, 635, 995–1003. [Google Scholar] [CrossRef]
- Ji, X.; Shen, Q.; Liu, F.; Ma, J.; Xu, G.; Wang, Y.; Wu, M. Antibiotic resistance gene abundances associated with antibiotics and heavy metals in animal manures and agricultural soils adjacent to feedlots in Shanghai; China. J. Hazard. Mater. 2012, 235–236, 178–185. [Google Scholar] [CrossRef]
- Yuan, W.; Tian, T.; Yang, Q.; Riaz, L. Transfer potentials of antibiotic resistance genes in Escherichia spp. strains from different sources. Chemosphere 2020, 246, 125736. [Google Scholar] [CrossRef]
- Qian, X.; Gu, J.; Sun, W.; Wang, X.J.; Su, J.Q.; Stedfeld, R. Diversity, abundance, and persistence of antibiotic resistance genes in various types of animal manure following industrial composting. J. Hazard. Mater. 2018, 344, 716–722. [Google Scholar] [CrossRef]
- Zhu, Y.G.; Johnson, T.A.; Su, J.Q.; Qiao, M.; Guo, G.X.; Stedtfeld, R.D.; Hashsham, S.A.; Tiedje, J.M. Diverse and abundant antibiotic resistance genes in Chinese swine farms. Proc. Natl. Acad. Sci. USA 2013, 110, 3435–3440. [Google Scholar] [CrossRef]
- Baker-Austin, C.; Wright, M.S.; Stepanauskas, R.; McArthur, J.V. Co-selection of antibiotic and metal resistance. Trends Microbiol. 2006, 14, 176–182. [Google Scholar] [CrossRef]
- Berg, J.; Thorsen, M.K.; Holm, P.E.; Jensen, J.; Nybroe, O.; Brandt, K.K. Cu exposure under field conditions coselects for antibiotic resistance as determined by a novel cultivation-independent bacterial community tolerance assay. Environ. Sci. Technol. 2010, 44, 8724–8728. [Google Scholar] [CrossRef]
- Wang, Y.; Gong, J.; Li, J.; Xin, Y.; Hao, Z.; Chen, C.; Li, H.; Wang, B.; Ding, M.; Li, W.; et al. Insights into bacterial diversity in compost: Core microbiome and prevalence of potential pathogenic bacteria. Sci. Total Environ. 2020, 718, 137304. [Google Scholar] [CrossRef]
- Manyi-Loh, C.E.; Mamphweli, S.N.; Meyer, E.L.; Makaka, G.; Simon, M.; Okoh, A.I. An Overview of the Control of Bacterial Pathogens in Cattle Manure. Int. J. Environ. Res. Public Health 2016, 13, 843. [Google Scholar] [CrossRef]
- Heuer, H.; Schmitt, H.; Smalla, K. Antibiotic resistance gene spread due to manure application on agricultural fields. Curr. Opin. Microbiol. 2011, 14, 236–243. [Google Scholar] [CrossRef]
- Chen, M.; Qiu, T.; Sun, Y.; Song, Y.; Wang, X.; Gao, M. Diversity of tetracycline- and erythromycin-resistant bacteria in aerosols and manures from four types of animal farms in China. Environ. Sci. Pollut. Res. Int. 2019, 26, 24213–24222. [Google Scholar] [CrossRef]
- Pachepsky, Y.A.; Sadeghi, A.M.; Bradford, S.A.; Shelton, D.R.; Guber, A.K.; Dao, T. Transport and fate of manure-borne pathogens: Modeling perspective. Agric. Water Manag. 2006, 86, 81–92. [Google Scholar] [CrossRef]
- Switt, A.I.; den Bakker, H.C.; Vongkamjan, K.; Hoelzer, K.; Warnick, L.D.; Cummings, K.J.; Wiedmann, M. Salmonella bacteriophage diversity reflects host diversity on dairy farms. Food Microbiol. 2013, 36, 275–285. [Google Scholar] [CrossRef]
- Pornsukarom, S.; Thakur, S. Assessing the Impact of Manure Application in Commercial Swine Farms on the Transmission of Antimicrobial Resistant Salmonella in the Environment. PLoS ONE 2016, 11, e0164621. [Google Scholar] [CrossRef]
- He, Y.; Yuan, Q.; Mathieu, J.; Stadler, L.; Senehi, N.; Sun, R.; Alvarez, P.J.J. Antibiotic resistance genes from livestock waste: Occurrence, dissemination, and treatment. NPJ Clean Water 2020, 3, 4. [Google Scholar] [CrossRef]
- Lin, H.; Chapman, S.J.; Freitag, T.E.; Kyle, C.; Ma, J.; Yang, Y.; Zhang, Z. Fate of tetracycline and sulfonamide resistance genes in a grassland soil amended with different organic fertilizers. Ecotoxicol. Environ. Saf. 2019, 170, 39–46. [Google Scholar] [CrossRef]
- Wang, F.; Han, W.; Chen, S.; Dong, W.; Qiao, M.; Hu, C.; Liu, B. Fifteen-Year Application of Manure and Chemical Fertilizers Differently Impacts Soil ARGs and Microbial Community Structure. Front. Microbiol. 2020, 11, 62. [Google Scholar] [CrossRef]
- Schwaiger, K.; Harms, K.; Holzel, C.; Meyer, K.; Karl, M.; Bauer, J. Tetracycline in liquid manure selects for co-occurrence of the resistance genes tet(M) and tet(L) in Enterococcus faecalis. Vet. Microbiol. 2009, 139, 386–392. [Google Scholar] [CrossRef]
- Binh, C.; Heuer, H.; Gomes, N.; Kaupenjohann, M.; Smalla, K. Similar bacterial community structure and high abundance of sulfonamide resistance genes in field-scale manures. In Manure: Management, Uses and Environmental Impacts; Dellaguardia, C.S., Ed.; Nova Science Publishers: Hauppauge, NY, USA, 2010; pp. 141–166. [Google Scholar]
- Binh, C.T.; Heuer, H.; Kaupenjohann, M.; Smalla, K. Piggery manure used for soil fertilization is a reservoir for transferable antibiotic resistance plasmids. FEMS Microbiol. Ecol. 2008, 66, 25–37. [Google Scholar] [CrossRef]
- Norpoth, A.; Langhammer, J.P.; Winkelmann, J.; Petersen, B.; Buning-Pfaue, H. [Drug residues from slurry and their effect on the development of resistance of E. coli isolates from swine]. Zent. Hyg. Umweltmed. 1989, 189, 151–163. [Google Scholar]
- Kim, K.-R.; Owens, G.; Kwon, S.-I.; So, K.-H.; Lee, D.-B.; Ok, Y.S. Occurrence and Environmental Fate of Veterinary Antibiotics in the Terrestrial Environment. Water Air Soil Pollut. 2011, 214, 163–174. [Google Scholar] [CrossRef]
- Cycon, M.; Mrozik, A.; Piotrowska-Seget, Z. Antibiotics in the Soil Environment-Degradation and Their Impact on Microbial Activity and Diversity. Front. Microbiol. 2019, 10, 338. [Google Scholar] [CrossRef]
- Manyi-Loh, C.; Mamphweli, S.; Meyer, E.; Okoh, A. Antibiotic Use in Agriculture and Its Consequential Resistance in Environmental Sources: Potential Public Health Implications. Molecules 2018, 23, 765. [Google Scholar] [CrossRef]
- Cheng, W.; Chen, H.; Su, C.; Yan, S. Abundance and persistence of antibiotic resistance genes in livestock farms: A comprehensive investigation in eastern China. Environ. Int. 2013, 61, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Marti, R.; Scott, A.; Tien, Y.C.; Murray, R.; Sabourin, L.; Zhang, Y.; Topp, E. Impact of manure fertilization on the abundance of antibiotic-resistant bacteria and frequency of detection of antibiotic resistance genes in soil and on vegetables at harvest. Appl. Environ. Microbiol. 2013, 79, 5701–5709. [Google Scholar] [CrossRef] [PubMed]
- Qian, X.; Sun, W.; Gu, J.; Wang, X.J.; Zhang, Y.J.; Duan, M.L.; Li, H.C.; Zhang, R.R. Reducing antibiotic resistance genes, integrons, and pathogens in dairy manure by continuous thermophilic composting. Bioresour. Technol. 2016, 220, 425–432. [Google Scholar] [CrossRef] [PubMed]
- Fahrenfeld, N.; Knowlton, K.; Krometis, L.A.; Hession, W.C.; Xia, K.; Lipscomb, E.; Libuit, K.; Green, B.L.; Pruden, A. Effect of Manure Application on Abundance of Antibiotic Resistance Genes and Their Attenuation Rates in Soil: Field-Scale Mass Balance Approach. Environ. Sci. Technol. 2014, 48, 2643–2650. [Google Scholar] [CrossRef] [PubMed]
- Van Boeckel, T.P.; Brower, C.; Gilbert, M.; Grenfell, B.T.; Levin, S.A.; Robinson, T.P.; Teillant, A.; Laxminarayan, R. Global trends in antimicrobial use in food animals. Proc. Natl. Acad. Sci. USA 2015, 112, 5649–5654. [Google Scholar] [CrossRef]
- Chen, Z.; Zhang, W.; Yang, L.; Stedtfeld, R.D.; Peng, A.; Gu, C.; Boyd, S.A.; Li, H. Antibiotic resistance genes and bacterial communities in cornfield and pasture soils receiving swine and dairy manures. Environ. Pollut. 2019, 248, 947–957. [Google Scholar] [CrossRef]
- Lanz, R.; Kuhnert, P.; Boerlin, P. Antimicrobial resistance and resistance gene determinants in clinical Escherichia coli from different animal species in Switzerland. Vet. Microbiol. 2003, 91, 73–84. [Google Scholar] [CrossRef]
- Fang, H.; Wang, H.; Cai, L.; Yu, Y. Prevalence of antibiotic resistance genes and bacterial pathogens in long-term manured greenhouse soils as revealed by metagenomic survey. Environ. Sci. Technol. 2015, 49, 1095–1104. [Google Scholar] [CrossRef]
- Whittle, G.; Whitehead, T.R.; Hamburger, N.; Shoemaker, N.B.; Cotta, M.A.; Salyers, A.A. Identification of a new ribosomal protection type of tetracycline resistance gene, tet(36), from swine manure pits. Appl. Environ. Microbiol. 2003, 69, 4151–4158. [Google Scholar] [CrossRef]
- Leclercq, S.O.; Wang, C.; Zhu, Y.; Wu, H.; Du, X.; Liu, Z.; Feng, J. Diversity of the Tetracycline Mobilome within a Chinese Pig Manure Sample. Appl. Environ. Microbiol. 2016, 82, 6454–6462. [Google Scholar] [CrossRef]
- Xiong, W.; Wang, M.; Dai, J.; Sun, Y.; Zeng, Z. Application of manure containing tetracyclines slowed down the dissipation of tet resistance genes and caused changes in the composition of soil bacteria. Ecotoxicol. Environ. Saf. 2018, 147, 455–460. [Google Scholar] [CrossRef]
- Ricker, N.; Trachsel, J.; Colgan, P.; Jones, J.; Choi, J.; Lee, J.; Coetzee, J.F.; Howe, A.; Brockmeier, S.L.; Loving, C.L.; et al. Toward Antibiotic Stewardship: Route of Antibiotic Administration Impacts the Microbiota and Resistance Gene Diversity in Swine Feces. Front. Vet. Sci. 2020, 7, 255. [Google Scholar] [CrossRef]
- Figueroa, R.A.; Leonard, A.; MacKay, A.A. Modeling tetracycline antibiotic sorption to clays. Environ. Sci. Technol. 2004, 38, 476–483. [Google Scholar] [CrossRef]
- Speer, B.S.; Shoemaker, N.B.; Salyers, A.A. Bacterial resistance to tetracycline: Mechanisms, transfer, and clinical significance. Clin. Microbiol. Rev. 1992, 5, 387–399. [Google Scholar] [CrossRef]
- Amador, P.; Fernandes, R.; Prudencio, C.; Duarte, I. Prevalence of Antibiotic Resistance Genes in Multidrug-Resistant Enterobacteriaceae on Portuguese Livestock Manure. Antibiotics 2019, 8, 23. [Google Scholar] [CrossRef]
- Kim, S.Y.; Kuppusamy, S.; Kim, J.H.; Yoon, Y.E.; Kim, K.R.; Lee, Y.B. Occurrence and diversity of tetracycline resistance genes in the agricultural soils of South Korea. Environ. Sci. Pollut. Res. Int. 2016, 23, 22190–22196. [Google Scholar] [CrossRef]
- Rahube, T.O.; Yost, C.K. Characterization of a mobile and multiple resistance plasmid isolated from swine manure and its detection in soil after manure application. J. Appl. Microbiol. 2012, 112, 1123–1133. [Google Scholar] [CrossRef]
- Heuer, H.; Kopmann, C.; Binh, C.T.; Top, E.M.; Smalla, K. Spreading antibiotic resistance through spread manure: Characteristics of a novel plasmid type with low %G+C content. Environ. Microbiol. 2009, 11, 937–949. [Google Scholar] [CrossRef]
- Kyselkova, M.; Jirout, J.; Vrchotova, N.; Schmitt, H.; Elhottova, D. Spread of tetracycline resistance genes at a conventional dairy farm. Front. Microbiol. 2015, 6, 536. [Google Scholar] [CrossRef]
- Byrne-Bailey, K.G.; Gaze, W.H.; Kay, P.; Boxall, A.B.; Hawkey, P.M.; Wellington, E.M. Prevalence of sulfonamide resistance genes in bacterial isolates from manured agricultural soils and pig slurry in the United Kingdom. Antimicrob. Agents Chemother. 2009, 53, 696–702. [Google Scholar] [CrossRef]
- Perreten, V.; Boerlin, P. A new sulfonamide resistance gene (sul3) in Escherichia coli is widespread in the pig population of Switzerland. Antimicrob. Agents Chemother. 2003, 47, 1169–1172. [Google Scholar] [CrossRef]
- Antunes, P.; Machado, J.; Sousa, J.C.; Peixe, L. Dissemination of sulfonamide resistance genes (sul1, sul2, and sul3) in Portuguese Salmonella enterica strains and relation with integrons. Antimicrob. Agents Chemother. 2005, 49, 836–839. [Google Scholar] [CrossRef]
- He, L.Y.; Liu, Y.S.; Su, H.C.; Zhao, J.L.; Liu, S.S.; Chen, J.; Liu, W.R.; Ying, G.G. Dissemination of antibiotic resistance genes in representative broiler feedlots environments: Identification of indicator ARGs and correlations with environmental variables. Environ. Sci. Technol. 2014, 48, 13120–13129. [Google Scholar] [CrossRef]
- Heuer, H.; Solehati, Q.; Zimmerling, U.; Kleineidam, K.; Schloter, M.; Muller, T.; Focks, A.; Thiele-Bruhn, S.; Smalla, K. Accumulation of sulfonamide resistance genes in arable soils due to repeated application of manure containing sulfadiazine. Appl. Environ. Microbiol. 2011, 77, 2527–2530. [Google Scholar] [CrossRef]
- Wang, N.; Yang, X.; Jiao, S.; Zhang, J.; Ye, B.; Gao, S. Sulfonamide-resistant bacteria and their resistance genes in soils fertilized with manures from Jiangsu Province, Southeastern China. PLoS ONE 2014, 9, e112626. [Google Scholar] [CrossRef]
- Wegst-Uhrich, S.R.; Navarro, D.A.; Zimmerman, L.; Aga, D.S. Assessing antibiotic sorption in soil: A literature review and new case studies on sulfonamides and macrolides. Chem. Cent. J. 2014, 8, 5. [Google Scholar] [CrossRef]
- Zhang, H.; Zhou, Y.; Huang, Y.; Wu, L.; Liu, X.; Luo, Y. Residues and risks of veterinary antibiotics in protected vegetable soils following application of different manures. Chemosphere 2016, 152, 229–237. [Google Scholar] [CrossRef]
- Amador, P.; Duarte, I.M.; Roberto da Costa, R.P.; Fernandes, R.; Prudêncio, C. Characterization of Antibiotic Resistance in Enterobacteriaceae From Agricultural Manure and Soil in Portugal. Soil Sci. 2018, 182, 292–301. [Google Scholar] [CrossRef]
- McKinney, C.W.; Loftin, K.A.; Meyer, M.T.; Davis, J.G.; Pruden, A. tet and sul antibiotic resistance genes in livestock lagoons of various operation type, configuration, and antibiotic occurrence. Environ. Sci. Technol. 2010, 44, 6102–6109. [Google Scholar] [CrossRef]
- Martinez-Carballo, E.; Gonzalez-Barreiro, C.; Scharf, S.; Gans, O. Environmental monitoring study of selected veterinary antibiotics in animal manure and soils in Austria. Environ. Pollut. 2007, 148, 570–579. [Google Scholar] [CrossRef] [PubMed]
- Wang, N.; Guo, X.; Yan, Z.; Wang, W.; Chen, B.; Ge, F.; Ye, B. A Comprehensive Analysis on Spread and Distribution Characteristic of Antibiotic Resistance Genes in Livestock Farms of Southeastern China. PLoS ONE 2016, 11, e0156889. [Google Scholar] [CrossRef] [PubMed]
- Dungan, R.S.; Strausbaugh, C.A.; Leytem, A.B. Survey of selected antibiotic resistance genes in agricultural and non-agricultural soils in south-central Idaho. FEMS Microbiol. Ecol. 2019, 95. [Google Scholar] [CrossRef] [PubMed]
- Binh, C.T.; Heuer, H.; Gomes, N.C.; Kotzerke, A.; Fulle, M.; Wilke, B.M.; Schloter, M.; Smalla, K. Short-term effects of amoxicillin on bacterial communities in manured soil. FEMS Microbiol. Ecol. 2007, 62, 290–302. [Google Scholar] [CrossRef]
- Oliver, J.P.; Gooch, C.A.; Lansing, S.; Schueler, J.; Hurst, J.J.; Sassoubre, L.; Crossette, E.M.; Aga, D.S. Invited review: Fate of antibiotic residues, antibiotic-resistant bacteria, and antibiotic resistance genes in US dairy manure management systems. J. Dairy Sci. 2020, 103, 1051–1071. [Google Scholar] [CrossRef] [PubMed]
- Wichmann, F.; Udikovic-Kolic, N.; Andrew, S.; Handelsman, J. Diverse antibiotic resistance genes in dairy cow manure. mBio 2014, 5, e01017. [Google Scholar] [CrossRef]
- Moodley, A.; Guardabassi, L. Transmission of IncN plasmids carrying blaCTX-M-1 between commensal Escherichia coli in pigs and farm workers. Antimicrob. Agents Chemother. 2009, 53, 1709–1711. [Google Scholar] [CrossRef]
- Lee, S.; Mir, R.A.; Park, S.H.; Kim, D.; Kim, H.Y.; Boughton, R.K.; Morris, J.G., Jr.; Jeong, K.C. Prevalence of extended-spectrum beta-lactamases in the local farm environment and livestock: Challenges to mitigate antimicrobial resistance. Crit. Rev. Microbiol. 2020, 46, 1–14. [Google Scholar] [CrossRef]
- Mu, Q.; Li, J.; Sun, Y.; Mao, D.; Wang, Q.; Luo, Y. Occurrence of sulfonamide-, tetracycline-, plasmid-mediated quinolone- and macrolide-resistance genes in livestock feedlots in Northern China. Environ. Sci. Pollut. Res. Int. 2015, 22, 6932–6940. [Google Scholar] [CrossRef]
- Luby, E.M.; Moorman, T.B.; Soupir, M.L. Fate and transport of tylosin-resistant bacteria and macrolide resistance genes in artificially drained agricultural fields receiving swine manure. Sci. Total Environ. 2016, 550, 1126–1133. [Google Scholar] [CrossRef]
- Garder, J.L.; Moorman, T.B.; Soupir, M.L. Transport and persistence of tylosin-resistant enterococci, genes, and tylosin in soil and drainage water from fields receiving Swine manure. J. Environ. Qual. 2014, 43, 1484–1493. [Google Scholar] [CrossRef]
- Chen, J.; Yu, Z.; Michel, F.C., Jr.; Wittum, T.; Morrison, M. Development and application of real-time PCR assays for quantification of erm genes conferring resistance to macrolides-lincosamides-streptogramin B in livestock manure and manure management systems. Appl. Environ. Microbiol. 2007, 73, 4407–4416. [Google Scholar] [CrossRef]
- Hoang, T.T.T.; Soupir, M.L.; Liu, P.; Bhandari, A. Occurrence of Tylosin-Resistant Enterococci in Swine Manure and Tile Drainage Systems under No-Till Management. Water Air Soil Pollut. 2013, 224, 1754. [Google Scholar] [CrossRef][Green Version]
- Van den Meersche, T.; Rasschaert, G.; Haesebrouck, F.; Van Coillie, E.; Herman, L.; Van Weyenberg, S.; Daeseleire, E.; Heyndrickx, M. Presence and fate of antibiotic residues, antibiotic resistance genes and zoonotic bacteria during biological swine manure treatment. Ecotoxicol. Environ. Saf. 2019, 175, 29–38. [Google Scholar] [CrossRef]
- Peng, S.; Feng, Y.; Wang, Y.; Guo, X.; Chu, H.; Lin, X. Prevalence of antibiotic resistance genes in soils after continually applied with different manure for 30 years. J. Hazard. Mater. 2017, 340, 16–25. [Google Scholar] [CrossRef]
- Macedo, G.; Hernandez-Leal, L.; van der Maas, P.; Heederik, D.; Mevius, D.; Schmitt, H. The impact of manure and soil texture on antimicrobial resistance gene levels in farmlands and adjacent ditches. Sci. Total Environ. 2020, 737, 139563. [Google Scholar] [CrossRef]
- Wang, L.; Wang, J.; Wang, J.; Zhu, L.; Yang, L.; Yang, R. Distribution characteristics of antibiotic resistant bacteria and genes in fresh and composted manures of livestock farms. Sci. Total Environ. 2019, 695, 133781. [Google Scholar] [CrossRef]
- Dungan, R.S.; McKinney, C.W.; Leytem, A.B. Tracking antibiotic resistance genes in soil irrigated with dairy wastewater. Sci. Total Environ. 2018, 635, 1477–1483. [Google Scholar] [CrossRef]
- Lima, T.; Domingues, S.; Da Silva, G.J. Plasmid-Mediated Colistin Resistance in Salmonella enterica: A Review. Microorganisms 2019, 7, 55. [Google Scholar] [CrossRef]
- Davis, C.A.; Janssen, E.M. Environmental fate processes of antimicrobial peptides daptomycin, bacitracins, and polymyxins. Environ. Int. 2020, 134, 105271. [Google Scholar] [CrossRef]
- Rhouma, M.; Beaudry, F.; Theriault, W.; Letellier, A. Colistin in Pig Production: Chemistry, Mechanism of Antibacterial Action, Microbial Resistance Emergence, and One Health Perspectives. Front. Microbiol. 2016, 7, 1789. [Google Scholar] [CrossRef]
- Wang, C.; Feng, Y.; Liu, L.; Wei, L.; Kang, M.; Zong, Z. Identification of novel mobile colistin resistance gene mcr-10. Emerg. Microbes Infect. 2020, 9, 508–516. [Google Scholar] [CrossRef]
- Xia, X.; Wang, Z.; Fu, Y.; Du, X.D.; Gao, B.; Zhou, Y.; He, J.; Wang, Y.; Shen, J.; Jiang, H.; et al. Association of colistin residues and manure treatment with the abundance of mcr-1 gene in swine feedlots. Environ. Int. 2019, 127, 361–370. [Google Scholar] [CrossRef]
- Brauer, A.; Telling, K.; Laht, M.; Kalmus, P.; Lutsar, I.; Remm, M.; Kisand, V.; Tenson, T. Plasmid with Colistin Resistance Gene mcr-1 in Extended-Spectrum-beta-Lactamase-Producing Escherichia coli Strains Isolated from Pig Slurry in Estonia. Antimicrob. Agents Chemother. 2016, 60, 6933–6936. [Google Scholar] [CrossRef]
- Gao, Y.; Lu, C.; Shen, D.; Liu, J.; Ma, Z.; Yang, B.; Ling, W.; Waigi, M.G. Elimination of the risks of colistin resistance gene (mcr-1) in livestock manure during composting. Environ. Int. 2019, 126, 61–68. [Google Scholar] [CrossRef]
- Zheng, B.; Huang, C.; Xu, H.; Guo, L.; Zhang, J.; Wang, X.; Jiang, X.; Yu, X.; Jin, L.; Li, X.; et al. Occurrence and Genomic Characterization of ESBL-Producing, MCR-1-Harboring Escherichia coli in Farming Soil. Front. Microbiol. 2017, 8, 2510. [Google Scholar] [CrossRef]
- Guenther, S.; Falgenhauer, L.; Semmler, T.; Imirzalioglu, C.; Chakraborty, T.; Roesler, U.; Roschanski, N. Environmental emission of multiresistant Escherichia coli carrying the colistin resistance gene mcr-1 from German swine farms. J. Antimicrob. Chemother. 2017, 72, 1289–1292. [Google Scholar] [CrossRef]
- Jacoby, G.A.; Strahilevitz, J.; Hooper, D.C. Plasmid-mediated quinolone resistance. Microbiol. Spectr. 2014, 2. [Google Scholar] [CrossRef]
- Villa, T.G.; Feijoo-Siota, L.; Sánchez-Pérez, A.; Rama, J.R.; Sieiro, C. Horizontal Gene Transfer in Bacteria, an Overview of the Mechanisms Involved. In Horizontal Gene Transfer: Breaking Borders Between Living Kingdoms; Villa, T.G., Viñas, M., Eds.; Springer International Publishing: Cham, Switzerland, 2019; pp. 3–76. [Google Scholar] [CrossRef]
- Guo, L.; Wei, D.; Zhang, X.; Wu, Y.; Li, Q.; Zhou, M.; Qu, J. Clinical features predicting mortality risk in patients with viral pneumonia: The MuLBSTA score. Front. Microbiol. 2019, 10, 2752. [Google Scholar] [CrossRef]
- Ochman, H.; Lerat, E.; Daubin, V. Examining bacterial species under the specter of gene transfer and exchange. Proc. Natl. Acad. Sci. USA 2005, 102 (Suppl. 1), 6595–6599. [Google Scholar] [CrossRef]
- Chee-Sanford, J.C.; Mackie, R.I.; Koike, S.; Krapac, I.G.; Lin, Y.F.; Yannarell, A.C.; Maxwell, S.; Aminov, R.I. Fate and transport of antibiotic residues and antibiotic resistance genes following land application of manure waste. J. Environ. Qual. 2009, 38, 1086–1108. [Google Scholar] [CrossRef] [PubMed]
- Dong, P.; Wang, H.; Fang, T.; Wang, Y.; Ye, Q. Assessment of extracellular antibiotic resistance genes (eARGs) in typical environmental samples and the transforming ability of eARG. Environ. Int. 2019, 125, 90–96. [Google Scholar] [CrossRef] [PubMed]
- Chen, Q.; An, X.; Li, H.; Su, J.; Ma, Y.; Zhu, Y.G. Long-term field application of sewage sludge increases the abundance of antibiotic resistance genes in soil. Environ. Int. 2016, 92–93, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Baker, M.; Hobman, J.L.; Dodd, C.E.; Ramsden, S.J.; Stekel, D.J. Mathematical modelling of antimicrobial resistance in agricultural waste highlights importance of gene transfer rate. FEMS Microbiol. Ecol. 2016, 92, fiw040. [Google Scholar] [CrossRef] [PubMed]
- Peng, H.; Gu, J.; Wang, X.; Wang, Q.; Sun, W.; Hu, T.; Guo, H.; Ma, J.; Bao, J. Insight into the fate of antibiotic resistance genes and bacterial community in co-composting green tea residues with swine manure. J. Environ. Manag. 2020, 266, 110581. [Google Scholar] [CrossRef] [PubMed]
- Knapp, C.W.; McCluskey, S.M.; Singh, B.K.; Campbell, C.D.; Hudson, G.; Graham, D.W. Antibiotic resistance gene abundances correlate with metal and geochemical conditions in archived Scottish soils. PLoS ONE 2011, 6, e27300. [Google Scholar] [CrossRef] [PubMed]
- Jechalke, S.; Heuer, H.; Siemens, J.; Amelung, W.; Smalla, K. Fate and effects of veterinary antibiotics in soil. Trends Microbiol. 2014, 22, 536–545. [Google Scholar] [CrossRef]
- Pornsukarom, S.; Thakur, S. Horizontal Dissemination of Antimicrobial Resistance Determinants in Multiple Salmonella Serotypes following Isolation from the Commercial Swine Operation Environment after Manure Application. Appl. Environ. Microbiol. 2017, 83. [Google Scholar] [CrossRef]
- Thanner, S.; Drissner, D.; Walsh, F. Antimicrobial Resistance in Agriculture. mBio 2016, 7, e02227-15. [Google Scholar] [CrossRef]
- Yanagiya, K.; Maejima, Y.; Nakata, H.; Tokuda, M.; Moriuchi, R.; Dohra, H.; Inoue, K.; Ohkuma, M.; Kimbara, K.; Shintani, M. Novel Self-Transmissible and Broad-Host-Range Plasmids Exogenously Captured From Anaerobic Granules or Cow Manure. Front. Microbiol. 2018, 9, 2602. [Google Scholar] [CrossRef]
- Tokuda, M.; Suzuki, H.; Yanagiya, K.; Yuki, M.; Inoue, K.; Ohkuma, M.; Kimbara, K.; Shintani, M. Determination of Plasmid pSN1216-29 Host Range and the Similarity in Oligonucleotide Composition Between Plasmid and Host Chromosomes. Front. Microbiol. 2020, 11, 1187. [Google Scholar] [CrossRef] [PubMed]
- Touati, M.; Hadjadj, L.; Berrazeg, M.; Baron, S.A.; Rolain, J.M. Emergence of Escherichia coli harbouring mcr-1 and mcr-3 genes in North West Algerian farmlands. J. Glob. Antimicrob. Resist. 2019, 21, 132–137. [Google Scholar] [CrossRef] [PubMed]
- Liu, P.; Jia, S.; He, X.; Zhang, X.; Ye, L. Different impacts of manure and chemical fertilizers on bacterial community structure and antibiotic resistance genes in arable soils. Chemosphere 2017, 188, 455–464. [Google Scholar] [CrossRef] [PubMed]
- De Leener, E.; Martel, A.; De Graef, E.M.; Top, J.; Butaye, P.; Haesebrouck, F.; Willems, R.; Decostere, A. Molecular analysis of human, porcine, and poultry Enterococcus faecium isolates and their erm(B) genes. Appl. Environ. Microbiol. 2005, 71, 2766–2770. [Google Scholar] [CrossRef] [PubMed]
- Alvarez-Narvaez, S.; Giguere, S.; Berghaus, L.J.; Dailey, C.; Vazquez-Boland, J.A. Horizontal Spread of Rhodococcus equi Macrolide Resistance Plasmid pRErm46 across Environmental Actinobacteria. Appl. Environ. Microbiol. 2020, 86. [Google Scholar] [CrossRef]
- Jechalke, S.; Kopmann, C.; Rosendahl, I.; Groeneweg, J.; Weichelt, V.; Krogerrecklenfort, E.; Brandes, N.; Nordwig, M.; Ding, G.C.; Siemens, J.; et al. Increased abundance and transferability of resistance genes after field application of manure from sulfadiazine-treated pigs. Appl. Environ. Microbiol. 2013, 79, 1704–1711. [Google Scholar] [CrossRef]
- Kyselkova, M.; Chrudimsky, T.; Husnik, F.; Chronakova, A.; Heuer, H.; Smalla, K.; Elhottova, D. Characterization of tet(Y)-carrying LowGC plasmids exogenously captured from cow manure at a conventional dairy farm. FEMS Microbiol. Ecol. 2016, 92, fiw075. [Google Scholar] [CrossRef]
- Toleman, M.A.; Bennett, P.M.; Walsh, T.R. ISCR elements: Novel gene-capturing systems of the 21st century? Microbiol. Mol. Biol. Rev. MMBR 2006, 70, 296–316. [Google Scholar] [CrossRef]
- Agersø, Y.; Jensen, L.B.; Givskov, M.; Roberts, M.C. The identification of a tetracycline resistance gene tet(M), on a Tn916-like transposon, in the Bacillus cereus group. FEMS Microbiol. Lett. 2002, 214, 251–256. [Google Scholar] [CrossRef]
- Hansen, L.H.; Johannesen, E.; Burmolle, M.; Sorensen, A.H.; Sorensen, S.J. Plasmid-encoded multidrug efflux pump conferring resistance to olaquindox in Escherichia coli. Antimicrob. Agents Chemother. 2004, 48, 3332–3337. [Google Scholar] [CrossRef]
- Hansen, L.H.; Sorensen, S.J.; Jorgensen, H.S.; Jensen, L.B. The prevalence of the OqxAB multidrug efflux pump amongst olaquindox-resistant Escherichia coli in pigs. Microb. Drug Resist. 2005, 11, 378–382. [Google Scholar] [CrossRef] [PubMed]
- Mann, N.H. The third age of phage. PLoS Biol. 2005, 3, e182. [Google Scholar] [CrossRef] [PubMed]
- Calero-Caceres, W.; Ye, M.; Balcazar, J.L. Bacteriophages as Environmental Reservoirs of Antibiotic Resistance. Trends Microbiol. 2019, 27, 570–577. [Google Scholar] [CrossRef]
- Batinovic, S.; Wassef, F.; Knowler, S.A.; Rice, D.T.F.; Stanton, C.R.; Rose, J.; Tucci, J.; Nittami, T.; Vinh, A.; Drummond, G.R.; et al. Bacteriophages in Natural and Artificial Environments. Pathogens 2019, 8, 100. [Google Scholar] [CrossRef] [PubMed]
- Niu, Y.D.; McAllister, T.A.; Xu, Y.; Johnson, R.P.; Stephens, T.P.; Stanford, K. Prevalence and impact of bacteriophages on the presence of Escherichia coli O157:H7 in feedlot cattle and their environment. Appl. Environ. Microbiol. 2009, 75, 1271–1278. [Google Scholar] [CrossRef] [PubMed]
- Ross, J.; Topp, E. Abundance of Antibiotic Resistance Genes in Bacteriophage following Soil Fertilization with Dairy Manure or Municipal Biosolids, and Evidence for Potential Transduction. Appl. Environ. Microbiol. 2015, 81, 7905–7913. [Google Scholar] [CrossRef]
- Evans, T.J.; Crow, M.A.; Williamson, N.R.; Orme, W.; Thomson, N.R.; Komitopoulou, E.; Salmond, G.P. Characterization of a broad-host-range flagellum-dependent phage that mediates high-efficiency generalized transduction in, and between, Serratia and Pantoea. Microbiology 2010, 156, 240–247. [Google Scholar] [CrossRef][Green Version]
- Thomas, C.M.; Nielsen, K.M. Mechanisms of, and barriers to, horizontal gene transfer between bacteria. Nat. Rev. Microbiol. 2005, 3, 711–721. [Google Scholar] [CrossRef]
- Domingues, S.; Nielsen, K. Horizontal Gene Transfer: Uptake of Extracellular DNA by Bacteria. Ref. Modul. Biomed. Sci. 2016. [Google Scholar] [CrossRef]
- Domingues, S.; Nielsen, K.M. Membrane vesicles and horizontal gene transfer in prokaryotes. Curr. Opin. Microbiol. 2017, 38, 16–21. [Google Scholar] [CrossRef]
- Soler, N.; Forterre, P. Vesiduction: The fourth way of HGT. Environ. Microbiol. 2020. [Google Scholar] [CrossRef] [PubMed]
- San Millan, A.; MacLean, R.C. Fitness Costs of Plasmids: A Limit to Plasmid Transmission. Microbiol. Spectr. 2017, 5. [Google Scholar] [CrossRef]
- Botelho, J.; Grosso, F.; Peixe, L. Antibiotic resistance in Pseudomonas aeruginosa—Mechanisms, epidemiology and evolution. Drug Resist. Updat. 2019, 44, 100640. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Cheng, W.; Li, B.; Xu, Y.; Zheng, X. The fate of antibiotic resistance genes during co-composting of swine manure with cauliflower and corn straw. Bioresour. Technol. 2020, 300, 122669. [Google Scholar] [CrossRef]
- Collignon, P.J.; McEwen, S.A. One Health-Its Importance in Helping to Better Control Antimicrobial Resistance. Trop. Med. Infect. Dis. 2019, 4, 22. [Google Scholar] [CrossRef] [PubMed]
- Fan, H.; Wu, S.; Woodley, J.; Zhuang, G.; Bai, Z.; Xu, S.; Wang, X.; Zhuang, X. Effective removal of antibiotic resistance genes and potential links with archaeal communities during vacuum-type composting and positive-pressure composting. J. Environ. Sci. 2020, 89, 277–286. [Google Scholar] [CrossRef]
- Esperon, F.; Albero, B.; Ugarte-Ruiz, M.; Dominguez, L.; Carballo, M.; Tadeo, J.L.; Del Mar Delgado, M.; Moreno, M.A.; de la Torre, A. Assessing the benefits of composting poultry manure in reducing antimicrobial residues, pathogenic bacteria, and antimicrobial resistance genes: A field-scale study. Environ. Sci. Pollut. Res. Int. 2020, 27, 27738–27749. [Google Scholar] [CrossRef]
- Kakimoto, T.; Osawa, T.; Funamizu, N. Antibiotic effect of amoxicillin on the feces composting process and reactivation of bacteria by intermittent feeding of feces. Bioresour. Technol. 2007, 98, 3555–3560. [Google Scholar] [CrossRef]
- Duan, M.; Zhang, Y.; Zhou, B.; Wang, Q.; Gu, J.; Liu, G.; Qin, Z.; Li, Z. Changes in antibiotic resistance genes and mobile genetic elements during cattle manure composting after inoculation with Bacillus subtilis. Bioresour. Technol. 2019, 292, 122011. [Google Scholar] [CrossRef]
- Li, H.; Duan, M.; Gu, J.; Zhang, Y.; Qian, X.; Ma, J.; Zhang, R.; Wang, X. Effects of bamboo charcoal on antibiotic resistance genes during chicken manure composting. Ecotoxicol. Environ. Saf. 2017, 140, 1–6. [Google Scholar] [CrossRef]
- Le Devendec, L.; Mourand, G.; Bougeard, S.; Leaustic, J.; Jouy, E.; Keita, A.; Couet, W.; Rousset, N.; Kempf, I. Impact of colistin sulfate treatment of broilers on the presence of resistant bacteria and resistance genes in stored or composted manure. Vet. Microbiol. 2016, 194, 98–106. [Google Scholar] [CrossRef] [PubMed]
- Youngquist, C.P.; Mitchell, S.M.; Cogger, C.G. Fate of Antibiotics and Antibiotic Resistance during Digestion and Composting: A Review. J. Environ. Qual. 2016, 45, 537–545. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Chen, M.; Sui, Q.; Tong, J.; Jiang, C.; Lu, X.; Zhang, Y.; Wei, Y. Impacts of addition of natural zeolite or a nitrification inhibitor on antibiotic resistance genes during sludge composting. Water Res. 2016, 91, 339–349. [Google Scholar] [CrossRef]
- Peng, S.; Li, H.; Song, D.; Lin, X.; Wang, Y. Influence of zeolite and superphosphate as additives on antibiotic resistance genes and bacterial communities during factory-scale chicken manure composting. Bioresour. Technol. 2018, 263, 393–401. [Google Scholar] [CrossRef] [PubMed]
- Cui, E.; Wu, Y.; Jiao, Y.; Zuo, Y.; Rensing, C.; Chen, H. The behavior of antibiotic resistance genes and arsenic influenced by biochar during different manure composting. Environ. Sci. Pollut. Res. Int. 2017, 24, 14484–14490. [Google Scholar] [CrossRef]
- Guo, A.; Gu, J.; Wang, X.; Zhang, R.; Yin, Y.; Sun, W.; Tuo, X.; Zhang, L. Effects of superabsorbent polymers on the abundances of antibiotic resistance genes, mobile genetic elements, and the bacterial community during swine manure composting. Bioresour. Technol. 2017, 244, 658–663. [Google Scholar] [CrossRef]
- Wang, R.; Zhang, J.; Sui, Q.; Wan, H.; Tong, J.; Chen, M.; Wei, Y.; Wei, D. Effect of red mud addition on tetracycline and copper resistance genes and microbial community during the full scale swine manure composting. Bioresour. Technol. 2016, 216, 1049–1057. [Google Scholar] [CrossRef]
- Flores-Orozco, D.; Patidar, R.; Levin, D.B.; Sparling, R.; Kumar, A.; Cicek, N. Effect of mesophilic anaerobic digestion on the resistome profile of dairy manure. Bioresour. Technol. 2020, 315, 123889. [Google Scholar] [CrossRef]
- Zhang, J.; Lu, T.; Shen, P.; Sui, Q.; Zhong, H.; Liu, J.; Tong, J.; Wei, Y. The role of substrate types and substrate microbial community on the fate of antibiotic resistance genes during anaerobic digestion. Chemosphere 2019, 229, 461–470. [Google Scholar] [CrossRef]
- Flores-Orozco, D.; Patidar, R.; Levin, D.B.; Sparling, R.; Kumar, A.; Cicek, N. Effect of ceftiofur on mesophilic anaerobic digestion of dairy manure and the reduction of the cephalosporin-resistance gene cmy-2. Bioresour. Technol. 2020, 301, 122729. [Google Scholar] [CrossRef]
- Sun, W.; Qian, X.; Gu, J.; Wang, X.J.; Duan, M.L. Mechanism and Effect of Temperature on Variations in Antibiotic Resistance Genes during Anaerobic Digestion of Dairy Manure. Sci. Rep. 2016, 6, 30237. [Google Scholar] [CrossRef] [PubMed]
- Joy, S.R.; Li, X.; Snow, D.D.; Gilley, J.E.; Woodbury, B.; Bartelt-Hunt, S.L. Fate of antimicrobials and antimicrobial resistance genes in simulated swine manure storage. Sci. Total Environ. 2014, 481, 69–74. [Google Scholar] [CrossRef]
- Zou, Y.; Xiao, Y.; Wang, H.; Fang, T.; Dong, P. New insight into fates of sulfonamide and tetracycline resistance genes and resistant bacteria during anaerobic digestion of manure at thermophilic and mesophilic temperatures. J. Hazard. Mater. 2020, 384, 121433. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.; Wilson, C.A.; Novak, J.T.; Riffat, R.; Aynur, S.; Murthy, S.; Pruden, A. Effect of various sludge digestion conditions on sulfonamide, macrolide, and tetracycline resistance genes and class I integrons. Environ. Sci. Technol. 2011, 45, 7855–7861. [Google Scholar] [CrossRef]
- Zhu, L.; Zhao, Y.; Yang, K.; Chen, J.; Zhou, H.; Chen, X.; Liu, Q.; Wei, Z. Host bacterial community of MGEs determines the risk of horizontal gene transfer during composting of different animal manures. Environ. Pollut. 2019, 250, 166–174. [Google Scholar] [CrossRef]
- Chuppava, B.; Keller, B.; Abd El-Wahab, A.; Surie, C.; Visscher, C. Resistance Reservoirs and Multi-Drug Resistance of Commensal Escherichia coli From Excreta and Manure Isolated in Broiler Houses With Different Flooring Designs. Front. Microbiol. 2019, 10, 2633. [Google Scholar] [CrossRef]
- European Medicines Agency (EMA). Reflection Paper on Antimicrobial Resistance in the Environment: Considerations for Current and Future Risk Assessment of Veterinary Medicinal Products Draft. 2018. Available online: https://www.ema.europa.eu/documents/scientific-guideline/draft-reflection-paper-antimicrobial-resistance-environment-considerations-current-future-risk_en.pdf (accessed on 29 July 2020).
- Zhang, Y.J.; Hu, H.W.; Chen, Q.L.; Singh, B.K.; Yan, H.; Chen, D.; He, J.Z. Transfer of antibiotic resistance from manure-amended soils to vegetable microbiomes. Environ. Int. 2019, 130, 104912. [Google Scholar] [CrossRef]
- Blaak, H.; van Hoek, A.H.; Veenman, C.; Docters van Leeuwen, A.E.; Lynch, G.; van Overbeek, W.M.; de Roda Husman, A.M. Extended spectrum B-lactamase- and constitutively AmpC-producing Enterobacteriaceae on fresh produce and in the agricultural environment. Int. J. Food Microbiol. 2014, 168–169, 8–16. [Google Scholar] [CrossRef]
- Food and Agriculture Organization of the United Nations (FAO). Drivers, Dynamics and Epidemiology of Antimicrobial Resistance in Animal Production. 2016. Available online: http://www.fao.org/3/a-i6209e.pdf (accessed on 29 July 2020).
- Marshall, B.M.; Levy, S.B. Food animals and antimicrobials: Impacts on human health. Clin. Microbiol. Rev. 2011, 24, 718–733. [Google Scholar] [CrossRef]
- Pepper, I.L.; Brooks, J.P.; Gerba, C.P. Antibiotic Resistant Bacteria in Municipal Wastes: Is There Reason for Concern? Environ. Sci. Technol. 2018, 52, 3949–3959. [Google Scholar] [CrossRef]
- Zhou, X.; Wang, J.; Lu, C.; Liao, Q.; Gudda, F.O.; Ling, W. Antibiotics in animal manure and manure-based fertilizers: Occurrence and ecological risk assessment. Chemosphere 2020, 255, 127006. [Google Scholar] [CrossRef] [PubMed]
- Wellington, E.M.; Boxall, A.B.; Cross, P.; Feil, E.J.; Gaze, W.H.; Hawkey, P.M.; Johnson-Rollings, A.S.; Jones, D.L.; Lee, N.M.; Otten, W.; et al. The role of the natural environment in the emergence of antibiotic resistance in gram-negative bacteria. Lancet Infect. Dis. 2013, 13, 155–165. [Google Scholar] [CrossRef]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).