Lab-Made Electronic Nose for Fast Detection of Listeria monocytogenes and Bacillus cereus
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Preparation
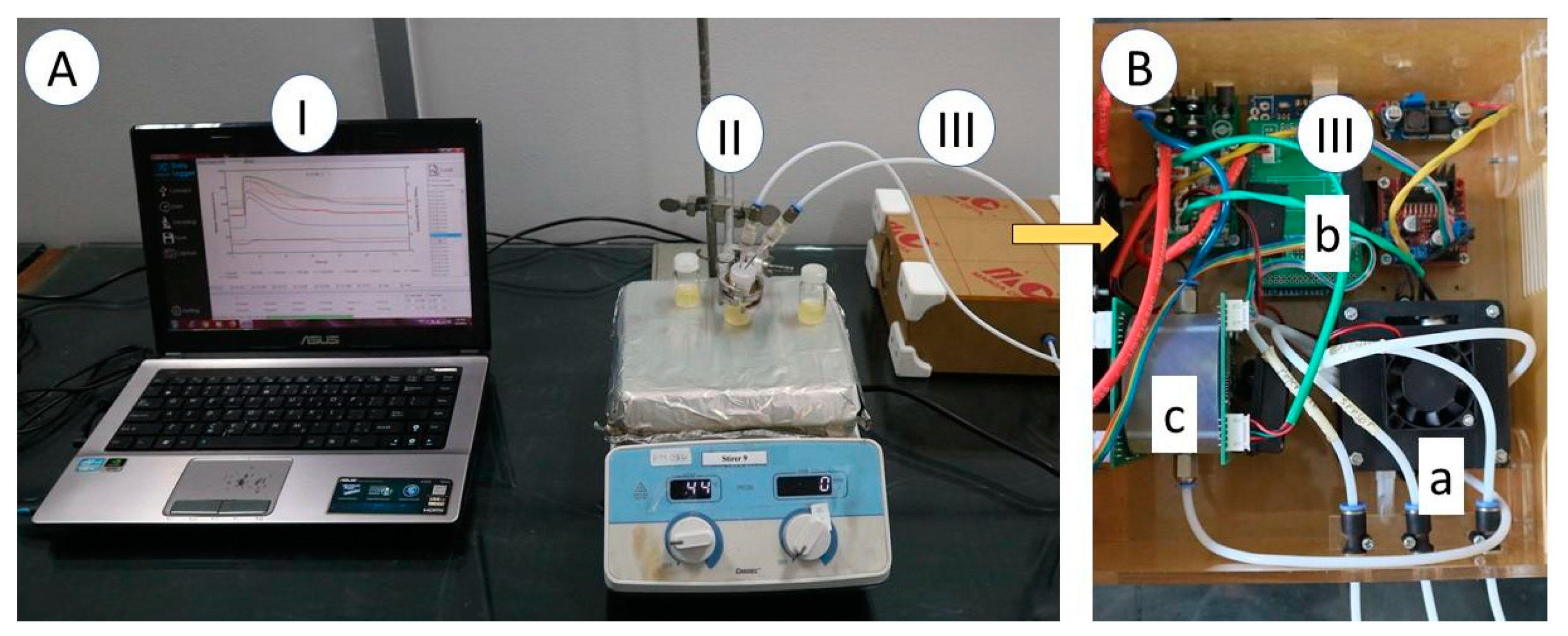
2.2. Electronic Nose Specification
2.3. Electronic Nose Measurement and Chemometric Analysis
3. Results
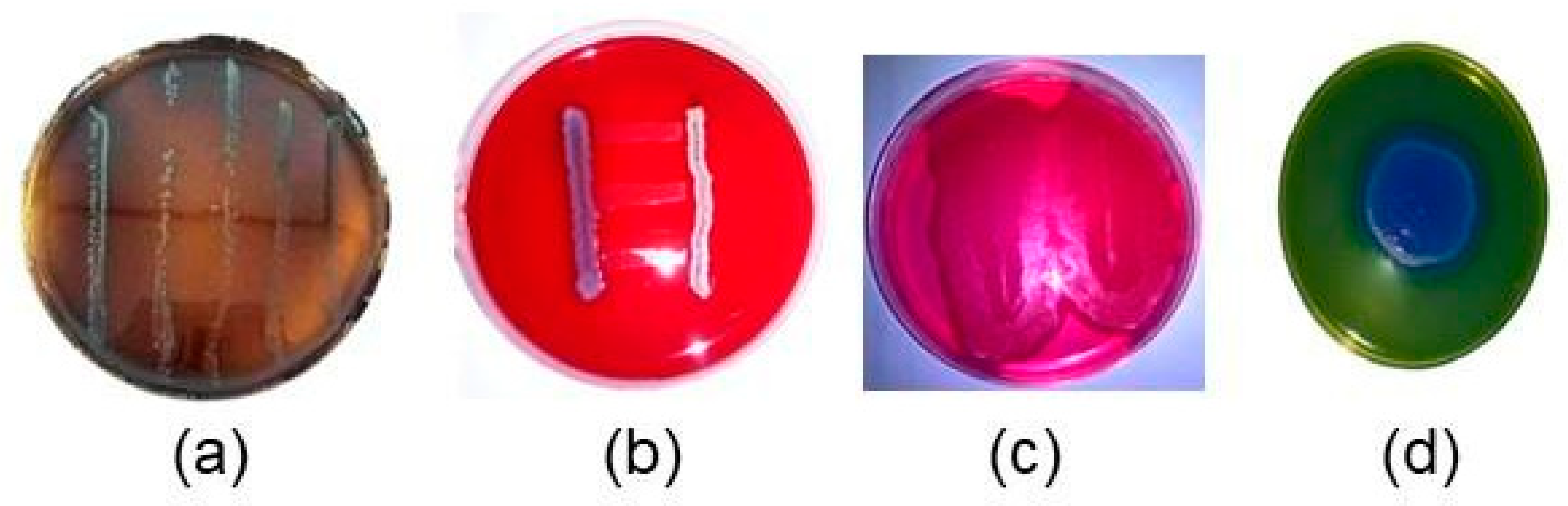
3.1. Bacteria re-Identification Colony Counting
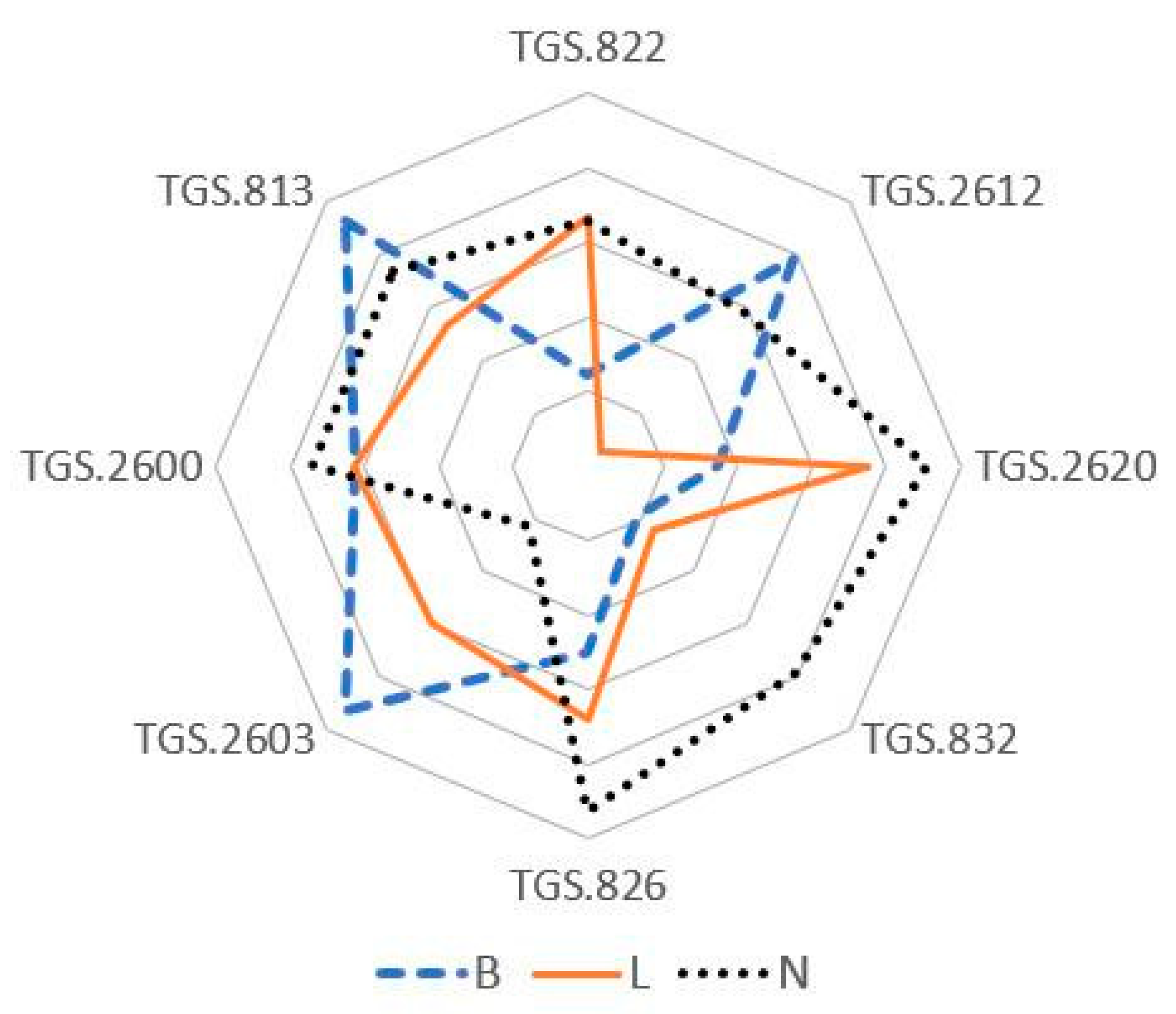
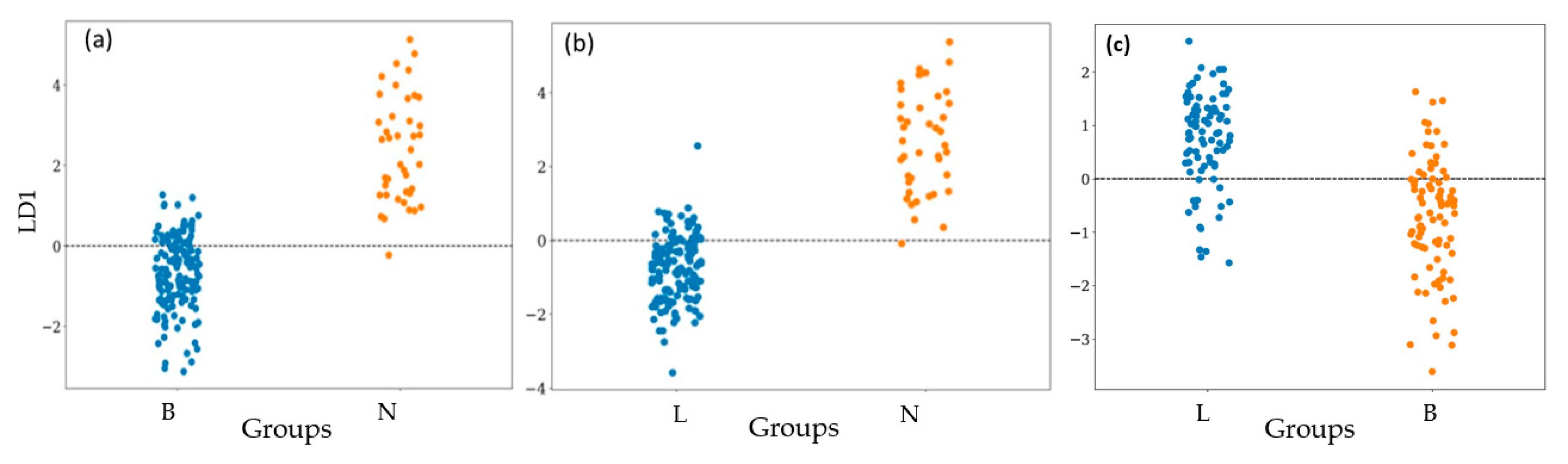
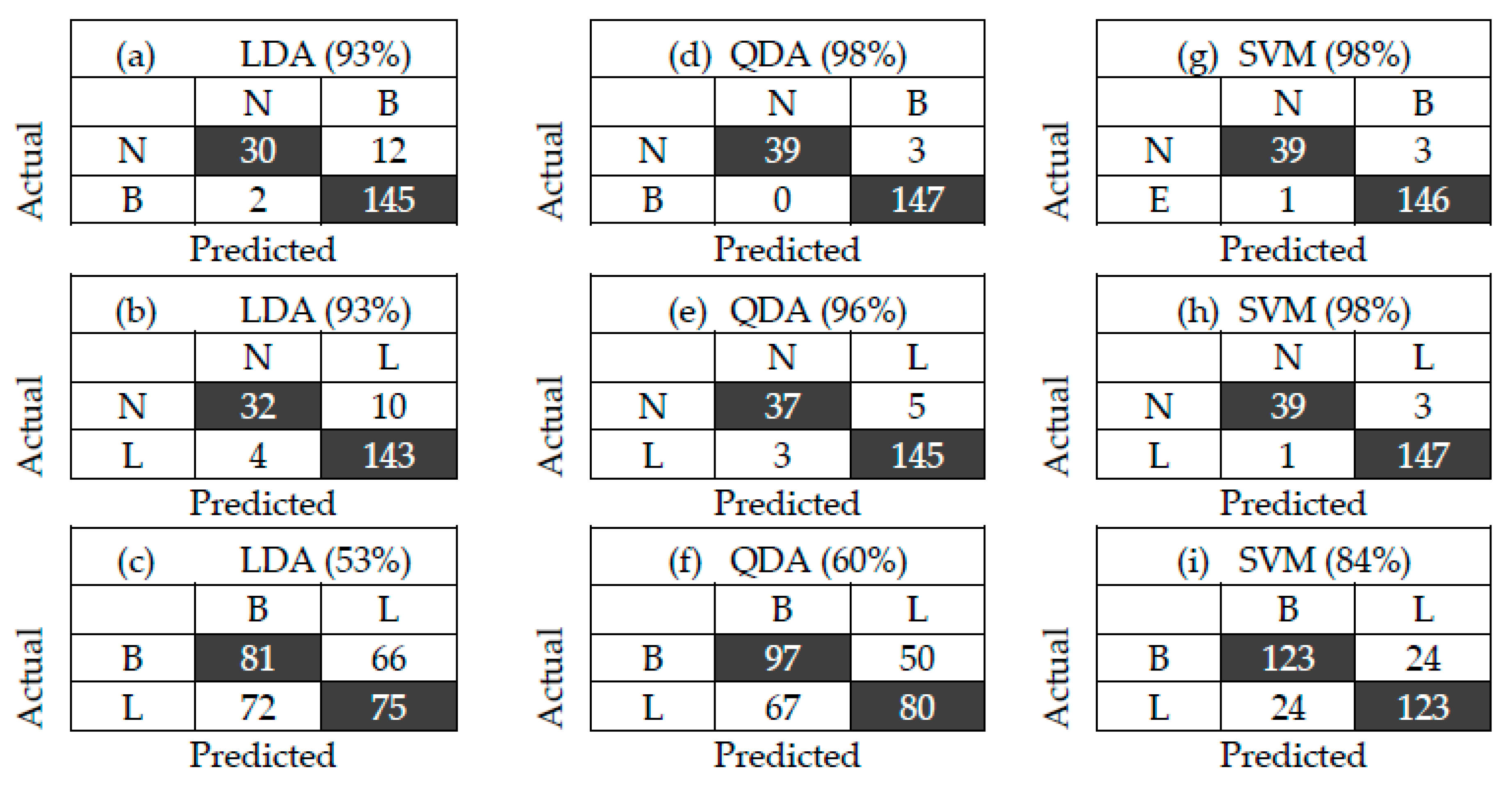
3.2. Bacterial Classification
4. Discussion
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Zhao, X.; Cui, Y.; Wang, J.; Wang, J. Preparation of Fluorescent Molecularly Imprinted Polymers via Pickering Emulsion Interfaces and the Application for Visual Sensing Analysis of Listeria monocytogenes. Polymers 2019, 11, 984. [Google Scholar] [CrossRef] [PubMed]
- Chlebicz, A.; Slizewska, K. Campylobacteriosis, Salmonellosis, Yersiniosis, and Listeriosis as Zoonotic Foodborne Diseases: A Review. Int. J. Environ. Res. Public Health 2018, 15, 863. [Google Scholar] [CrossRef] [PubMed]
- Radhakrishnan, R.; Poltronieri, P. Fluorescence-Free Biosensor Methods in Detection of Food Pathogens with a Special Focus on Listeria monocytogenes. Biosensors 2017, 7, 63. [Google Scholar] [CrossRef] [PubMed]
- Latha, C.; Sunil, B.; Kumar, V.J.; Anu, C.J.; Deepa, J. Evaluation of various cultural enrichment methods for the detection of selected food borne bacterial pathogens. Vet. World 2014, 7, 172–176. [Google Scholar] [CrossRef][Green Version]
- Stambach, N.R.; Carr, S.A.; Cox, C.R.; Voorhees, K.J. Rapid Detection of Listeria by Bacteriophage Amplification and SERS-Lateral Flow Immunochromatography. Viruses 2015, 7, 6631–6641. [Google Scholar] [CrossRef] [PubMed]
- Brugère-Picoux, J. Ovine listeriosis. Small Rumin. Res. 2008, 76, 12–20. [Google Scholar] [CrossRef]
- Liu, A.; Shen, L.; Zeng, Z.; Sun, M.; Liu, Y.; Liu, S. A Minireview of the Methods for Listeria monocytogenes Detection. Food Anal. Methods 2018, 11, 215–223. [Google Scholar] [CrossRef]
- Tewari, A.; Abdullah, S. Bacillus cereus food poisoning: International and Indian perspective. J. Food Sci. Technol. 2015, 52, 2500–2511. [Google Scholar] [CrossRef]
- Griffiths, M.W.; Schraft, H. Bacillus cereus Food Poisoning, 3rd ed.; Elsevier Inc.: Amsterdam, The Netherlands, 2017; ISBN 9780123850072. pp. 395–400. [Google Scholar]
- Velusamy, V.; Arshak, K.; Korostynska, O.; Oliwa, K.; Adley, C. An overview of foodborne pathogen detection: In the perspective of biosensors. Biotechnol. Adv. 2010, 28, 232–254. [Google Scholar] [CrossRef]
- Khan, J.A.; Rathore, R.S.; Khan, S.; Ahmad, I. In vitro detection of pathogenic Listeria monocytogenes from food sources by conventional, molecular and cell culture method. Braz. J. Microbiol. 2013, 44, 751–758. [Google Scholar] [CrossRef]
- Sakamoto, S.; Putalun, W.; Vimolmangkang, S.; Phoolcharoen, W.; Shoyama, Y.; Tanaka, H.; Morimoto, S. Enzyme-linked immunosorbent assay for the quantitative/qualitative analysis of plant secondary metabolites. J. Nat. Med. 2018, 72, 32–42. [Google Scholar] [CrossRef]
- Wang, Y.; Wang, Y.; Ma, A.; Li, D.; Luo, L.; Liu, D.; Hu, S. The Novel Multiple Inner Primers-Loop-Mediated Isothermal Amplification (MIP-LAMP) for Rapid Detection and Differentiation of Listeria monocytogenes. Molecules 2015, 20, 21515–21531. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Lv, P.; Wang, L.; Guo, A.; Ma, M.; Qi, X. Application of high resolution pyrolysis gas chromatography / mass spectrometry (HRPGC/MS) for detecting Listeria monocytogenes. J. Chromatogr. B 2014, 971, 107–111. [Google Scholar] [CrossRef] [PubMed]
- Gębicki, J.; Szulczyński, B. Discrimination of selected fungi species based on their odour profile using prototypes of electronic nose instruments. Meas. J. Int. Meas. Confed. 2018, 116, 307–313. [Google Scholar] [CrossRef]
- Yusuf, N.; Omar, M.I.; Zakaria, A.; Jeffree, A.I.; Thriumani, R.; Abdullah, A.A.; Shakaff, A.Y.M.; Masnan, M.J.; Yeap, E.J.; Othman, A.; et al. Evaluation of E-nose technology for detection of the causative bacteria in different culture media on diabetic foot infection. In Proceedings of the 2014 IEEE Conference on Biomedical Engineering and Sciences, Miri, Sarawak, 8–10 December 2014; pp. 67–70. [Google Scholar]
- Tubia, I.; Prasad, K.; Pérez-Lorenzo, E.; Abadín, C.; Zumárraga, M.; Oyanguren, I.; Barbero, F.; Paredes, J.; Arana, S. Beverage spoilage yeast detection methods and control technologies: A review of Brettanomyces. Int. J. Food Microbiol. 2018, 283, 65–76. [Google Scholar] [CrossRef] [PubMed]
- Siripatrawan, U. Rapid differentiation between E. coli and Salmonella Typhimurium using metal oxide sensors integrated with pattern recognition. Sens. Actuators B Chem. 2008, 133, 414–419. [Google Scholar] [CrossRef]
- Balasubramanian, S.; Amamcharla, J.; Shin, J. Possible Application of Electronic Nose Systems for Meat Safety: An Overview; Elsevier Inc.: Amsterdam, The Netherlands, 2016; pp. 59–70. ISBN 9780128002438. [Google Scholar]
- Triyana, K.; Subekti, M.T.; Aji, P.; Hidayat, S.N.; Rohman, A. Development of Electronic Nose with Low-Cost Dynamic Headspace for Classifying Vegetable Oils and Animal Fats. Appl. Mech. Mater. 2015, 771, 50–54. [Google Scholar] [CrossRef]
- Yu, Y.; Sun, X.; Liu, Y.; Pan, Y.; Zhao, Y. Odor Fingerprinting of Listeria monocytogenes Recognized by SPME-GC / MS and E-nose. Can. J. Microbiol. 2015, 61, 1–6. [Google Scholar] [CrossRef]
- Dutta, R.; Hines, E.L.; Gardner, J.W.; Boilot, P. Bacteria classification using Cyranose 320 electronic nose. Biomed. Eng. Online 2002, 7, 1–7. [Google Scholar]
- Green, G.C.; Chan, A.D.C.; Dan, H.; Lin, M. Using a metal oxide sensor (MOS) -based electronic nose for discrimination of bacteria based on individual colonies in suspension. Sens. Actuators B Chem. 2011, 152, 21–28. [Google Scholar] [CrossRef]
- Hidayat, S.N.; Triyana, K.; Fauzan, I.; Julian, T. The Electronic Nose Coupled with Chemometric Tools for Discriminating the Quality of Black Tea Samples In Situ. Chemosensors 2019, 7, 29. [Google Scholar] [CrossRef]
- Mathakiya, R.A.; Roy, A.; Nayak, J.B. Characterization of Listeria monocytogenes isolates by CAMP test. Vet. World 2011, 4, 301–303. [Google Scholar] [CrossRef]
- Markey, B.; Finola, L.; Archambault, M.; Cullinane, A.; Maguire, D. Clinical Veterinary Microbiology, 2nd ed.; Mosby Elsevier: Amsterdam, The Netherlands, 2013; ISBN 9780723432371. [Google Scholar]
- Chen, J.; Tang, J.; Shi, H.; Tang, C.; Zhang, R. Characteristics of volatile organic compounds produced from five pathogenic bacteria by headspace-solid phase micro-extraction/gas chromatography-mass spectrometry. J. Basic Microbiol. 2017, 57, 228–237. [Google Scholar] [CrossRef] [PubMed]
- Deshmukh, Y.; Khare, P.; Patra, D.D.; Nadaf, A.B. HS-SPME-GC-FID method for detection and quantification of Bacillus cereus ATCC 10702 mediated 2-acetyl-1-pyrroline. Biotechnol. Prog. 2014, 30, 1356–1363. [Google Scholar] [CrossRef]
- Elgaali, H.; Hamilton-Kemp, T.; Newman, M.C.; Collins, R.W.; Archbold, D.D. Comparison of long-chain alcohols and other volatile compounds emitted from food-borne and related Gram positive and Gram negative bacteria. J. Basic Microbiol. 2002, 42, 373–380. [Google Scholar] [CrossRef]
- Filipiak, W.; Sponring, A.; Filipiak, A.; Baur, M.; Clemens, A.; Wiesenhofer, H.; Margesin, R.; Nagis, M.; Troppmair, J. Interpretation of Breath Analysis Data; Elsevier: Amsterdam, The Netherlands, 2013; ISBN 9780444626134. pp. 470–505. [Google Scholar]
- Talaro, K.P.; Chess, B. Foundations in Microbiology, 8th ed.; McGraw-Hill Companies, Inc.: New York, NY, USA, 2012; ISBN 9780073375298. [Google Scholar]
- Benedict, R.C.; Partridge, T.; Wells, D.; Buchanan, R.L. Bacillus cereus: Aerobic Growth Kinetics. J. Food Prot. 1993, 56, 211–214. [Google Scholar] [CrossRef]
- Rogers, K. Biochemistry, Cells, and Life Bacteria and Viruses; Britannica Educational Publishing: New York, NY, USA, 2011; ISBN 9781615303762. [Google Scholar]
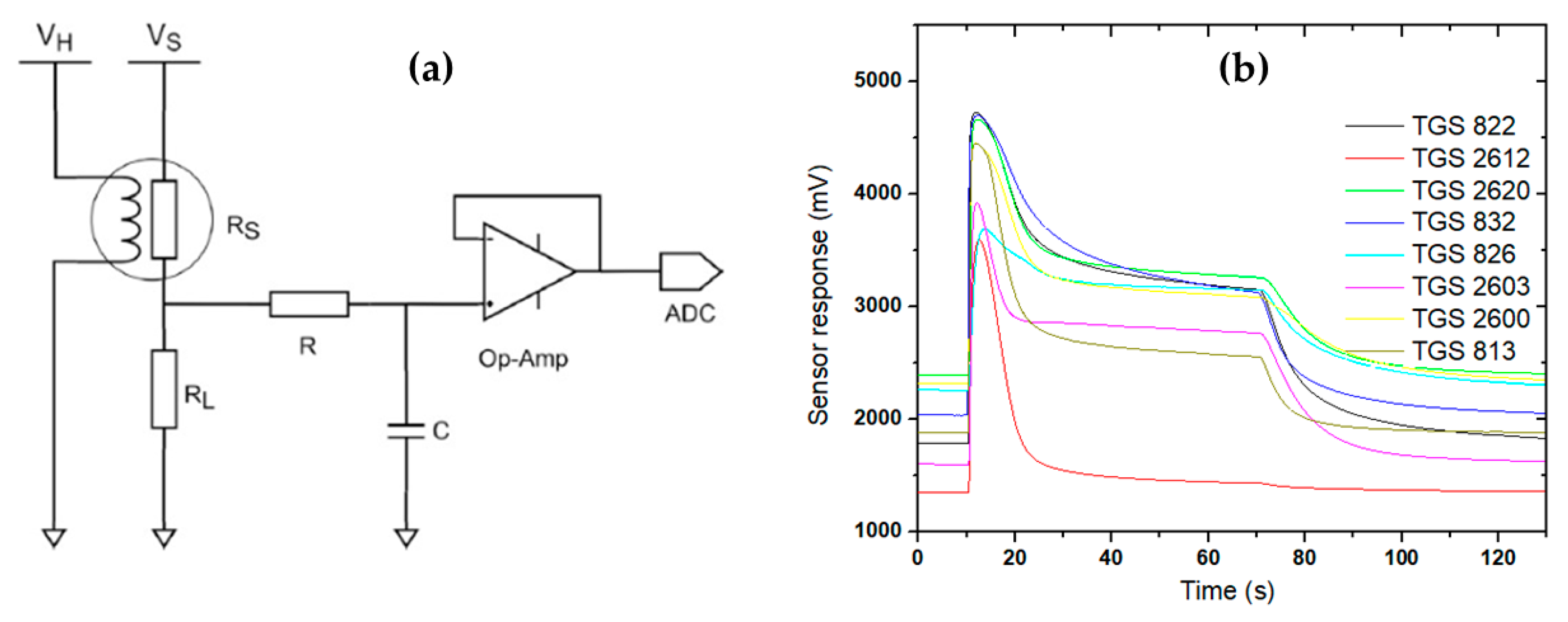
| Type of Sensor | Targeted Volatile Compounds |
|---|---|
| TGS813 | Methane, ethanol, propane, isobutane, hydrogen, and carbon monoxide |
| TGS822 | Ethanol, acetone, benzene, n-hexane, isobutane, carbon monoxide, and methane |
| TGS823 | Combustible gas, i.e., ethanol |
| TGS826 | Ammonia |
| TGS2600 | Methane, carbon monoxide, isobutane, ethanol, and hydrogen |
| TGS2603 | Hydrogen, H2S, ethanol, methyl mercaptan, and trimethylamine |
| TGS2612 | Ethanol, methane, isobutane, and propane |
| TGS2620 | Methane, carbon monoxide, isobutane, hydrogen, and ethanol |
| Test | L. monocytogenes | B. cereus |
|---|---|---|
| Catalase | + | + |
| Sulfur Indole Motility (SIM) | + | + |
| CAMP | + | na |
| Voges–Proskouer | + | + |
| Carbohydrate | ||
| Glucose | + | + |
| Maltose | na | + |
| Mannitol | − | − |
| Sucrose | + | + |
| Rhamnose | + | na |
| Xylose | − | na |
| Gram staining | + | + |
| Spore staining | na | + |
| MVOCs | Functional Groups | Active Sensors |
|---|---|---|
| Listeria monocytogenes | ||
| 2-undecanone | methyl vinyl ketone | TGS 822 |
| 2-nonanone/1-undecene | acrylic alkanes | TGS 813, TGS 2612, TGS 2620, TGS 2600 |
| dimethyl trisulfide | 2,3,4-trithiapentane | TGS 813, TGS 2612, TGS 2620, TGS 2600 |
| aldehydes | aldehyde | TGS 823, TGS 2600, TGS 2603 |
| ketones | ketones | TGS 822 |
| 3-methyl-butanal | butanal | TGS 823, TGS 2600, TGS 2603 |
| acetone | propanone | TGS 813, TGS 2612, TGS 2620, TGS 2600 |
| 2-methyl-butane | isopentane | TGS 813, TGS 2612, TGS 2620, TGS 2600 |
| 3-hydroxy-2-butanone | methyl acetoin | TGS 813, TGS 2612, TGS 2620, TGS 2600 |
| Bacillus cereus | ||
| 2-undecanone | metal ketones | TGS 822 |
| dimethylsulfide | 2,3,4-trithiapentane | TGS 813, TGS 2612, TGS 2620, TGS 2600 |
| 4-hydroxy-2-butanone | methyl acetone | TGS 822 |
| ethyl acetate | ester | TGS 813, TGS 2612, TGS 2620, TGS 2600 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Astantri, P.F.; Prakoso, W.S.A.; Triyana, K.; Untari, T.; Airin, C.M.; Astuti, P. Lab-Made Electronic Nose for Fast Detection of Listeria monocytogenes and Bacillus cereus. Vet. Sci. 2020, 7, 20. https://doi.org/10.3390/vetsci7010020
Astantri PF, Prakoso WSA, Triyana K, Untari T, Airin CM, Astuti P. Lab-Made Electronic Nose for Fast Detection of Listeria monocytogenes and Bacillus cereus. Veterinary Sciences. 2020; 7(1):20. https://doi.org/10.3390/vetsci7010020
Chicago/Turabian StyleAstantri, Prima Febri, Wredha Sandhi Ardha Prakoso, Kuwat Triyana, Tri Untari, Claude Mona Airin, and Pudji Astuti. 2020. "Lab-Made Electronic Nose for Fast Detection of Listeria monocytogenes and Bacillus cereus" Veterinary Sciences 7, no. 1: 20. https://doi.org/10.3390/vetsci7010020
APA StyleAstantri, P. F., Prakoso, W. S. A., Triyana, K., Untari, T., Airin, C. M., & Astuti, P. (2020). Lab-Made Electronic Nose for Fast Detection of Listeria monocytogenes and Bacillus cereus. Veterinary Sciences, 7(1), 20. https://doi.org/10.3390/vetsci7010020






