Abstract
Molecular typing of bacterial isolates provides a powerful approach for distinguishing Mycobacterium bovis (M. bovis) genotypes. It is known that M. bovis strain virulence plays a role in prevalence and spread of the disease, suggesting that strain virulence and prevailing genotypes are associated. However, it is not well understood whether strain virulence correlates with particular genotypes. In this study, we assessed the in vitro intracellular growth of 18 M. bovis isolates in bovine macrophages as an indicator of bacterial virulence and sought a relationship with the genotype identified by spoligotyping. We found 14 different spoligotypes—11 were already known and three spoligotypes had never been reported before. We identified 2 clusters that were phylogenetically related, containing 10 and 6 strains, respectively, and 2 orphan strains. Intracellular growth and phagocytic rates of 18 M. bovis strains were heterogeneous. Our results suggest that M. bovis intracellular growth and phagocytosis are independent of the bacterial lineage identified by spoligotyping.
1. Introduction
Bovine tuberculosis (BTB) is an infectious disease originated by the presence of Mycobacterium bovis in cattle. BTB generates high economic losses in livestock and affects animal health policies. This entity has spread worldwide, within a broad range of hosts, including humans, livestock, and wildlife [1]. According to the available disease timelines in the http.mbovis.org information database, M. bovis has been isolated from countries in all continents, except Antarctica [2].
M. bovis strains differ depending on the host they are infecting and the geographic region they are located in. Molecular typing of bacterial isolates based on polymorphisms in genomic DNA (genotyping) provides a powerful approach in distinguishing M. bovis strains, and may further yet provide valuable insights into the maintenance and transmission of infection [3]. The most common epidemiological molecular typing methods for members of the Mycobacterium tuberculosis complex are: (a) insertion sequence 6110-based restriction fragment length polymorphism (IS6110-RFLP), (b) spacer oligonucleotide typing (spoligotyping), and (c) the analysis of the copy number of mycobacterial interspersed repetitive unit-variable number tandem repeats (MIRU-VNTRs). Spoligotyping is the most commonly used tool for M. bovis genotyping because it is easy to perform and has the capability to discriminate between strains, since it excludes other species from the M. tuberculosis complex [4,5].
Mycobacterium tuberculosis strains differ in the way they invade, multiply, and persist in the host’s tissues. For example, the Euro-American lineage of Mycobacterium tuberculosis is less capable of producing meningeal tuberculosis than those strains predominantly originating in Asia [6]. The Beijing strains grow significantly faster in human macrophages and have a higher number of colony-forming units than those of non-Beijing strains, and are correlated with more severe lung lesions in mouse infections [7,8]. When we combine all of this data, it is suggestive of an association between specific genotypes and virulence.
The macrophages derived from peripheral blood have been use as a key of innate immunity to tuberculosis and a phenotypic marker of cattle susceptibility to M. bovis infection. Macrophages from different animals had differential intracellular control of M. bovis strains, depending on virulence. Our previous study showed differences in the bacterial survival rate of 65% and 100% for the avirulent M. bovis BCG and the virulent field isolate known as “El Paso”, respectively [9,10]. Macrophages from animals that better control bacterial replication were found to produce a higher oxidative burst, having greater bacteriostatic activity, producing higher concentrations of nitric oxide, and having a higher pro-inflammatory cytokine gene expression [11,12,13]. Results from our laboratory demonstrated that the M. bovis strain virulence classification obtained by a well-characterized mouse model of progressive pulmonary tuberculosis [14] was also replicable in the bovine macrophage in vitro model. Virulent strains had higher intracellular growth than the attenuated ones [15]. Therefore, bacterial intracellular survival and replication in an in vitro macrophage infection assay may be considered as a correlate of virulence [8,16,17].
According to literature, some M. bovis genotypes are more predominant in certain geographical regions than others within a cattle population [18]. We hypothesize that virulence influences the persistence and prevalence of strains, and that virulence is related to lineage. However, it is not well understood whether strain virulence correlates directly with specific genotypes. In this study, we aimed to assess intracellular growth of M. bovis isolates in macrophages of naturally resistant cattle as an indicator of bacterial virulence, and evaluate the possible correlation with the strain genotype identified by spoligotyping. Our results demonstrated that strains belonging to the same phylogenetically related clusters showed a differential intracellular survival/replication rate, suggesting that spacer composition does not influence M. bovis intracellular growth in bovine macrophages.
2. Methods
2.1. Ethic Statement
All animal procedures were performed according to the Facultad de Medicina Veterinaria y Zootecnia from the Universidad Nacional Autónoma de México (FMVZ-UNAM) board’s statements on animal research, based on the Mexican law on animal studies. Ethical approval for this study was obtained from the FMVZ-UNAM Institutional Committee for Care and Use of Experimental Animals (CICUAE) (JAGP-2002).
2.2. Macrophage Culture
Venous peripheral blood was obtained from healthy adult cattle, from a tuberculosis-free herd housed at the facilities of the Research and Teaching Center (CEPIPSA) of the Universidad Nacional Autónoma de México (UNAM). Macrophages were obtained from peripheral blood mononuclear cells (PBMC), as described before [11]. Blood was collected from the jugular vein into 60 mL syringes containing an acid-citrate-dextrose solution and was centrifuged at 1000 ×g for 30 min. Buffy coats were diluted in 30 mL of citrated PBS, layered onto 15 mL of Percoll (Pharmacia, Uppsala, Sweden) at a specific density of 1.077, and centrifuged at 1200 ×g for 25 min. PBMC were then removed from the interface between the plasma and Percoll solution, pooled, diluted in 50 mL of citrated phosphate buffered saline (PBS), and centrifuged at 500 ×g for 15 min. The cell pellets were then washed three times with citrated PBS at 500 ×g for 10 min, suspended in Roswell Park Memorial Institute (RPMI) (Gibco, New York, NY, USA) supplemented with 5 mM L-glutamine (Gibco, New York, NY, USA), 5 mM non-essential amino acids, and 5 mM sodium pyruvate (Gibco, New York, NY, USA) (CRPMI) containing 4% autologous serum to facilitate adherence, and cultured overnight at 37 °C and 5% CO2. Non-adherent cells were then removed by three washes with prewarmed PBS, and adherent monocytes were cultured, as described previously, in CRPMI plus 12% autologous serum for 12 days until they differentiated to macrophages. Flasks were chilled on ice for 45 min and macrophages were harvested by repeated gentle pipetting.
2.3. Mycobacterium Bovis Strains
The strains used in this study are listed in Table 1. Strains were obtained by convenience sampling performed by official veterinarians in the field. All strains were isolated from visible lesions observed at the slaughterhouse. Strains from the same geographical region came from different farms. Strains were grown in Middlebrook 7H11 medium (Difco Laboratories, Detroit, MI, USA) with oleica acid, dextrose and catalase (OADC) (Becton and Dickinson, Sparks, NV, USA) from 18 to 21 days and then were seeded for 19 days in 7H9 broth supplemented with 0.05% Tween 80 and OADC (Becton and Dickinson, Sparks, NV, USA). Bacteria were suspended in CRPMI, passed twice through a 27-gauge needle, and sonicated for 30 s in order to disrupt the clumps. Aliquots of 1 mL were stored at −80 °C. Inoculums were titrated by plating colony-forming unit (CFU) serial dilutions on Middlebrook 7H11 (Difco Laboratories, Detroit, MI, USA) plus 10% of OADC.

Table 1.
M. bovis strains used in this study. Data summarize spoligotype identity and strain origin. Nil (spoligotype not reported before). Strain control: M. bovis field strain SB2084 and Bacillus Calmette-Guerin (BCG) Montreal.
2.4. Spoligotyping
DNA was obtained by proteinase K-guanidine hydrochloride method. Spoligotyping was performed following a standard protocol (Kamerbeek et al., 1997). Briefly, bacilli DNA was amplified with AmpliTaq DNA polymerase (PerkinElmer, Norwalk, CT, USA) in a 50 μL PCR mix containing 5 μL of 10 × reaction buffer (100 μM Tris–HCl, pH 8.3, 500 M KCl, 15 μM MgCl2, 0.01% (w/v) gelatin, 0.2 μM of dNTP, 20 pmol each primer DRa (5′ -GGT TTT GGG TCT GAC GAC- 3′) marked with biotin at the 5′ DRb (5′-CCG AGA GGG GAC GGA AAC-3′), and 10 ng of mycobacterial DNA. The mixture was heated in a Gene Amp PCR system 2400 (PerkinElmer, Norwalk, CT, USA) at 96 °C for 3 min and subjected to 30 cycles at 96 °C for 1 min, 55 °C for 1 min, and 72 °C for 40 s, until reaching 72 °C for 10 min. The amplified DNA was visualized after electrophoresis in a Sodium Dodecyl sulfate ( SDS) polyacrylamide 12% gel stained with silver nitrate at 120 V for 90 min. Twenty microliters of PCR product was added to 150 μL of 2 × SSPE-0.1% SDS (15 mM sodium chloride, 1 mM sodium phosphate, 1mM EDTA, 0.1% SDS) and heat denatured at 99 °C for 10 min, and applied to a nylon membrane to which 37 spacer sequences from M. tuberculosis H37Rv and 6 spacer sequences from M. bovis BCG were covalently bound (ISOGEN, Maarssen, The Netherlands). This membrane was then placed in a miniblotter MN45 (Immunetics; Cambridge, MA, USA) in such a way that the slots were perpendicular to the line pattern of the previously applied oligonucleotides. The membranes were then incubated in 10 mL of 2 × SSPE-0.5% SDS plus 5 μL of streptavidin-peroxidase conjugate for 45 min at 42 °C. For detection of hybridizing DNA, enhanced chemiluminescence (ECL) detection liquid (Amersham Biosciences; Piscataway, NJ, USA) was used, followed by exposure to X-ray film (Kodak, Rochester, NY, USA) for 12 min.
2.5. Bactericidal Assay
The bactericidal assay was performed as described previously [11]. Briefly, macrophage monolayers in 60-microwell tissue culture plates (Nunc, Roskilde, Denmark) were infected with each strain of M. bovis at a multiplicity of infection (MOI) of 10:1, centrifuged at 200 ×g for 10 min, and incubated at 37 °C with 95% humidity and 5% CO2 for 4 h (the time previously determined to allow macrophage phagocytosis of mycobacteria). After this time (considered 0 h post-infection), cells were washed 5 times to remove extracellular bacteria with 5 μL of fresh CRPMI and incubated for 24 h at 37 °C. To calculate mycobacteria intracellular growth, the number of intracellular CFU at the end of the assay (24 h) was divided by the total number of CFU at the beginning (0 h) and expressed as percentage. CFU were recovered, serial diluted, plated onto Middlebrook 7H11 plus 10% OADC agar, and cultured for 21 days for counting after lysis of macrophages with 0.5% Tween 20 at 0 h and 24 h post-infection. We also estimated the average number of mycobacteria per macrophage after phagocytosis by dividing the total number of CFU at 0 h by the total number of seeded macrophages. The results are the average of three independent experiments, each one with three internal replicates.
2.6. Statistical Analysis
A data analysis composed of matrix 1 or matrix 0 was built on the bases of presence (1) or absence (0), respectively, of fragments hybridized in the spoligotyping. Unweighted pairwise group method (UPGMA) analysis was performed to build the dendrogram with multivariate statistical analysis, using squared Euclidean distance with the hierarchical cluster option and nearest neighbor.
Comparisons within rates of phagocytosis and intracellular growth of all M. bovis strains were performed using one-way ANOVA tests, as well as Tukey’s post hoc test for multiple comparisons between all strains and within spolygotype clusters. Fisher’s least significant difference test was done for comparisons of each strain rate of intracellular growth versus 100%, the cutoff reference value that signifies a strain’s replication into the macrophage. A p value of 0.05 or less was considered statistically significant.
3. Results
3.1. Spoligotyping Identified Strains Already Present in the Region and Three New Spoligotypes
We analyzed 18 different M. bovis strains isolated from tuberculous lesions of dairy cattle and found 14 different spoligotypes, among them were three spoligotypes that had never been before reported. Spoligotyping results are summarized in Table 1. M. bovis spoligotypes identified in this study correspond to spoligotypes reported previously in Mexico. Pattern information of the new strains was submitted to the M. bovis database strains 9524 (SB2353), QP129, 9930, 9926 (SB2352), and 9918 (SB2351). Several of the identified spoligotypes had been described previously in other countries, such as Argentina (SB0145 and SB0140) and the United Kingdom (SB0669, SB0673, and SB0140). The UPGMA agglomerative hierarchical clustering algorithm analysis identified 2 defined clusters, showing a dissimilarity ratio (0–5). Cluster one comprises 10 strains, ordered according to their kin relationship by the spoligopattern (T3, T41C, 9920, CO3, CO5, 9912, 9917, 9563, 9914, 9918). The second cluster is composed of 6 strains, in order of their relationship (9926, QP129, 9930, 9524, 9916, 9922). Two M. bovis strains (9927 and 163QR) are not related to any others; thus, they do not belong to any cluster (Figure 1).
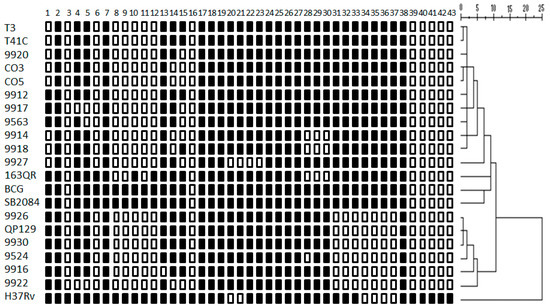
Figure 1.
Tree of spoligotype patterns of M. bovis strains. The unweighted pairwise group method analysis (UPGMA) was performed with multivariate statistical package software using squared Euclidean distance with the hierarchical cluster option and nearest neighbor. The agglomerative hierarchical clustering algorithm analysis identified 2 defined clusters, showing a dissimilarity ratio (0–3). M. bovis field strain SB2084, BCG Montreal strain, and Mycobacterium tuberculosis H37Rv international reference strain were used as controls.
3.2. Mycobacterium Bovis Strains Showed Dissimilar Phagocytosis and Intracellular Survival/Replication Rates
Macrophage phagocytic rates of M. bovis strains showed a dissimilar pattern. They were grouped into five groups based on the statistical differences shown in the ANOVA test (Figure 2, Table 2). Phagocytic rates were as follows: the first group consists of 4 strains with less than one mycobacteria per macrophage (0.16 to 0.87); the second group comprises 6 strains with more than one and less than 2 mycobacteria per macrophage (1.2 to 1.76); the third group consists of 2 strains with 2.21 and 2.53; the fourth group includes 5 strains with more than 3 and less than 4 mycobacteria per macrophage (3.22 to 3.68); and the fifth group includes one strain with 7.43 mycobacteria per macrophage (Figure 2). Based on the intracellular survival/replication rate, strains were grouped in three blocks according to the statistical differences shown by the ANOVA test. The first group consists of one strain (1/18) with less than 35.6% intracellular survival/replication; interestingly enough, this strain has an intracellular survival/replication pattern similar to the avirulent M. bovis BCG. The second group is the largest in number, comprising 14/18 strains, with intracellular survival/replication between 53% to 129.3%. The last group had 3/18 strains, and had an intracellular survival/replication rate between 146% and 158.6% (Figure 3). We defined the analysis by spoligotype clusters in order to seek a relation between the genotype and their capability of surviving inside macrophages (suggesting virulence), but were unable to identify such a relation. Strains belonging to the same spoligotype cluster had different phagocytic and intracellular survival/replication rates between them (p ≤ 0.05).
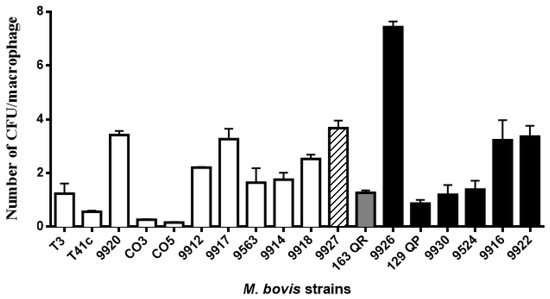
Figure 2.
Macrophage phagocytic rates of M. bovis strains showed a dissimilar pattern. Macrophage monolayers were infected with M. bovis at a multiplicity of infection (MOI) ratio of 10:1 for 4 h. Bacterial phagocytosis was calculated by plating serial dilutions of the live intracellular bacteria released from the macrophages after treatment with 0.5% Tween 20. Total number of colony-forming units was divided by the total number of macrophages to obtain an average bacterial concentration per macrophage. Phagocytic rates were grouped into different blocks based on statistical differences. The color pattern of the bar indicates the group to which it belongs. Results are the average of three independent experiments, each one with three internal replicates.

Table 2.
Macrophage phagocytic and intracellular growth rates. Data summarize percentages of bacterial phagocytosis per macrophage and the intracellular replication. In both cases the strains were grouped according to statistical differences using Tukey’s test.
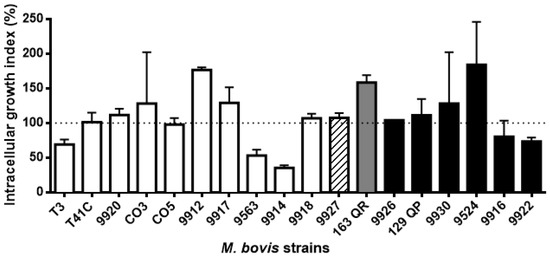
Figure 3.
M. bovis intracellular survival/replication rates showed a high degree of variation among strains. Macrophage monolayers were infected with M. bovis at a multiplicity of infection (MOI) ratio of 10:1 for 4 h; this was considered time 0 h. The cells were harvested at 0 h and 24 h post-infection for analysis. Bacterial growth was calculated as the ratio of the total number of intracellular bacteria at the end of the assay to the total number of bacteria at the start the assay, expressed as percentage. Results are the average of three independent experiments, each one with three internal replicates. Statistical analysis was performed using Tukey’s test. A p value of 0.05 or less was considered statistically significant. The color pattern of each bar indicates the group to which it belongs.
3.3. Spacer Composition Does Not Influence M. bovis Intracellular Survival/Replication in Bovine Macrophages
Strains belonging to the same spoligotype cluster and even to the same spoligotype have different intracellular survival/replication rates. Statistical analysis did not show a specific pattern of association for intracellular survival/replication and phagocytosis with bacterial spoligotype identity. Our results suggest that M. bovis intracellular survival/replication and phagocytosis are independent of the bacterial spoligotype.
4. Discussion
Most of the molecular tools used to classify the Mycobacterium tuberculosis complex isolates rely on sequences that, per se, do not code for virulence factors. For instance, spoligotyping is based on identification of the direct repeat (DR) locus. The DR region is composed of multiple direct variant repeats (DVRs), each of which is composed of a 36-bp direct repeat (DR) plus a non-repetitive spacer sequence of a similar size. Maintenance of this region in the M. tuberculosis complex and the strong sequence conservation of the DRs and spacers among clinical isolates are features that suggest a biological function of this region [19,20,21]. At the present time, the function of the DR region is not known; however, it may play a role in regulating gene expression, and therefore influence, to some extent, the bacteria’s ability to survive the killing machine that is the macrophage.
Our results identified two spoligotype clusters. Strains belonging to each cluster have a small variation in their spacer composition; in some cases, only one spacer is missing, suggesting that the influence of the DR locus in bacterial performance may be similar. In the present study, we used macrophages from a donor with a phenotype of natural disease resistance to intracellular bacterial pathogens, in order to evaluate intracellular survival/replication as a means of identifying virulence of M. bovis strains. Previous results from our laboratory linked the resistance phenotype with some immunological markers, such as nitric oxide. Our experiments not only explore the nitric oxide production on resting macrophages, but also on classical and alternative activated macrophages. Macrophages from resistant donors produced more nitric oxide than susceptible cells. Our previous study demonstrated that nitric oxide is a major determinant of bacterial survival/replication in macrophages [11], [12].
According to our results, phagocytic efficiency and intracellular replication are independent variables within the spoligotype clusters, while strains with similar phagocytic index had different intracellular survival/replication rates. Higher intracellular survival/replication indicates that bacteria possess mechanisms to avoid the macrophage killing machinery; therefore, it may be postulated that the bacteria harbor higher virulence. Our working hypothesis proposed that strains with similar intracellular survival/replication may belong to the same spoligotype cluster; however, our observations did not find any evidence that connected both features. Moreover, strains in the same cluster and even with the same spoligotype showed a differential intracellular survival/replication, suggesting that spoligotype spacer composition could not be associated with the strain’s ability to survive in macrophages. Similar results from a field study support our observations, suggesting that virulence is divergent among strains belonging to the same spoligotype. A macroscopical lesion score showed a variation from 3 to 43 in the degree of tissue damage induced by more than 60 strains isolated from cattle that correspond to spoligotype SB0140 [18,22]. The presence of strains belonging to a particular spoligotype in cattle populations may indicate that bacteria possess a basic group of virulence genes that they can pass among different individuals; however, it is not indicative of the full repertoire of virulence factors that each strain possess. In addition, there is variation in the genetic composition of the host that is not contemplated in field studies. In our in vitro study, we evaluated intracellular survival/replication using macrophages from the same donor, providing us a more comparable setting. A possible pitfall in our study is the limited number of strains included, however, studies with a higher number of strains also observed similar results [18]. In a closely related study, 3 strains from the Beijing family with a dissimilar IS6110 pattern had a similar infective performance when they were inoculated in mice. To summarize, these data indicate that genotype patterns do not correlate with bacterial virulence performance. Further analysis, such as next-generation sequencing, indicates strong association between genotype and virulence [23]. M. bovis spoligotypes identified in this study correspond to spoligotypes reported previously in Mexico, with the exception of SB1498. We also found 3 spoligotype patterns that had not been reported before. All of the spoligotypes lack spacer number 11, suggesting that those strains are part of the European 1 clonal complex (EU1), which is believed to have its origin in the United Kingdom. In addition, it has been hypothesized that this group of strains is derived from the SB0140 spoligotype [24,25,26]. Strains from the same geographical region came from different farms, suggesting that the spoligotype represented is prevalent in the region. However, in some other instances, strains from the same region represent different spoligotypes, suggesting a high spoligotype variation in one geographical area. Our study is by no means an epidemiological survey; however, our results correspond with previous studies that showed high spoligotype diversity among isolates in Mexico [27,28,29,30].
5. Conclusions
Our findings showed that spacer composition similarity is not associated with the bacterial ability to survive inside macrophages from a donor with capacities to control intracellular growth of M. bovis.
Author Contributions
Conceptualization, J.A.G.-P., C.A.-D. and A.B.-G.; methodology, A.B.-G., H.E.-S. and C.R.-T.; software, H.E.-S., A.B.-G. and C.R.-T.; validation, A.B.-G. and C.R.-T.; formal analysis, A.B.-G. and H.E.-S.; investigation, A.B.-G., and C.R.-T.; resources, J.A.G.-P.; data curation, J.A.G.-P., H.E.-S. and A.B.-G.; writing original draft preparation, J.A.G.-P.; writing—review and editing, J.A.G.-P. and A.B.-G.; visualization, J.A.G.-P.; supervision, J.A.G.-P.; project administration, J.A.G.-P.; funding acquisition, J.A.G.-P.
Funding
This research was funded by Grant CB-167488 from CONACYT (Consejo Nacional de Ciencia y Tecnología). Alejandro Benítez-Guzmán is a recipient of the Posdoc-DGAPA (Dirección General de Asuntos del Personal Académico) fellowship.
Acknowledgments
We acknowledge revision of the English language usage, performed by Diego Gutiérrez Fernández.
Conflicts of Interest
The authors declare no conflict of interest.
References
- White, P.C.L.; Böhm, M.; Marion, G.; Hutchings, M.R. Control of bovine tuberculosis in British livestock: There is no “silver bullet”. Trends Microbiol. 2008, 16, 420–427. [Google Scholar] [CrossRef] [PubMed]
- Smith, N.H. The global distribution and phylogeography of Mycobacterium bovis clonal complexes. Infect. Genet. Evol. 2012, 12, 857–865. [Google Scholar] [CrossRef] [PubMed]
- Boniotti, M.B.; Goria, M.; Loda, D.; Garrone, A.; Benedetto, A.; Mondo, A.; Tisato, E.; Zanoni, M.; Zoppi, S.; Dondo, A.; et al. Pacciarini, Molecular typing of Mycobacterium bovis strains isolated in Italy from 2000 to 2006 and evaluation of variable-number tandem repeats for geographically optimized genotyping. J. Clin. Microbiol. 2009, 47, 636–644. [Google Scholar] [CrossRef] [PubMed]
- Duarte, E.L.; Domingos, M.; Amado, A.; Cunha, M.V.; Botelho, A. MIRU-VNTR typing adds discriminatory value to groups of Mycobacterium bovis and Mycobacterium caprae strains defined by spoligotyping. Vet. Microbiol. 2010, 143, 299–306. [Google Scholar] [CrossRef]
- Mbovis.org. Mycobacterium bovis Molecular Typing Database. 2002. Available online: http://www.mbovis.org/.
- Caws, M.; Thwaites, G.; Dunstan, S.; Hawn, T.R.; Lan, N.T.N.; Thuong, N.T.T.; Stepniewska, K.; Huyen, M.N.T.; Bang, N.D.; Loc, T.H.; et al. The influence of host and bacterial genotype on the development of disseminated disease with Mycobacterium tuberculosis. PLoS Pathog. 2008, 4, e1000034. [Google Scholar] [CrossRef]
- Zhang, M.; Gong, J.; Yang, Z.; Samten, B.; Cave, M.D.; Barnes, P.F. Enhanced capacity of a widespread strain of Mycobacterium tuberculosis to grow in human macrophages. J. Infect. Dis. 1999, 179, 1213–1217. [Google Scholar] [CrossRef]
- Theus, S.; Eisenach, K.; Fomukong, N.; Silver, R.F.; Cave, M.D. Beijing family Mycobacterium tuberculosis strains differ in their intracellular growth in THP-1 macrophages SUMMARY. Int. J. 2007, 11, 1087–1093. [Google Scholar]
- Qureshi, T.; Templeton, J.W.; Adams, L.G. Intracellular survival of Brucella abortus, Mycobacterium bovis BCG, Salmonella dublin, and Salmonella typhimurium in macrophages from cattle genetically resistant to Brucella abortus. Vet. Immunol. Immunopathol. 1996, 50, 55–65. [Google Scholar] [CrossRef]
- Gutiérrez-pabello, J.A.; Adams, L.G. Survival of Mycobacterium bovis in macrophages from cattle naturally resistant and susceptible to intracellular pathogens. Vet. México 2003, 34, 277–281. [Google Scholar]
- Esquivel-Solís, H.; Vallecillo, A.J.; Benítez-Guzmán, A.; Adams, L.G.; López-Vidal, Y.; Gutiérrez-Pabello, J.A. Nitric oxide not apoptosis mediates differential killing of Mycobacterium bovis in bovine macrophages. PLoS ONE 2013, 8, e63464. [Google Scholar] [CrossRef]
- Castillo-Velázquez, U.; Aranday-Cortés, E.; Gutiérrez-Pabello, J.A. Alternative activation modifies macrophage resistance to Mycobacterium bovis. Vet. Microbiol. 2011, 151, 51–59. [Google Scholar] [CrossRef]
- Harmon, B.G.; Adams, L.G.; Templeton, J.W.; Smith, R. Macrophage function in mammary glands of Brucella abortus-infected cows and cows that resisted infection after inoculation of Brucella abortus. Am. J. Vet. Res. 1989, 50, 459–465. [Google Scholar] [PubMed]
- León, D.A.; Zumárraga, M.J.; Oropeza, R.J.; Gioffré, A.K.; Bernardelli, A.; Estévez, H.O.; Cataldi, A.A.; Pando, R.H. Mycobacterium bovis with different genotypes and from different hosts induce dissimilar immunopathological lesions in a mouse model of tuberculosis. Clin. Exp. Immunol. 2009, 157, 139–147. [Google Scholar] [CrossRef] [PubMed]
- Alfonseca-Silva, E.; Hernández-Pando, R.; Gutiérrez-Pabello, J.A. Mycobacterium bovis-infected macrophages from resistant and susceptible cattle exhibited a differential pro-inflammatory gene expression profile depending on strain virulence. Vet. Immunol. Immunopathol. 2015, 176, 34–43. [Google Scholar] [CrossRef]
- Theus, S.A.; Cave, M.D.; Eisenach, K.; Walrath, J.; Lee, H.; Mackay, W.; Whalen, C.; Silver, R.F. Differences in the growth of paired Ugandan isolates of Mycobacterium tuberculosis within human mononuclear phagocytes correlate with epidemiological evidence of strain virulence. Infect. Immun. 2006, 74, 6865–6876. [Google Scholar] [CrossRef]
- Wong, K.C.; Leong, W.M.; Law, H.K.W.; Ip, K.F.; Lam, J.T.H.; Yuen, K.Y.; Ho, P.L.; Tse, W.S.; Weng, X.H.; Zhang, W.H.; et al. Molecular characterization of clinical isolates of Mycobacterium tuberculosis and their association with phenotypic virulence in human macrophages. Clin. Vaccine Immunol. 2007, 14, 1279–1284. [Google Scholar] [CrossRef]
- Garbaccio, S.; Macias, A.; Shimizu, E.; Paolicchi, F.; Pezzone, N.; Magnano, G.; Zapata, L.; Abdala, A.; Tarabla, H.; Peyru, M.; et al. Association between spoligotype-VNTR types and virulence of Mycobacterium bovis in cattle. Virulence 2014, 5, 297–302. [Google Scholar] [CrossRef]
- Fang, Z.; Doig, C.; Kenna, D.T.; Smittipat, N.; Palittapongarnpim, P.; Watt, B.; Forbes, K.J. IS6110-mediated deletions of wild-type chromosomes of Mycobacterium tuberculosis. J. Bacteriol. 1999, 181, 1014–1020. [Google Scholar]
- Groenen, P.M.; Bunschoten, A.E.; Soolingen, D.V.; Errtbden, J.D.V. Nature of DNA polymorphism in the direct repeat cluster of Mycobacterium tuberculosis; application for strain differentiation by a novel typing method. Mol. Microbiol. 1993, 10, 1057–1065. [Google Scholar] [CrossRef]
- Van Embden, J.D.A.; van Gorkom, T.; Kremer, K.; Jansen, R.; van der Zeijst, B.A.M.; Schouls, L.M. Genetic variation and evolutionary origin of the direct repeat locus of Mycobacterium tuberculosis complex bacteria. J. Bacteriol. 2000, 182, 2393–2401. [Google Scholar] [CrossRef]
- Wright, D.M.; Allen, A.R.; Mallon, T.R.; McDowell, S.W.J.; Bishop, S.C.; Glass, E.J.; Bermingham, M.L.; Woolliams, J.A.; Skuce, R.A. Field-isolated genotypes of Mycobacterium bovis vary in virulence and influence case pathology but do not affect outbreak size. PLoS ONE 2013, 8, e74503. [Google Scholar] [CrossRef] [PubMed]
- Abdelaal, H.F.M.; Spalink, D.; Amer, A.; Steinberg, H.; Hashish, E.A.; Nasr, E.A.; Talaat, A.M. Genomic polymorphism associated with the emergence of virulent isolates of Mycobacterium bovis in the Nile Delta. Sci. Rep. 2019, 9, 1–15. [Google Scholar] [CrossRef] [PubMed]
- JReyes, A.G.; Casanova, L.G.; Torres, C.R.; Gallegos, S.L.S.; Alarcón, G.J.C.; Pezzat, M.M.; Martínez, O.P.; Chávez, C.E.; Suazo, F.M. Population structure of Mycobacterium bovis isolates from cattle in Mexico. Prev. Vet. Med. 2012, 106, 1–8. [Google Scholar] [CrossRef]
- Rodwell, T.C.; Kapasi, A.J.; Moore, M.; Milian-Suazo, F.; Harris, B.; Guerrero, L.P.; Moser, K.; Strathdee, S.A.; Garfein, R.S. Tracing the origins of Mycobacterium bovis tuberculosis in humans in the USA to cattle in Mexico using spoligotyping. Int. J. Infect. Dis. 2010, 14, e129–e135. [Google Scholar] [CrossRef] [PubMed]
- MZumárraga, J.; Arriaga, C.; Barandiaran, S.; Cobos-Marín, L.; de Waard, J.; Estrada-Garcia, I.; Figueiredo, T.; Figueroa, A.; Giménez, F.; Gomes, H.M.; et al. Understanding the relationship between Mycobacterium bovis spoligotypes from cattle in Latin American Countries. Res. Vet. Sci. 2013, 94, 9–21. [Google Scholar] [CrossRef] [PubMed]
- Milian-Suazo, F.; Harris, B.; Díaz, C.A.; Torres, C.R.; Stuber, T.; Ojeda, G.A.; Loredo, A.M.; Soria, M.P.; Payeur, J.B. Molecular epidemiology of Mycobacterium bovis: Usefulness in international trade. Prev. Vet. Med. 2008, 87, 261–271. [Google Scholar] [CrossRef]
- Cobos-Marín, L.; Montes-Vargas, J.; Zumarraga, M.; Cataldi, A.; Romano, M.I.; Estrada-Garcia, I.; Gonzalez-y-Merchand, J.A. Spoligotype analysis of Mycobacterium bovis isolates from Northern México. Can. J. Microbiol. 2005, 51, 996–1000. [Google Scholar] [CrossRef]
- Santillan-Flores, M.A.; Flores, J.; Arriaga-Diaz, C.; Romero-Torres, C.; Suárez-Güemes, F.; Espitia, C. Polymorphism of the PE domain of PE/PE_PGRS sequences in clinical isolates of Mycobacterium bovis in Mexico. Vet. Microbiol. 2006, 115, 364–369. [Google Scholar] [CrossRef]
- Milián-Suazo, F.; Banda-Ruíz, V.; Ramírez-Casillas, C.; Arriaga-Díaz, C. Genotyping of Mycobacterium bovis by geographic location within Mexico. Prev. Vet. Med. 2002, 55, 255–264. [Google Scholar] [CrossRef]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).