Host miR-146a-3p Facilitates Replication of Infectious Hematopoietic Necrosis Virus by Targeting WNT3a and CCND1
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Cells and Virus
2.2. Reagents and Antibodies
2.3. Virus Infection and Titer Determination
2.4. miRNA Mimics and Inhibitors
2.5. Plasmid Construction
2.6. Cell Transfection
2.7. RNA Interference
2.8. Quantitative Reverse Transcription-PCR (qRT-PCR)
2.9. Luciferase Reporter Assays
2.10. Immunoblot
2.11. Statistical Analysis
3. Results
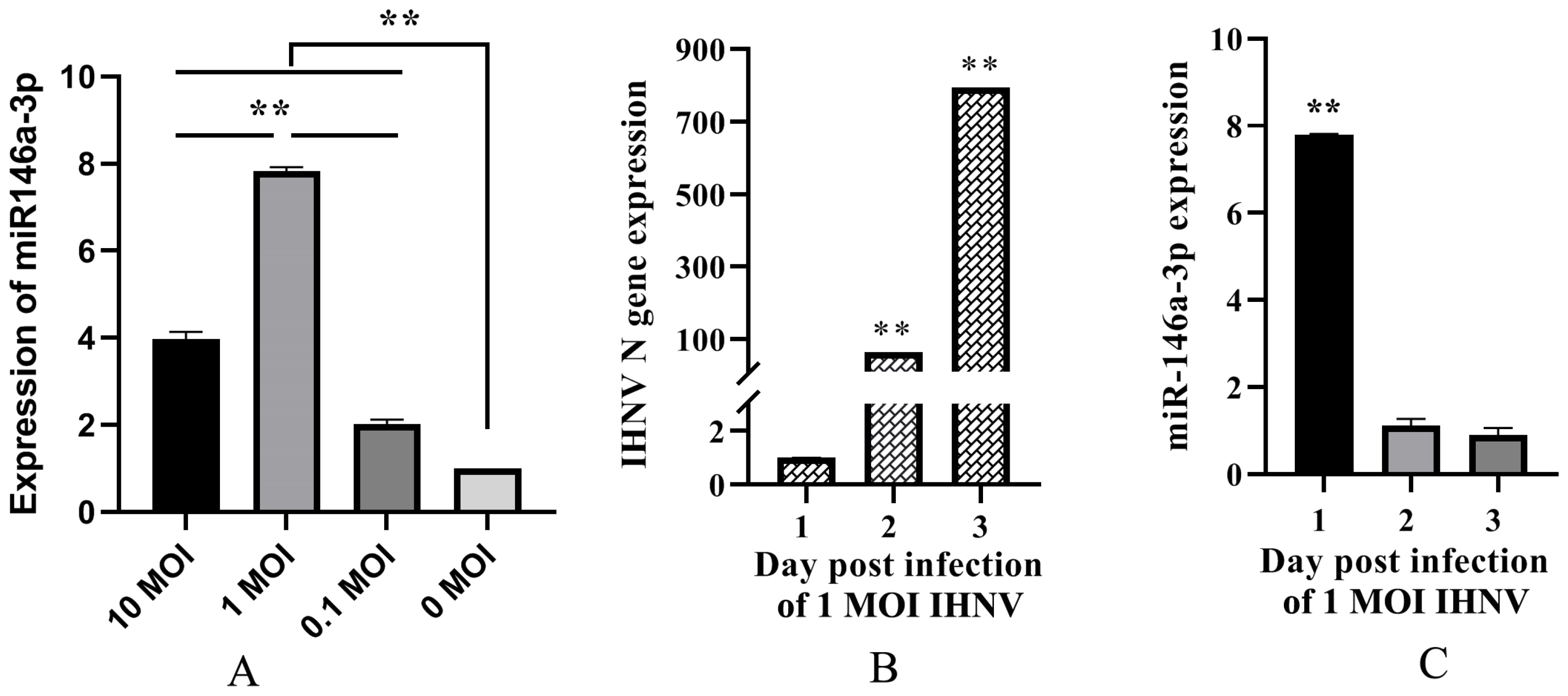
3.1. IHNV Infection Upregulated the Expression of miR-146a-3p
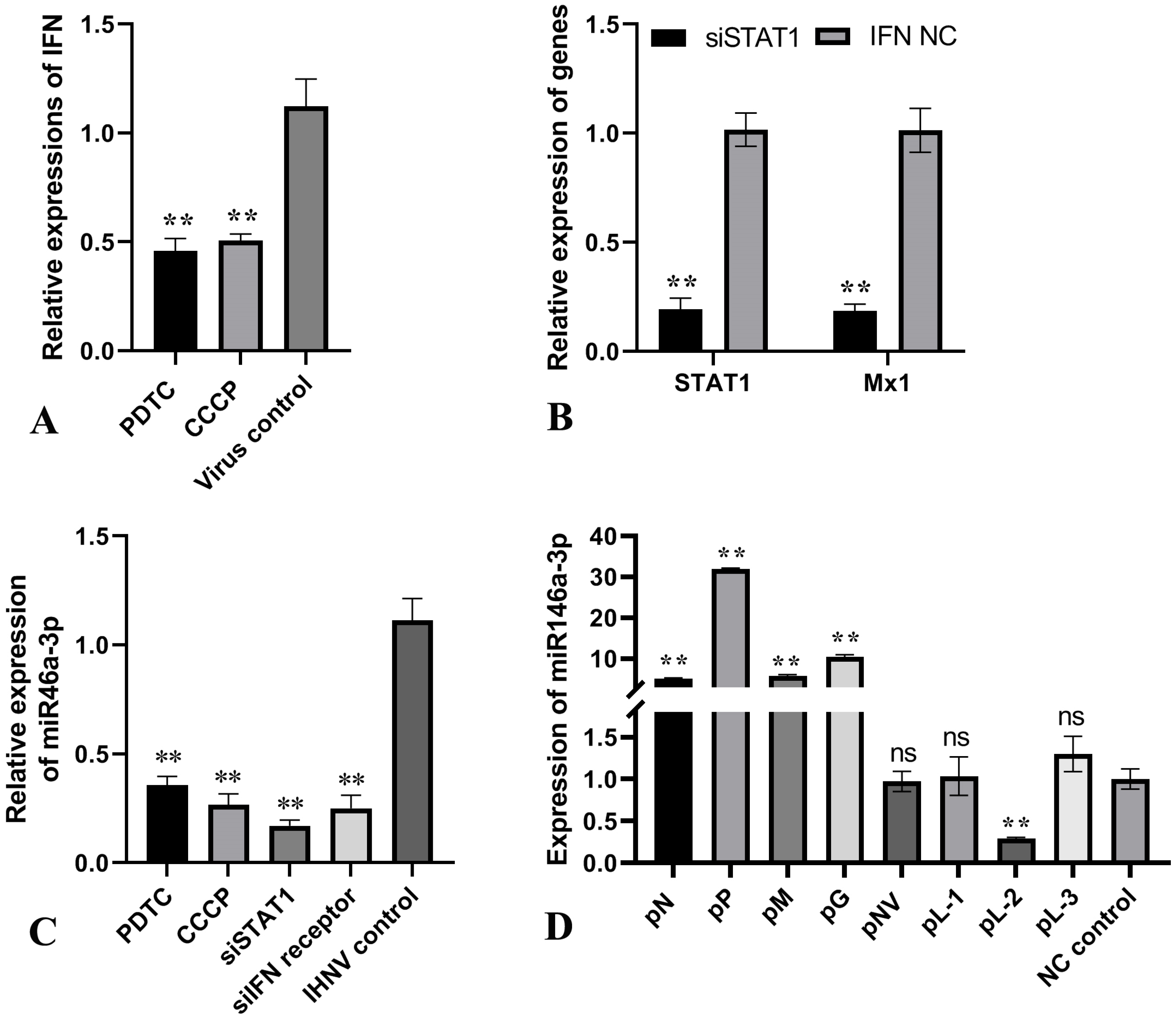
3.2. The Induction of miR-146a-3p Was Associated with IHNV Inducing IFN Response
3.3. Identification of the Potential Target Genes of miR-146a-3p
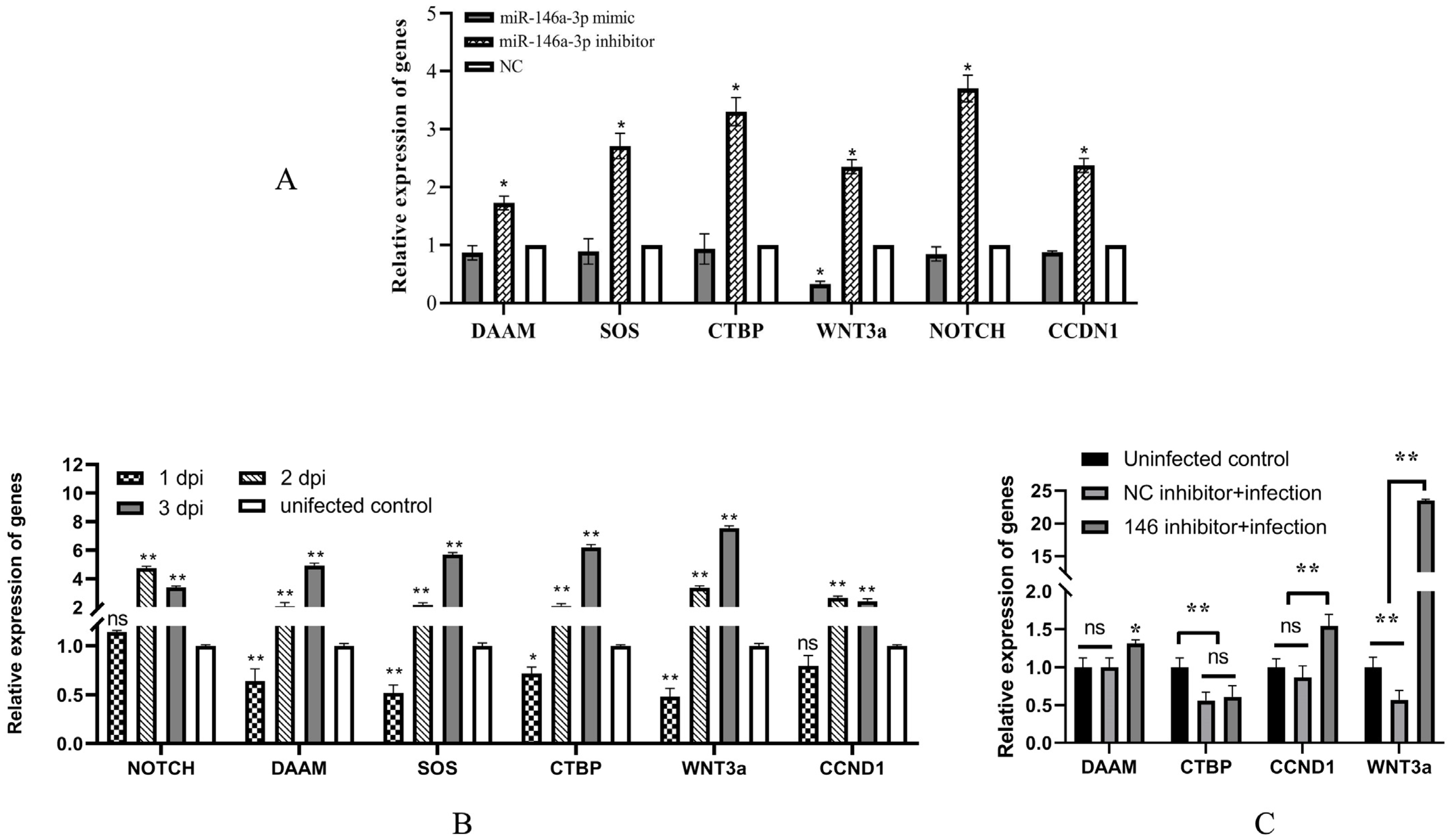
3.4. IHNV Downregulates the Expression of Potential Target Genes at Early Time after Infection
3.5. miR-146a-3p Participates in the Repression of Potential Target Genes by IHNV
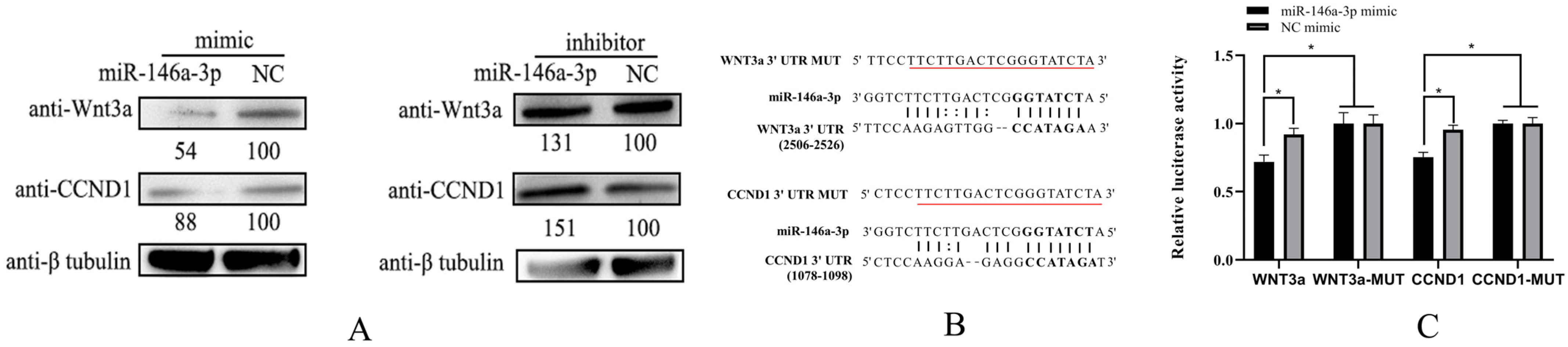
3.6. WNT3a and CCND1 Are the Target Genes of miR-146a-3p
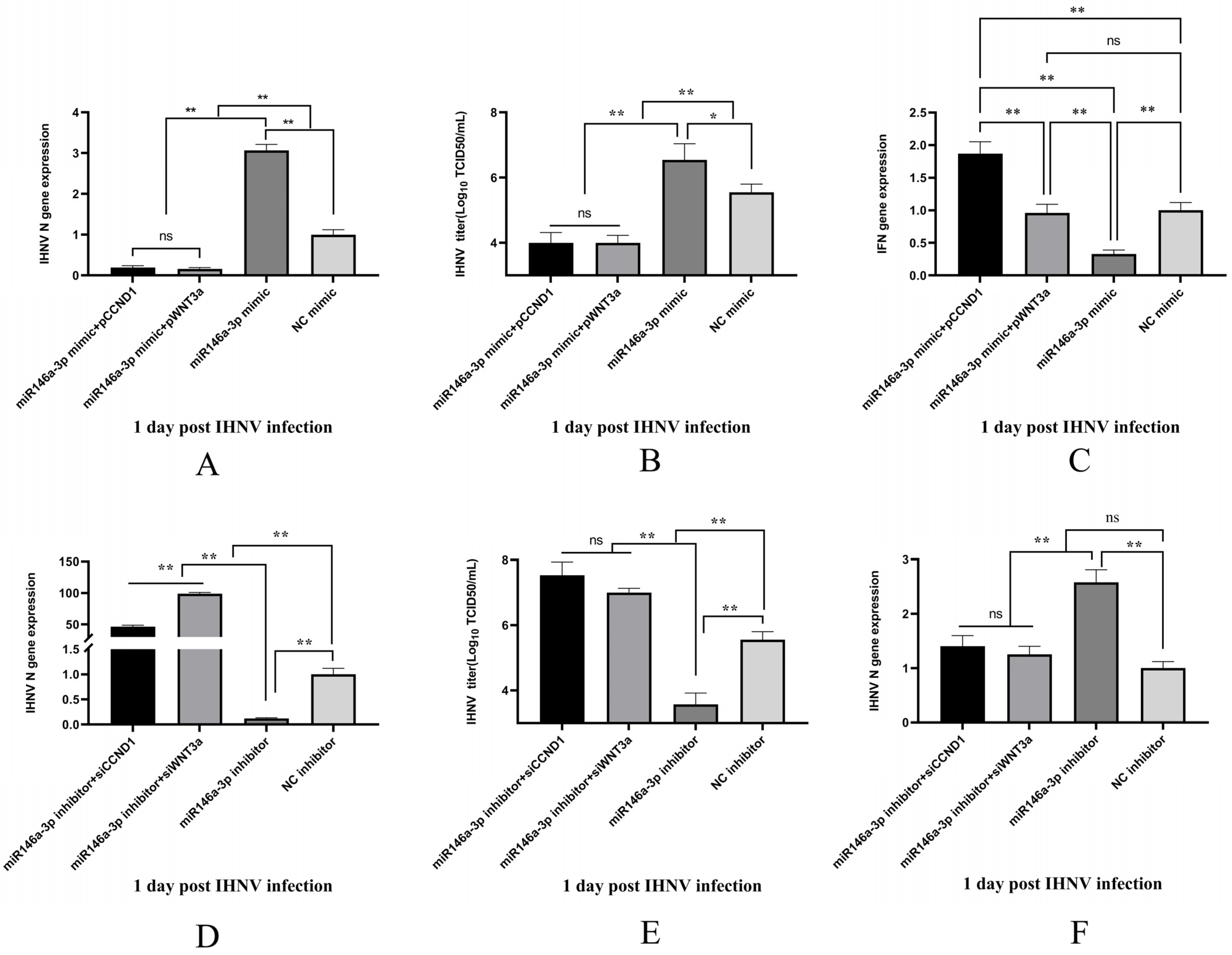
3.7. The Roles of miR-146a-3p in IHNV Infection Were Dependent on WNT3a and CCND1
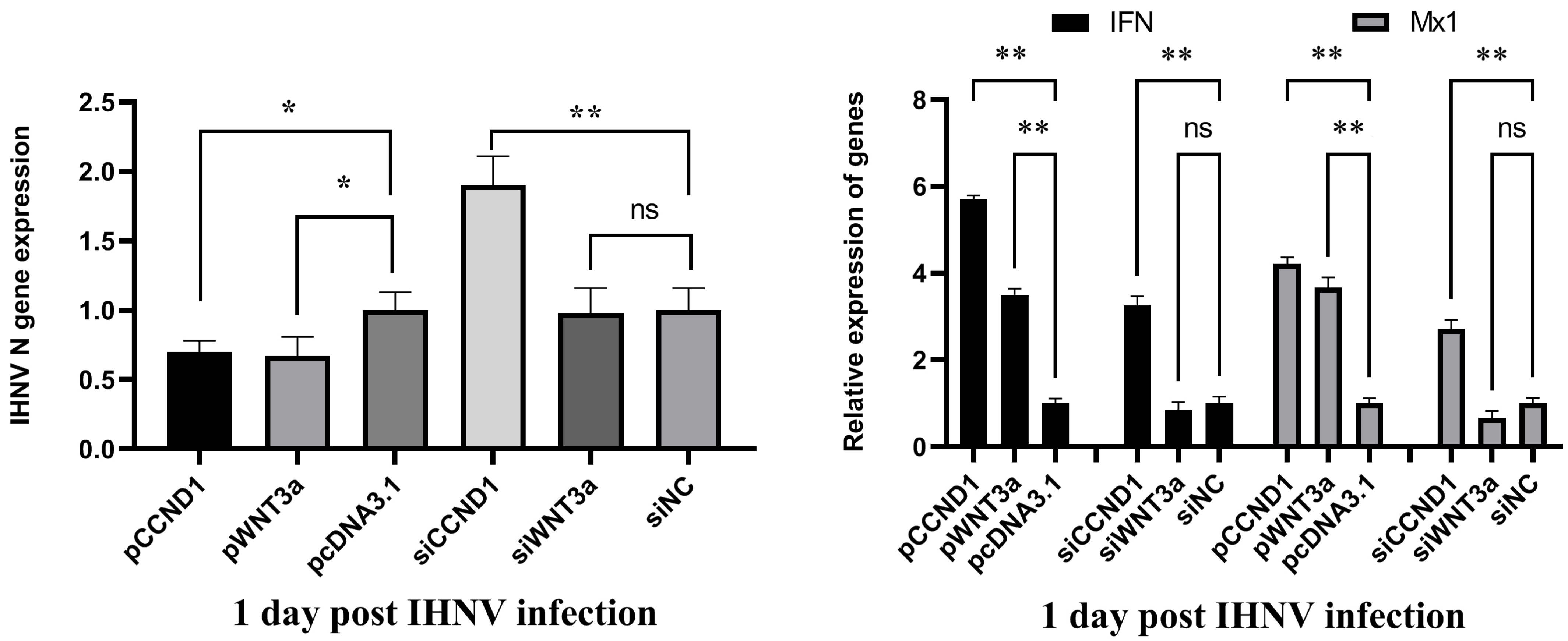
3.8. WNT3a and CCND1 Inhibited IHNV Replication
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- FAO. The state of world fisheries and aquaculture 2022. In Towards Blue Transformation; FAO: Rome, Italy, 2022. [Google Scholar]
- Dixon, P.; Paley, R.; Alegria-Moran, R.; Oidtmann, B. Epidemiological characteristics of infectious hematopoietic necrosis virus (ihnv): A review. Vet. Res. 2016, 47, 63. [Google Scholar] [CrossRef] [PubMed]
- Louboutin, L.; Cabon, J.; Vigouroux, E.; Morin, T.; Danion, M. Comparative analysis of the course of infection and the immune response in rainbow trout (oncorhynchus mykiss) infected with the 5 genotypes of infectious hematopoietic necrosis virus. Virology 2021, 552, 20–31. [Google Scholar] [CrossRef] [PubMed]
- Muller, A.; Sutherland, B.J.G.; Koop, B.; Johnson, S.C.; Garver, K.A. Infectious hematopoietic necrosis virus (ihnv) persistence in sockeye salmon: Influence on brain transcriptome and subsequent response to the viral mimic poly(i:C). BMC Genom. 2015, 16, 634. [Google Scholar] [CrossRef]
- Cao, Y.; Zhang, Q.; Xu, L.; Li, S.; Wang, D.; Zhao, J.; Liu, H.; Feng, J.; Lu, T. Effects of different cytokines on immune responses of rainbow trout in a virus DNA vaccination model. Oncotarget 2017, 8, 112222–112235. [Google Scholar] [CrossRef] [PubMed]
- Cao, Y.; Xu, L.; LaPatra, S.E.; Zhao, J.; Liu, M.; Liu, H.; Lu, T.; Zhang, Q. The kinetics and protection of the antiviral state induced by recombinant iifn1a in rainbow trout against infectious hematopoietic necrosis virus. Mol. Immunol. 2016, 76, 55–61. [Google Scholar] [CrossRef]
- Corbeil, S.; LaPatra, S.E.; Anderson, E.D.; Kurath, G. Nanogram quantities of a DNA vaccine protect rainbow trout fry against heterologous strains of infectious hematopoietic necrosis virus. Vaccine 2000, 18, 2817–2824. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.X.; Zhou, Y.; Lu, L.F.; Lu, X.B.; Ni, B.; Liu, M.X.; Guan, H.X.; Li, S.; Zhang, Y.A.; Ouyang, S. Infectious hematopoietic necrosis virus n protein suppresses fish ifn1 production by targeting the mita. Fish. Shellfish. Immunol. 2020, 97, 523–530. [Google Scholar] [CrossRef] [PubMed]
- Biacchesi, S.; Merour, E.; Chevret, D.; Lamoureux, A.; Bernard, J.; Bremont, M. Nv proteins of fish novirhabdovirus recruit cellular ppm1bb protein phosphatase and antagonize rig-i-mediated ifn induction. Sci. Rep. 2017, 7, 44025. [Google Scholar] [CrossRef]
- Chiou, P.P.; Kim, C.H.; Ormonde, P.; Leong, J.A. Infectious hematopoietic necrosis virus matrix protein inhibits host-directed gene expression and induces morphological changes of apoptosis in cell cultures. J. Virol. 2000, 74, 7619–7627. [Google Scholar] [CrossRef] [PubMed]
- Gorgoglione, B.; Ringiesn, J.L.; Pham, L.H.; Shepherd, B.S.; Leaman, D.W. Comparative effects of novirhabdovirus genes on modulating constitutive transcription and innate antiviral responses, in different teleost host cell types. Virol. J. 2020, 17, 110. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Gui, J.-F. Virus genomes and virus-host interactions in aquaculture animals. Sci. China Life Sci. 2015, 58, 156–169. [Google Scholar] [CrossRef] [PubMed]
- Mehta, A.; Baltimore, D. Micrornas as regulatory elements in immune system logic. Nat. Rev. Immunol. 2016, 16, 279–294. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Tu, J.G.; Zhang, Y.A. Microrna regulation of viral replication in teleost fish: A review. Rev. Aquacult. 2021, 13, 1367–1378. [Google Scholar] [CrossRef]
- Chu, Q.; Xu, T.J.; Zheng, W.W.; Chang, R.J.; Zhang, L. Long noncoding rna marl regulates antiviral responses through suppression mir-122-dependent mavs downregulation in lower vertebrates. PLoS Pathog. 2020, 16, e1008670. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Chu, Q.; Chang, R.; Xu, T. Inducible microrna-217 inhibits nf-κb– and irf3-driven immune responses in lower vertebrates through targeting tak1. J. Immunol. 2020, 205, 1620–1632. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.C.; Zhou, Z.J.; Sun, L. Pol-mir-731, a teleost mirna upregulated by megalocytivirus, negatively regulates virus-induced type i interferon response, apoptosis, and cell cycle arrest. Sci. Rep. 2016, 6, 28354. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Wang, Q.; Liu, A.Q.; Zhang, C.; Liu, L.H.; Lu, L.F.; Tu, J.G.; Zhang, Y.A. Microrna mir-155 inhibits cyprinid herpesvirus 3 replication via regulating ampk-mavs-ifn axis. Dev. Comp. Immunol. 2022, 129, 104335. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Feng, S.; Zhang, W.; Chen, N.; Hegazy, A.M.; Chen, W.; Liu, X.; Zhao, L.; Li, J.; Lin, L.; et al. Microrna mir-214 inhibits snakehead vesiculovirus replication by promoting ifn-α expression via targeting host adenosine 5′-monophosphate-activated protein kinase. Front. Immunol. 2017, 8, 1775. [Google Scholar] [CrossRef] [PubMed]
- Cao, Y.; Wang, D.; Li, S.; Xu, L.; Zhao, J.; Liu, H.; Lu, T.; Zhang, Q. Identification and analysis of differentially expressed micrornas in rainbow trout (oncorhynchus mykiss) responding to infectious hematopoietic necrosis virus infection. Dev. Comp. Immunol. 2018, 88, 28–36. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, T.G.M.; Batz, L.; Bonet, L.; Carl, U.; Holzapfel, G.; Kiem, K.; Matulewicz, K.; Niermeier, D.; Schuchardt, I.; Stanar, K. Development of the twin-strep-tag® and its application for purification of recombinant proteins from cell culture supernatants. Protein Expr. Purif. 2013, 92, 54–61. [Google Scholar] [CrossRef] [PubMed]
- Hou, J.; Wang, P.; Lin, L.; Liu, X.; Ma, F.; An, H.; Wang, Z.; Cao, X. Microrna-146a feedback inhibits rig-i-dependent type i ifn production in macrophages by targeting traf6, irak1, and irak2. J. Immunol. 2009, 183, 2150–2158. [Google Scholar] [CrossRef] [PubMed]
- Németh, Z.H.; Deitch, E.A.; Szabó, C.; Haskó, G. Pyrrolidinedithiocarbamate inhibits nf-kappab activation and il-8 production in intestinal epithelial cells. Immunol. Lett. 2003, 85, 41–46. [Google Scholar] [CrossRef] [PubMed]
- Kwon, D.; Park, E.; Sesaki, H.; Kang, S.J. Carbonyl cyanide 3-chlorophenylhydrazone (cccp) suppresses sting-mediated DNA sensing pathway through inducing mitochondrial fission. Biochem. Biophys. Res. Commun. 2017, 493, 737–743. [Google Scholar] [CrossRef] [PubMed]
- Skalsky, R.L.; Cullen, B.R. Viruses, micrornas, and host interactions. Annu. Rev. Microbiol. 2010, 64, 123–141. [Google Scholar] [CrossRef] [PubMed]
- Hum, C.; Loiselle, J.; Ahmed, N.; Shaw, T.A.; Toudic, C.; Pezacki, J.P. Microrna mimics or inhibitors as antiviral therapeutic approaches against COVID-19. Drugs 2021, 81, 517–531. [Google Scholar] [CrossRef] [PubMed]
- Saba, R.; Sorensen, D.L.; Booth, S.A. Microrna-146a: A dominant, negative regulator of the innate immune response. Front. Immunol. 2014, 5, 578. [Google Scholar] [CrossRef]
- Tang, Y.; Luo, X.; Cui, H.; Ni, X.; Yuan, M.; Guo, Y.; Huang, X.; Zhou, H.; de Vries, N.; Tak, P.P.; et al. Microrna-146a contributes to abnormal activation of the type i interferon pathway in human lupus by targeting the key signaling proteins. Arthritis Rheum. 2009, 60, 1065–1075. [Google Scholar] [CrossRef] [PubMed]
- Bruscella, P.; Bottini, S.; Baudesson, C.; Pawlotsky, J.M.; Feray, C.; Trabucchi, M. Viruses and mirnas: More friends than foes. Front. Microbiol. 2017, 8, 824. [Google Scholar] [CrossRef] [PubMed]
- Pedersen, I.M.; Cheng, G.; Wieland, S.; Volinia, S.; Croce, C.M.; Chisari, F.V.; David, M. Interferon modulation of cellular micrornas as an antiviral mechanism. Nature 2007, 449, 919–922. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Hou, J.; Lin, L.; Wang, C.M.; Liu, X.G.; Li, D.; Ma, F.; Wang, Z.G.; Cao, X.T. Inducible microrna-155 feedback promotes type i ifn signaling in antiviral innate immunity by targeting suppressor of cytokine signaling 1. J. Immunol. 2010, 185, 6226–6233. [Google Scholar] [CrossRef] [PubMed]
- He, M.; Ding, N.Z.; He, C.Q. Novirhabdoviruses versus fish innate immunity: A review. Virus Res. 2021, 304, 198525. [Google Scholar] [CrossRef] [PubMed]
- Girardit, E.; Lopez, P.; Pfeffer, S. On the importance of host micrornas during viral infection. Front. Genet. 2018, 9, 439. [Google Scholar] [CrossRef] [PubMed]
- John, B.; Enright, A.J.; Aravin, A.; Tuschl, T.; Sander, C.; Marks, D.S. Human microrna targets. PLoS Biol. 2004, 2, 1862–1879. [Google Scholar] [CrossRef] [PubMed]
- Ljungberg, J.K.; Kling, J.C.; Tran, T.T.; Blumenthal, A. Functions of the wnt signaling network in shaping host responses to infection. Front. Immunol. 2019, 10, 2521. [Google Scholar] [CrossRef] [PubMed]
- Yap, L.F.; Ahmad, M.; Zabidi, M.M.; Chu, T.L.; Chai, S.J.; Lee, H.M.; Lim, P.V.; Wei, W.; Dawson, C.; Teo, S.H.; et al. Oncogenic effects of wnt5a in epstein-barr virus-associated nasopharyngeal carcinoma. Int. J. Oncol. 2014, 44, 1774–1780. [Google Scholar] [CrossRef] [PubMed]
- Zhu, X.; Wen, L.; Sheng, S.; Wang, W.; Xiao, Q.; Qu, M.; Hu, Y.; Liu, C.; He, K. Porcine circovirus-like virus p1 inhibits wnt signaling pathway in vivo and in vitro. Front. Microbiol. 2018, 9, 390. [Google Scholar] [CrossRef] [PubMed]
- Hao, H.P.; Wen, L.B.; Li, J.R.; Wang, Y.; Ni, B.; Wang, R.; Wang, X.; Sun, M.X.; Fan, H.J.; Mao, X. Licl inhibits prrsv infection by enhancing wnt/β-catenin pathway and suppressing inflammatory responses. Antiviral Res. 2015, 117, 99–109. [Google Scholar] [CrossRef] [PubMed]
- More, S.; Yang, X.; Zhu, Z.; Bamunuarachchi, G.; Guo, Y.; Huang, C.; Bailey, K.; Metcalf, J.P.; Liu, L. Regulation of influenza virus replication by wnt/β-catenin signaling. PLoS ONE 2018, 13, e0191010. [Google Scholar] [CrossRef] [PubMed]
- Baril, M.; Es-Saad, S.; Chatel-Chaix, L.; Fink, K.; Pham, T.N.Q.; Raymond, V.A.; Audette, K.; Guenier, A.-S.; Duchaine, J.; Servant, M.J.; et al. Genome-wide rnai screen reveals a new role of a wnt/ctnnb1 signaling pathway as negative regulator of virus-induced innate immune responses. PLoS Pathog. 2013, 9, e1003416. [Google Scholar] [CrossRef] [PubMed]
| Gene Name | Primer | Sequences (5′ to 3′) | Application |
|---|---|---|---|
| NOTCH | NOTCH-1 | AGGCCAACAGGTAAAACGATCAGAT | qRT-PCR |
| NOTCH-2 | CAGTGTCAGAACCCCACCTCACGAT | ||
| DAAM | DAAM-1 | ATCAGGAGTGACTCTCTAAGCTATT | |
| DAAM-2 | TCCGCCAGGGTCACTGCGTGTGT | ||
| SOS | SOS-1 | ATTCTCCCAGGTTTCTTCCCAAC | |
| SOS-2 | TGGTACACTGATATTGCCGTCAG | ||
| CTBP | CTBP-1 | GGCAAATCAGAGTTGGACAGGGA | |
| CTBP-2 | CTGTAATGGACCTCTGTGAGTCT | ||
| WNT3a | WNT3a-1 | ACAATCTTCTATCAATGCCTCGTCG | |
| WNT3a-2 | AACTCACTTTAGACTCTCTGACCAC | ||
| CCND1 | CCND1-1 | ACGGACACCAAGAGCATGGATGA | |
| CCND1-2 | CCAGACCAGTCCCTGTGCTATGA | ||
| IFN-I | IFN-I-1 | AGAATGCCCCAGTCCTTTTCC | |
| IFN-I-2 | GACTTTGTCCTCAAACTCAGCATCA | ||
| Mx | Mx-1 | GGTTGTGCCATGCAACGTT | |
| Mx-2 | GGCTTGGTCAGGATGCCTAAT | ||
| IHNV N | IHNV N-1 | TGTGCATGAAGTCAGTGGTGG | |
| IHNV N-2 | CCTGCTCATCATGACACCGTA | ||
| β-actin | β-actin-1 | GCCGGCCGCGACCTCACAGACTAC | |
| β-actin-2 | CGGCCGTGGTGGTGAAGCTGTAAC | ||
| miR-146a-3p | miR-146a-3p-1 | TCGGCAGGATCTATGGGCTCAGT | |
| 18S rRNA | 18S rRNA-1 | CGGAGGTTCGAAGACGATCA | |
| 18S rRNA-2 | TCGCTAGTTGGCATCGTTTAT | ||
| WNT3a-UTR | WNT3a-U1 | CCGCTCGAGACAATCTTCTATCAATGCCTCGTCG | Reporter plasmid |
| WNT3a-U2 | GCGTCGACAACTCACTTTAGACTCTCTGACCAC | ||
| WNT3a-MUT | WNT3a-mU1 | TAGATACCCGAGTCAAGAAGACCACTTTCTTCAAAGAGCTTGCAG | |
| WNT3a-mU2 | GGTCTTCTTGACTCGGGTATCTACGATAGATGATAAAAAACAAAGT | ||
| CCND1-UTR | CCND1-U1 | CCGCTCGAGACGGACACCAAGAGCATGGATGA | |
| CCND1-U2 | GCGTCGACCCAGACCAGTCCCTGTGCTATGA | ||
| CCND1-MUT | CCND1-mU1 | TAGATACCCGAGTCAAGAAGACCTGCACCAGACAACACTGTCCCA | |
| CCND1-mU2 | GGTCTTCTTGACTCGGGTATCTAGGAGGAGAGGGCTTCGGGGAGA | ||
| N | pN-1 | GGATCCACAAGCGCACTCAGAGAGA | Expression plasmid construction |
| pN-2 | GAATTCTCAGTGGAATGAGTCGGAGT | ||
| P | pP-1 | GGTACCTCAGATGGAGAAGGAGAAC | |
| pP-2 | GAATTCCTATTGACCCTGCTTCATGC | ||
| M | pM-1 | GGATCCTCCATTTTCAAGAGAGCA | |
| pM-2 | GGAATTCCTATTTTTCCTTCCCCCGT | ||
| G | pG-1 | GGATCCGACACCATGATCACCACT | |
| pG-2 | GAATTCTTAGGACCGGTTTGCCAG | ||
| NV | pNV-1 | GGATCCGACCACCGTGAAATAAAC | |
| pNV-2 | GAATTCCTATCTGGGATAAGCAAG | ||
| L1 | pL1-1 | GGTACCGACTTCTTCGATCTCGACA | |
| pL1-2 | GGATCCCTACATGACGCGTTCTACCCT | ||
| L2 | pL2-1 | GGATCCCAGAAAACAGCGCTCACCCA | |
| pL2-2 | GAATTCCTATTCCATGGGCATTGAGTA | ||
| L3 | pL3-1 | GGTACCTCACAACGGCTCCTCCAC | |
| pL3-2 | GAATTCCTATGGTTCGCCTAGTGG | ||
| WNT3a | pWNT3a-1 | GGATCCGCCACCATGTTTTGCGAAACTTTTTT | |
| pWNT3a-2 | CTCGAGTCATTTGCAGGTGTGAACAT | ||
| CCND1 | pCCND1-1 | GGATCCGCCACCATGGAACGTCAGTTGCTGTG | |
| pCCND1-2 | CTCGAGTCAGATGTTCACATCTCTCA |
| Gene_ID | KEGG | KEGG_name | Fold_change | Pathway |
|---|---|---|---|---|
| 110538410 | K02599 | NOTCH | 0.81 | Notch signaling pathway |
| 110505746 | K04512 | DAAM | 0.84 | Wnt signaling pathway |
| 110493792 | K03099 | SOS | 0.88 | MAPK signaling pathway |
| 110504746 | K04496 | CTBP | 0.90 | Wnt signaling pathway |
| 110530975 | K00312 | WNT3a | 0.16 | Wnt signaling pathway |
| 110499952 | K04503 | CCND1 | 0.83 | Wnt signaling pathway |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Huang, J.; Zheng, S.; Li, Q.; Zhao, H.; Zhou, X.; Yang, Y.; Zhang, W.; Cao, Y. Host miR-146a-3p Facilitates Replication of Infectious Hematopoietic Necrosis Virus by Targeting WNT3a and CCND1. Vet. Sci. 2024, 11, 204. https://doi.org/10.3390/vetsci11050204
Huang J, Zheng S, Li Q, Zhao H, Zhou X, Yang Y, Zhang W, Cao Y. Host miR-146a-3p Facilitates Replication of Infectious Hematopoietic Necrosis Virus by Targeting WNT3a and CCND1. Veterinary Sciences. 2024; 11(5):204. https://doi.org/10.3390/vetsci11050204
Chicago/Turabian StyleHuang, Jingwen, Shihao Zheng, Qiuji Li, Hongying Zhao, Xinyue Zhou, Yutong Yang, Wenlong Zhang, and Yongsheng Cao. 2024. "Host miR-146a-3p Facilitates Replication of Infectious Hematopoietic Necrosis Virus by Targeting WNT3a and CCND1" Veterinary Sciences 11, no. 5: 204. https://doi.org/10.3390/vetsci11050204
APA StyleHuang, J., Zheng, S., Li, Q., Zhao, H., Zhou, X., Yang, Y., Zhang, W., & Cao, Y. (2024). Host miR-146a-3p Facilitates Replication of Infectious Hematopoietic Necrosis Virus by Targeting WNT3a and CCND1. Veterinary Sciences, 11(5), 204. https://doi.org/10.3390/vetsci11050204






