Molecular Detection of Candidatus Anaplasma camelii in Naturally Infected Dromedary Camels (Camelus dromedarius) in Abu Dhabi Emirate, United Arab Emirates, 2019–2023
Abstract
Simple Summary
Abstract
1. Introduction
2. Methods
2.1. Sampling
2.2. DNA Extraction
2.3. Polymerase Chain Reaction
2.4. Sanger Sequencing
2.5. Sequence Alignment and Phylogenetic Analysis
3. Results
3.1. Clinical Status of the Sampled Animals
3.2. PCR Data
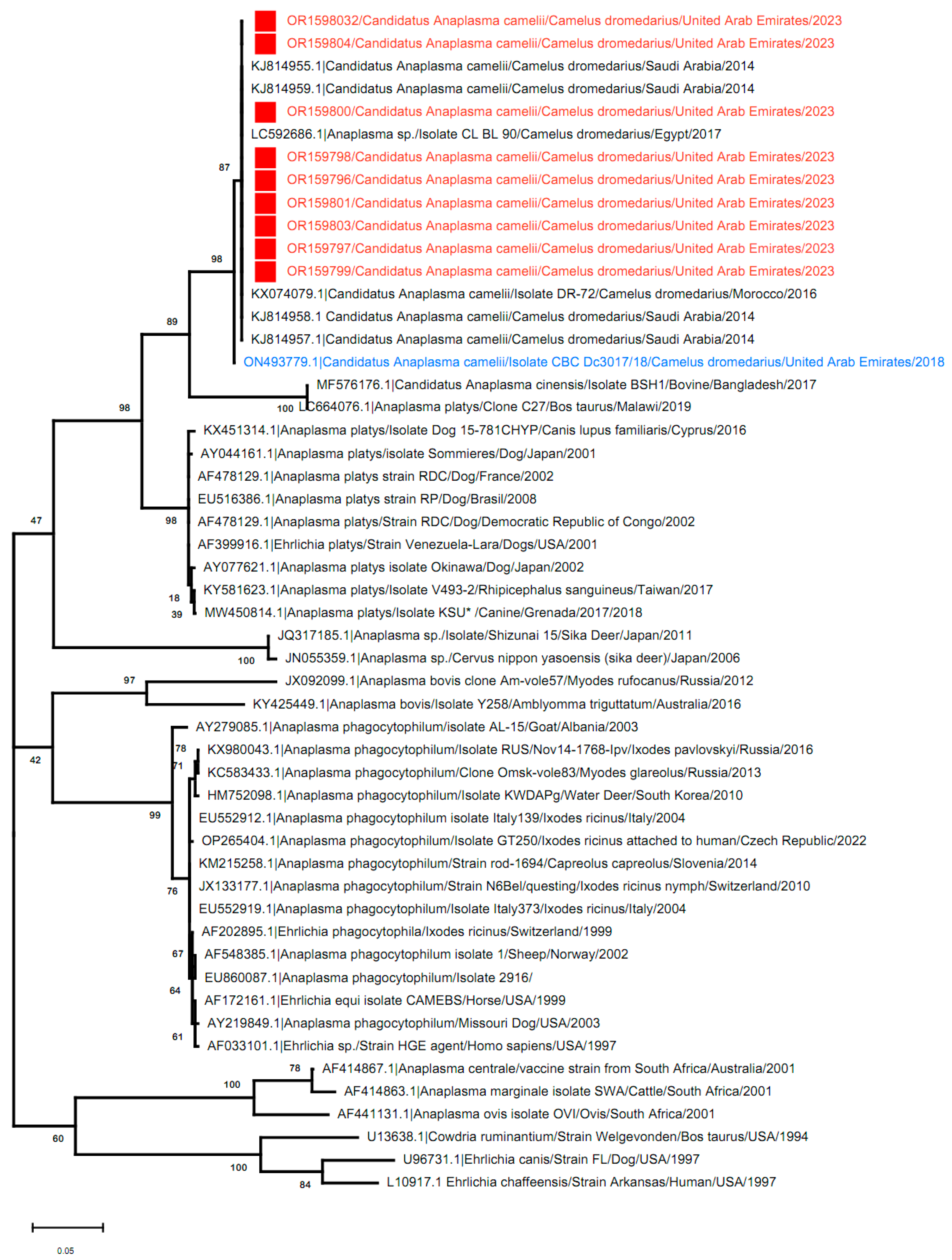
3.3. Phylogenetic Analysis
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Tefera, M.; Gebreah, F. A study on the productivity and diseases of camels in eastern Ethiopia. Trop. Anim. Health Prod. 2001, 33, 265–274. [Google Scholar] [CrossRef]
- Blackwell, P. East Africa’s Pastoralist Emergency: Is climate change the straw that breaks the camel’s back? Third World Q. 2010, 31, 1321–1338. [Google Scholar] [CrossRef]
- McAdam, A.; Sharpe, A.H. Robbins & Cotran Pathological Basis of Disease, 8th ed.; Kumar, V., Abbas, A.B., Fausto, N., Aster, J.C., Eds.; Infectious Diseases; Elsevier: Philadelphia, PA, USA, 2010; pp. 331–398. [Google Scholar]
- Mubashir, M.; Tariq, M.; Khan, M.S.; Safdar, M.; Özaslan, M.; Imran, M.; Ullah, Q.; Siddique, F.; Junejo, Y. Review on anaplasmosis in different ruminants. Zeugma Biol. Sci. 2022, 3, 32–45. [Google Scholar]
- Dunning Hotopp, J.C.; Lin, M.; Madupu, R.; Crabtree, J.; Angiuoli, S.V.; Eisen, J.; Seshadri, R.; Ren, Q.; Wu, M.; Utterback, T.R. Comparative genomics of emerging human ehrlichiosis agents. PLoS Genet. 2006, 2, e21. [Google Scholar]
- Hove, P.; Khumalo, Z.T.; Chaisi, M.E.; Oosthuizen, M.C.; Brayton, K.A.; Collins, N.E. Detection and Characterisation of Anaplasma marginale and A. centrale in South Africa. Vet. Sci. 2018, 5, 26. [Google Scholar] [CrossRef]
- Karshima, S.N.; Ahmed, M.I.; Mohammed, K.M.; Pam, V.A.; Momoh-Abdullateef, H.; Gwimi, B.P. Worldwide meta-analysis on Anaplasma phagocytophilum infections in animal reservoirs: Prevalence, distribution and reservoir diversity. Vet. Parasitol. Reg. Stud. Rep. 2023, 38, 100830. [Google Scholar] [CrossRef] [PubMed]
- Kolo, A. Anaplasma Species in Africa—A Century of Discovery: A Review on Molecular Epidemiology, Genetic Diversity, and Control. Pathogens 2023, 12, 702. [Google Scholar] [CrossRef]
- Cabezas-Cruz, A.; Gallois, M.; Fontugne, M.; Allain, E.; Denoual, M.; Moutailler, S.; Devillers, E.; Zientara, S.; Memmi, M.; Chauvin, A. Epidemiology and genetic diversity of Anaplasma ovis in goats in Corsica, France. Parasites Vectors 2019, 12, 3. [Google Scholar] [CrossRef]
- Iqbal, N.; Mukhtar, M.U.; Yang, J.; Sajid, M.S.; Niu, Q.; Guan, G.; Liu, Z.; Yin, H. First molecular evidence of Anaplasma bovis and Anaplasma phagocytophilum in bovine from central Punjab, Pakistan. Pathogens 2019, 8, 155. [Google Scholar] [CrossRef] [PubMed]
- Chochlakis, D.; Ioannou, I.; Sharif, L.; Kokkini, S.; Hristophi, N.; Dimitriou, T.; Tselentis, Y.; Psaroulaki, A. Prevalence of Anaplasma sp. in goats and sheep in Cyprus. Vector-Borne Zoonotic Dis. 2009, 9, 457–463. [Google Scholar] [CrossRef]
- Ismail, N.; Bloch, K.C.; McBride, J.W. Human ehrlichiosis and anaplasmosis. Clin. Lab. Med. 2010, 30, 261–292. [Google Scholar] [CrossRef] [PubMed]
- Bianchessi, L.; Rocchi, M.S.; Maley, M.; Allen, K.; Ballingall, K.; Turin, L. Presence of Anaplasma phagocytophilum Ecotype I in UK Ruminants and Associated Zoonotic Risk. Pathogens 2023, 12, 216. [Google Scholar] [CrossRef] [PubMed]
- Lbacha, H.A.; Zouagui, Z.; Alali, S.; Rhalem, A.; Petit, E.; Ducrotoy, M.J.; Boulouis, H.-J.; Maillard, R. “Candidatus anaplasma camelii” in one-humped camels (Camelus dromedarius) in Morocco: A novel and emerging anaplasma species? Infect. Dis. Poverty 2017, 6, 67–74. [Google Scholar] [CrossRef]
- Bahrami, S.; Hamidinejat, H.; Tafreshi, A.R.G. First molecular detection of Anaplasma phagocytophilum in dromedaries (Camelus dromedarius). J. Zoo Wildl. Med. 2018, 49, 844–848. [Google Scholar]
- El Tigani-Asil, E.T.A.; Blanda, V.; Abdelwahab, G.E.; Hammadi, Z.M.A.; Habeeba, S.; Khalafalla, A.I.; Alhosani, M.A.; La Russa, F.; Migliore, S.; Torina, A. Molecular Investigation on Tick-Borne Hemoparasites and Coxiella burnetii in Dromedary Camels (Camelus dromedarius) in Al Dhafra Region of Abu Dhabi, UAE. Animals 2021, 11, 666. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Yang, J.; Chen, Z.; Qin, G.; Li, Y.; Li, Q.; Liu, J.; Liu, Z.; Guan, G.; Yin, H. Anaplasma infection of Bactrian camels (Camelus bactrianus) and ticks in Xinjiang, China. Parasites Vectors 2015, 8, 313. [Google Scholar] [CrossRef] [PubMed]
- Lorusso, V.; Wijnveld, M.; Latrofa, M.S.; Fajinmi, A.; Majekodunmi, A.O.; Dogo, A.G.; Igweh, A.C.; Otranto, D.; Jongejan, F.; Welburn, S.C. Canine and ovine tick-borne pathogens in camels, Nigeria. Vet. Parasitol. 2016, 228, 90–92. [Google Scholar] [CrossRef]
- Noaman, V. Molecular detection of novel genetic variants associated to Anaplasma ovis among dromedary camels in Iran. Arch. Razi Inst. 2018, 73, 11–18. [Google Scholar]
- Mahmoud, H.Y.; Ali, A.O.; Tanaka, T. Molecular detection and characterization of Anaplasma marginale infecting cattle, buffalo, and camel populations in southern Egypt. Front. Vet. Sci. 2023, 10, 1169323. [Google Scholar] [CrossRef]
- Bastos, A.D.; Mohammed, O.B.; Bennett, N.C.; Petevinos, C.; Alagaili, A.N. Molecular detection of novel Anaplasmataceae closely related to Anaplasma platys and Ehrlichia canis in the dromedary camel (Camelus dromedarius). Vet. Microbiol. 2015, 179, 310–314. [Google Scholar] [CrossRef]
- Alshahrani, M.; Alanazi, A.; Alouffi, A.; Abdullah, H.; Allam, A.; Mahmoud, M.; Abdel-Shafy, S.; Alfaifi, M.; Alkhathami, A. Molecular detection of Candidatus Anaplasma camelii in camels (Camelus dromedarius) from Asir Province, Saudi Arabia. Trop. Biomed. 2020, 37, 587–598. [Google Scholar]
- Moradi, Z.; Ebrahimzadeh, E.; Shayan, P.; Zarghami, F. Morphological and Molecular Investigation of Anaplasma Infection in Dromedary Camel (Camelus dromedarius) in Bushehr Province, Iran. Iran. J. Vet. Med. 2021, 15, 295–300. [Google Scholar]
- Bargul, J.L.; Kidambasi, K.O.; Getahun, M.N.; Villinger, J.; Copeland, R.S.; Muema, J.M.; Carrington, M.; Masiga, D.K. Transmission of ‘Candidatus Anaplasma camelii’to mice and rabbits by camel-specific keds, Hippobosca camelina. PLoS Neglected Trop. Dis. 2021, 15, e0009671. [Google Scholar] [CrossRef] [PubMed]
- Getange, D.; Bargul, J.L.; Kanduma, E.; Collins, M.; Bodha, B.; Denge, D.; Chiuya, T.; Githaka, N.; Younan, M.; Fèvre, E.M. Ticks and tick-borne pathogens associated with dromedary camels (Camelus dromedarius) in northern Kenya. Microorganisms 2021, 9, 1414. [Google Scholar] [CrossRef] [PubMed]
- El-Naga, T.R.A.; Barghash, S. Blood parasites in camels (Camelus dromedarius) in Northern West Coast of Egypt. J. Bacteriol. Parasitol 2016, 7, 258. [Google Scholar]
- Wernery, U.; Kaaden, O.R. Infectious Diseases in Camelids; Georg Thieme Verlag: New York, NY, USA, 2002. [Google Scholar]
- Ismail, N.; McBride, J.W. Tick-borne emerging infections: Ehrlichiosis and anaplasmosis. Clin. Lab. Med. 2017, 37, 317–340. [Google Scholar] [CrossRef] [PubMed]
- Dumler, J.S.; Barbet, A.F.; Bekker, C.; Dasch, G.A.; Palmer, G.H.; Ray, S.C.; Rikihisa, Y.; Rurangirwa, F.R. Reorganization of genera in the families Rickettsiaceae and Anaplasmataceae in the order Rickettsiales: Unification of some species of Ehrlichia with Anaplasma, Cowdria with Ehrlichia and Ehrlichia with Neorickettsia, descriptions of six new species combinations and designation of Ehrlichia equi and ‘HGE agent’ as subjective synonyms of Ehrlichia phagocytophila. Int. J. Syst. Evol. Microbiol. 2001, 51, 2145–2165. [Google Scholar]
- Silaghi, C.; Santos, A.S.; Gomes, J.; Christova, I.; Matei, I.A.; Walder, G.; Domingos, A.; Bell-Sakyi, L.; Sprong, H.; Von Loewenich, F.D. Guidelines for the direct detection of Anaplasma spp. in diagnosis and epidemiological studies. Vector-Borne Zoonotic Dis. 2017, 17, 12–22. [Google Scholar] [CrossRef] [PubMed]
- Parvizi, O. Overview of Anaplasmosis in Arab Countries in North Africa and the Middle East, and Optimizing a Commercial c-ELISA for Camels. PhD Thesis, Free University of Berlin, Berling, Germany, 2020. [Google Scholar]
- von Fricken, M.E.; Lkhagvatseren, S.; Boldbaatar, B.; Nymadawa, P.; Weppelmann, T.A.; Baigalmaa, B.-O.; Anderson, B.D.; Reller, M.E.; Lantos, P.M.; Gray, G.C. Estimated seroprevalence of Anaplasma spp. and spotted fever group Rickettsia exposure among herders and livestock in Mongolia. Acta Trop. 2018, 177, 179–185. [Google Scholar] [CrossRef] [PubMed]
- Yoo, J.; Chung, J.-H.; Kim, C.-M.; Yun, N.R.; Kim, D.-M. Asymptomatic-anaplasmosis confirmation using genetic and serological tests and possible coinfection with spotted fever group Rickettsia: A case report. BMC Infect. Dis. 2020, 20, 458. [Google Scholar] [CrossRef]
- Ismael, A.; Swelum, A.-A.; Khalaf, A.; Alowaimer, A. First evidence of natural anaplasmosis in Camelus dromedarius in Saudi Arabia. J. Camel Pract. Res. 2016, 23, 95–100. [Google Scholar] [CrossRef]
- Wernery, U.; Pfister, K.; Marina, R.; Hakimudin, F.; Silaghi, C. No evidence of Mycoplasma haemolamae and Anaplasma marginale in anaemic dromedaries in the United Arab Emirates. J. Camel Pract. Res. 2014, 21, 5–8. [Google Scholar] [CrossRef]
- Ybañez, A.P.; Matsumoto, K.; Kishimoto, T.; Inokuma, H. Molecular analyses of a potentially novel Anaplasma species closely related to Anaplasma phagocytophilum detected in sika deer (Cervus nippon yesoensis) in Japan. Vet. Microbiol. 2012, 157, 232–236. [Google Scholar] [CrossRef]
- Onyiche, T.E.; Răileanu, C.; Tauchmann, O.; Fischer, S.; Vasić, A.; Schäfer, M.; Biu, A.A.; Ogo, N.I.; Thekisoe, O.; Silaghi, C. Prevalence and molecular characterization of ticks and tick-borne pathogens of one-humped camels (Camelus dromedarius) in Nigeria. Parasites Vectors 2020, 13, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Larkin, M.A.; Blackshields, G.; Brown, N.P.; Chenna, R.; McGettigan, P.A.; McWilliam, H.; Valentin, F.; Wallace, I.M.; Wilm, A.; Lopez, R. Clustal W and Clustal X version 2.0. Bioinformatics 2007, 23, 2947–2948. [Google Scholar] [CrossRef]
- Kimura, M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 1980, 16, 111–120. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- Ghafar, M.W.; Shobrak, M.Y. Molecular detection and characterization of Anaplasma phagocytophilum, the causative agent of human granulocytic anaplasmosis, in some animals suspected to be competent reservoirs in Taif district, Kingdom of Saudi Arabia. Life Sci. J. 2014, 11, 63–69. [Google Scholar]
- Ybanez, A.P.; Matsumoto, K.; Kishimoto, T.; Yokoyama, N.; Inokuma, H. Dual presence of Anaplasma phagocytophilum and its closely related Anaplasma sp. in ixodid ticks in Hokkaido, Japan, and their specific molecular detection. J. Vet. Med. Sci. 2012, 74, 1551–1560. [Google Scholar] [CrossRef]
- Al-Deeb, M.A.; Muzaffar, S.B.; Abu-Zeid, Y.A.; Enan, M.R.; Karim, S. First record of a spotted fever group Rickettsia sp. and Theileria annulata in Hyalomma dromedarii (Acari: Ixodidae) ticks in the United Arab Emirates. Fla. Entomol. 2015, 98, 135–139. [Google Scholar] [CrossRef]
- Khan, A.S.; Maupin, G.O.; Rollin, P.E.; Noor, A.M.; Shurie, H.; Shalabi, A.; Wasef, S.; Haddad, Y.; Sadek, R.; Ijaz, K. An outbreak of Crimean-Congo hemorrhagic fever in the United Arab Emirates, 1994–1995. Am. J. Trop. Med. Hyg. 1997, 57, 519–525. [Google Scholar] [CrossRef]
- Caudill, M.T.; Brayton, K.A. The use and limitations of the 16S rRNA sequence for species classification of Anaplasma samples. Microorganisms 2022, 10, 605. [Google Scholar] [CrossRef] [PubMed]
- Said, M.B.; Belkahia, H.; Selmi, R.; Messadi, L. Computational selection of minimum length groESL operon required for Anaplasma species attribution and strain diversity analysis. Mol. Cell. Probes 2019, 48, 101467. [Google Scholar] [CrossRef]
- Fassi-Fehri, M.M. Diseases of camels. Rev. Sci. Tech. Off. Int. Epiz. 1987, 6, 337–354. [Google Scholar]
- Wernery, U.; Kinne, J.; Schuster, R.K. Camelids Infectious Disorders; OIE: Paris, France, 2014; pp. 74–78. [Google Scholar]
- Alsaad, K. Clinical, hematological and biochemical studies of anaplasmosis in Arabian one-humped camels (Camelus dromedarius). J. Anim. Vet. Adv 2009, 8, 2106–2109. [Google Scholar]
- Sudan, V.; Sharma, R.; Borah, M. Subclinical anaplasmosis in camel (Camelus dromedarius) and its successful therapeutic management. J. Parasit. Dis. 2014, 38, 163–165. [Google Scholar] [CrossRef] [PubMed]
- Ben Said, M.; Belkahia, H.; Sayahi, L.; Aloui, M.; Jemli, M.; Hadj Mohamed, B.; Sassi, L.; Darghouth, M.; Djaïem, A.; Bayoudh, M. First serological study of the prevalence of Anaplasma phagocytophilum in dromedary (Camelus dromedarius) in Tunisia. Bull. Soc. Pathol. Exot. 2014, 107, 1–6. [Google Scholar] [CrossRef]
- Munderloh, U.G.; Jauron, S.D.; Fingerle, V.; Leitritz, L.; Hayes, S.F.; Hautman, J.M.; Nelson, C.M.; Huberty, B.W.; Kurtti, T.J.; Ahlstrand, G.G. Invasion and intracellular development of the human granulocytic ehrlichiosis agent in tick cell culture. J. Clin. Microbiol. 1999, 37, 2518–2524. [Google Scholar] [CrossRef] [PubMed]
 Orange color = Al Dhafra region,
Orange color = Al Dhafra region,  Light green color = Abu Dhabi region and
Light green color = Abu Dhabi region and  Light purple represents Al Ain region.
Light purple represents Al Ain region.
 Orange color = Al Dhafra region,
Orange color = Al Dhafra region,  Light green color = Abu Dhabi region and
Light green color = Abu Dhabi region and  Light purple represents Al Ain region.
Light purple represents Al Ain region.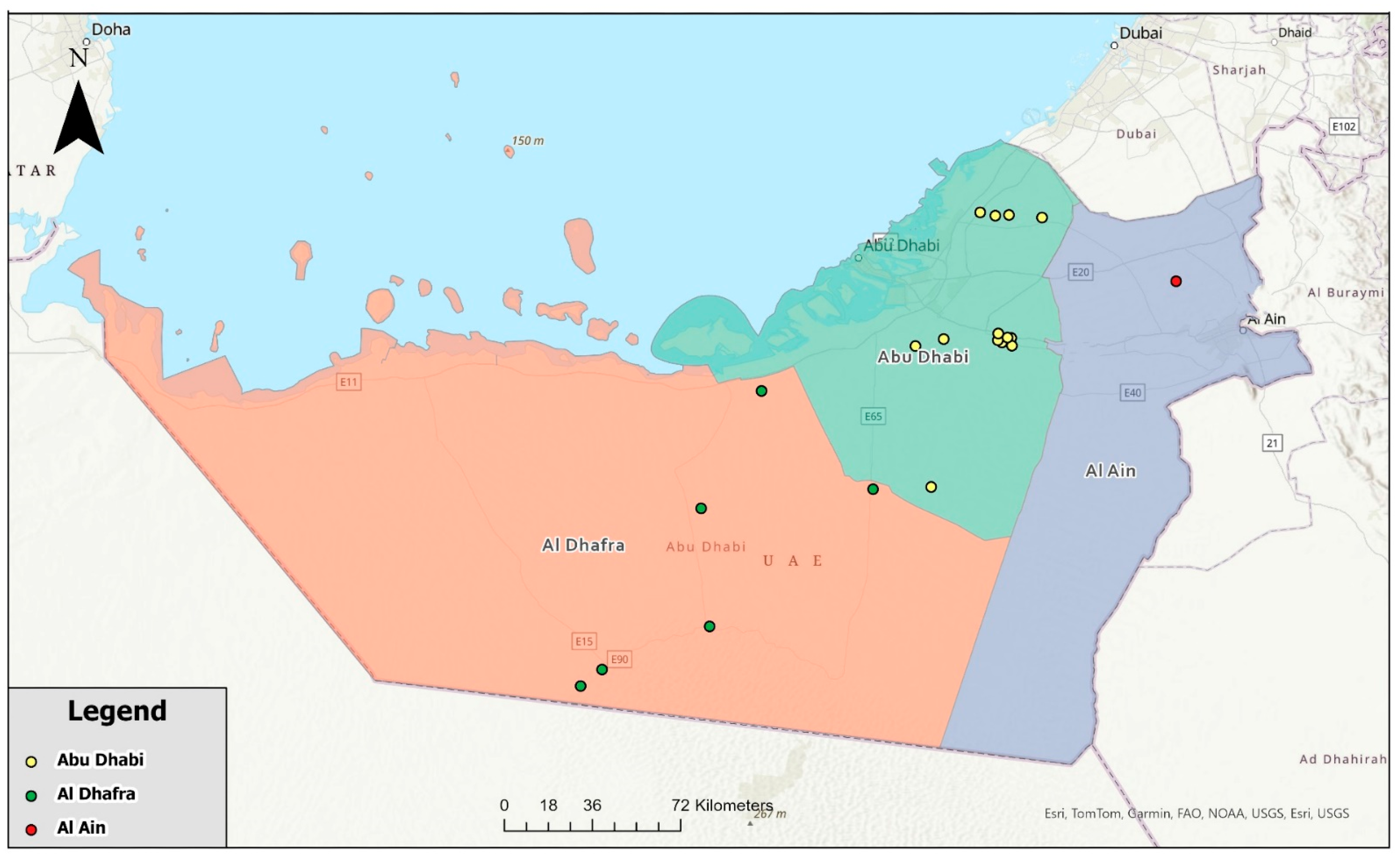
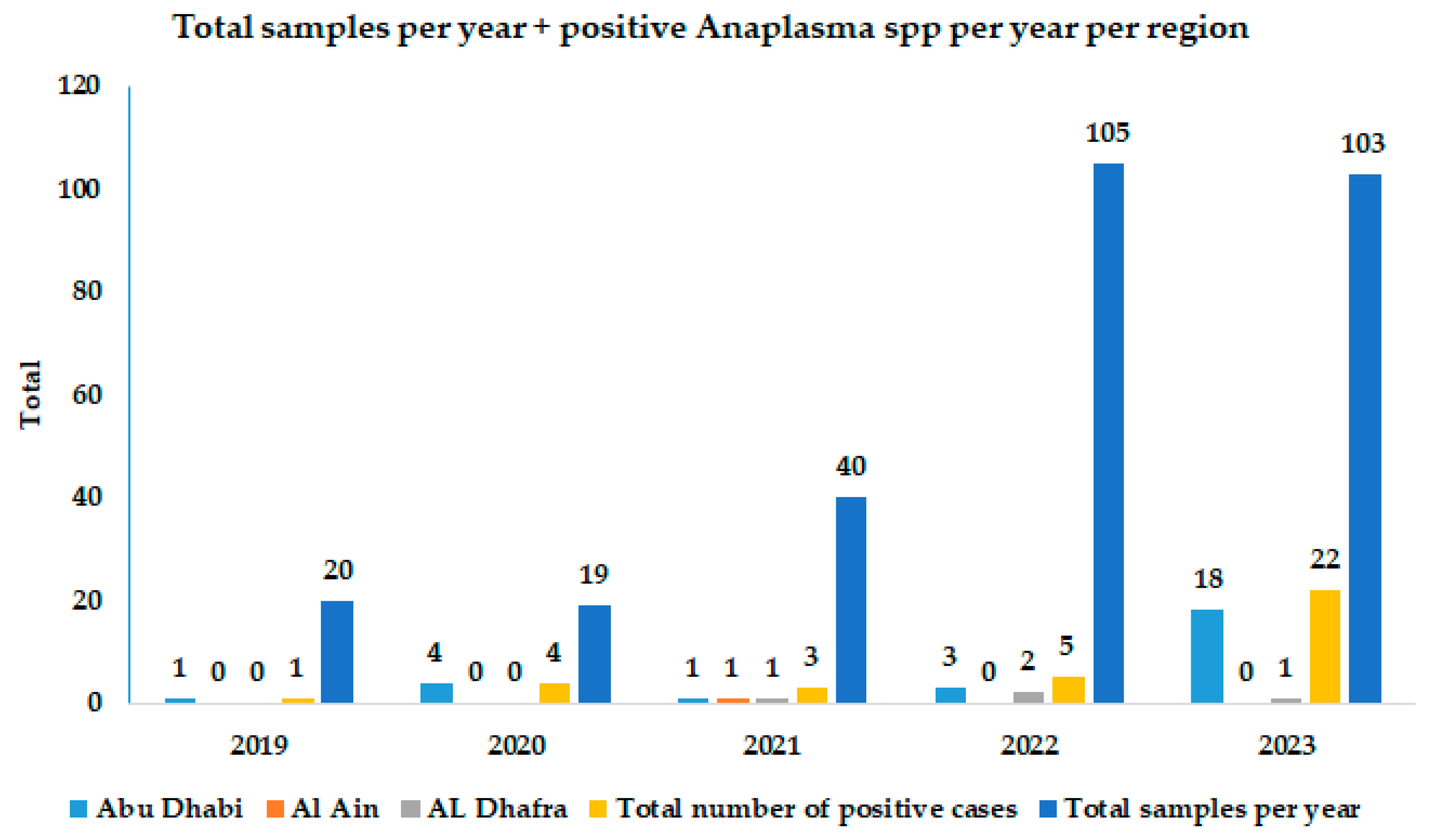

| Year | Positive Samples/Total per Region | Cumulative Positive Samples per Year | GenBank Accession Number | ||
|---|---|---|---|---|---|
| Abu Dhabi | Al Ain | Al Dhafra | |||
| 2019 | 1/5 (20%) | 0 | 0/15 (0%) | 1/20 (5%) | - |
| 2020 | 4/18 (22.2%) | 0 | 0/1 (0%) | 4/19 (21.1%) | - |
| 2021 | 1/27 (3.7%) | 1/12 (8.3%) | 1/1 (100%) | 3/40 (7.5%) | - |
| 2022 | 3/62 (4.8%) | 2/29 (6.9%) | 0/14 (0%) | 5/105 (4.8%) | - |
| 2023 | 18/74 (24.3%) | 0 | 4/29 (13.8%) | 22/103 (21.4%) | OR159796 To OR159804 |
| Total positive samples per region | 27/186 (14.5%) | 3/41 (4.3%) | 5/60 (8.3%) | ||
| Cumulative positive samples | 35/287 (12.2%) | ||||
| Total samples sequenced | 9 | - | - | 9/35 (25.7%) | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ishag, H.Z.A.; Habeeba, S.; El Tigani-Asil, E.T.A.; Yuosf, M.F.; Al Hammadi, Z.M.A.H.; Commey, A.N.O.; Bin Hraiz, H.T.A.A.; Shah, A.A.M.; Khalafalla, A.I. Molecular Detection of Candidatus Anaplasma camelii in Naturally Infected Dromedary Camels (Camelus dromedarius) in Abu Dhabi Emirate, United Arab Emirates, 2019–2023. Vet. Sci. 2024, 11, 123. https://doi.org/10.3390/vetsci11030123
Ishag HZA, Habeeba S, El Tigani-Asil ETA, Yuosf MF, Al Hammadi ZMAH, Commey ANO, Bin Hraiz HTAA, Shah AAM, Khalafalla AI. Molecular Detection of Candidatus Anaplasma camelii in Naturally Infected Dromedary Camels (Camelus dromedarius) in Abu Dhabi Emirate, United Arab Emirates, 2019–2023. Veterinary Sciences. 2024; 11(3):123. https://doi.org/10.3390/vetsci11030123
Chicago/Turabian StyleIshag, Hassan Zackaria Ali, Shameem Habeeba, El Tigani Ahmed El Tigani-Asil, Mohd Farouk Yuosf, Zulaikha Mohamed Abdel Hameed Al Hammadi, Abraham Nii Okai Commey, Hashel Talal Aboud Amer Bin Hraiz, Asma Abdi Mohamed Shah, and Abdelmalik Ibrahim Khalafalla. 2024. "Molecular Detection of Candidatus Anaplasma camelii in Naturally Infected Dromedary Camels (Camelus dromedarius) in Abu Dhabi Emirate, United Arab Emirates, 2019–2023" Veterinary Sciences 11, no. 3: 123. https://doi.org/10.3390/vetsci11030123
APA StyleIshag, H. Z. A., Habeeba, S., El Tigani-Asil, E. T. A., Yuosf, M. F., Al Hammadi, Z. M. A. H., Commey, A. N. O., Bin Hraiz, H. T. A. A., Shah, A. A. M., & Khalafalla, A. I. (2024). Molecular Detection of Candidatus Anaplasma camelii in Naturally Infected Dromedary Camels (Camelus dromedarius) in Abu Dhabi Emirate, United Arab Emirates, 2019–2023. Veterinary Sciences, 11(3), 123. https://doi.org/10.3390/vetsci11030123






