Multidrug-Resistant Commensal and Infection-Causing Staphylococcus spp. Isolated from Companion Animals in the Valencia Region
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Experimental Design
2.2. Epidemiological Data Collection
2.3. Sample Collection
2.4. Staphylococcus Isolation
2.5. Antimicrobial Susceptibility Testing
2.6. Statistical Analysis
3. Results
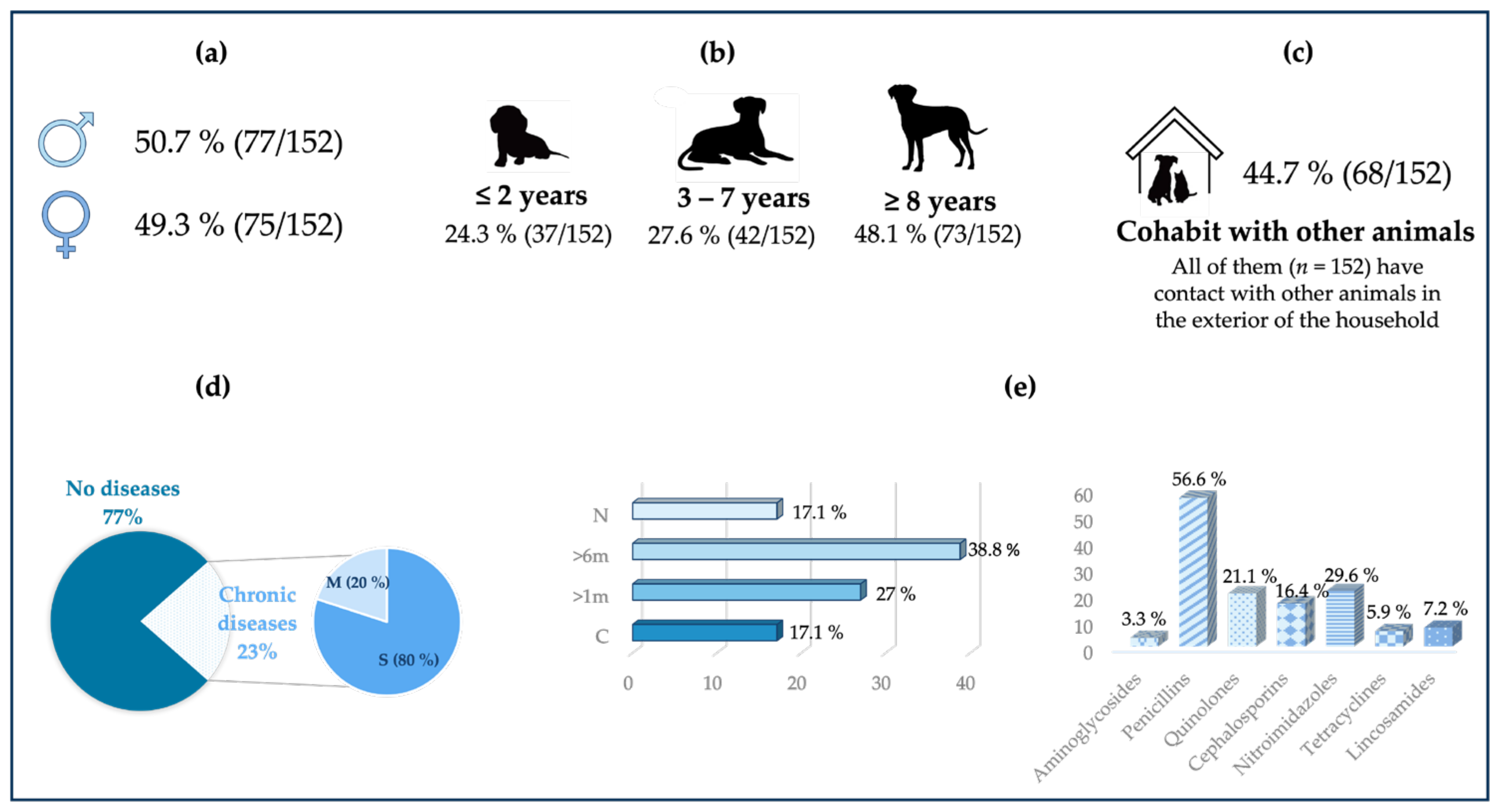
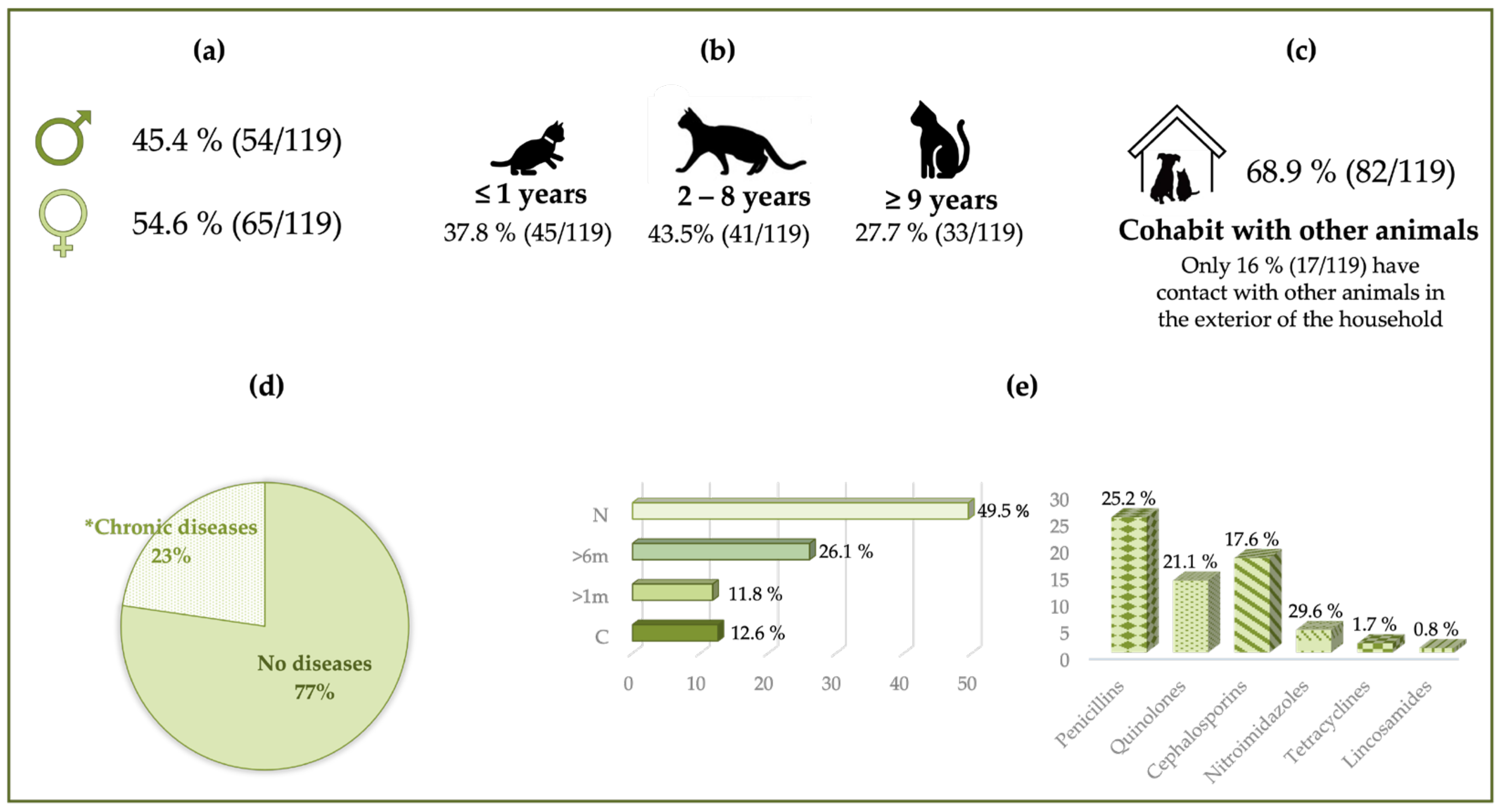
3.1. Epidemiological Data
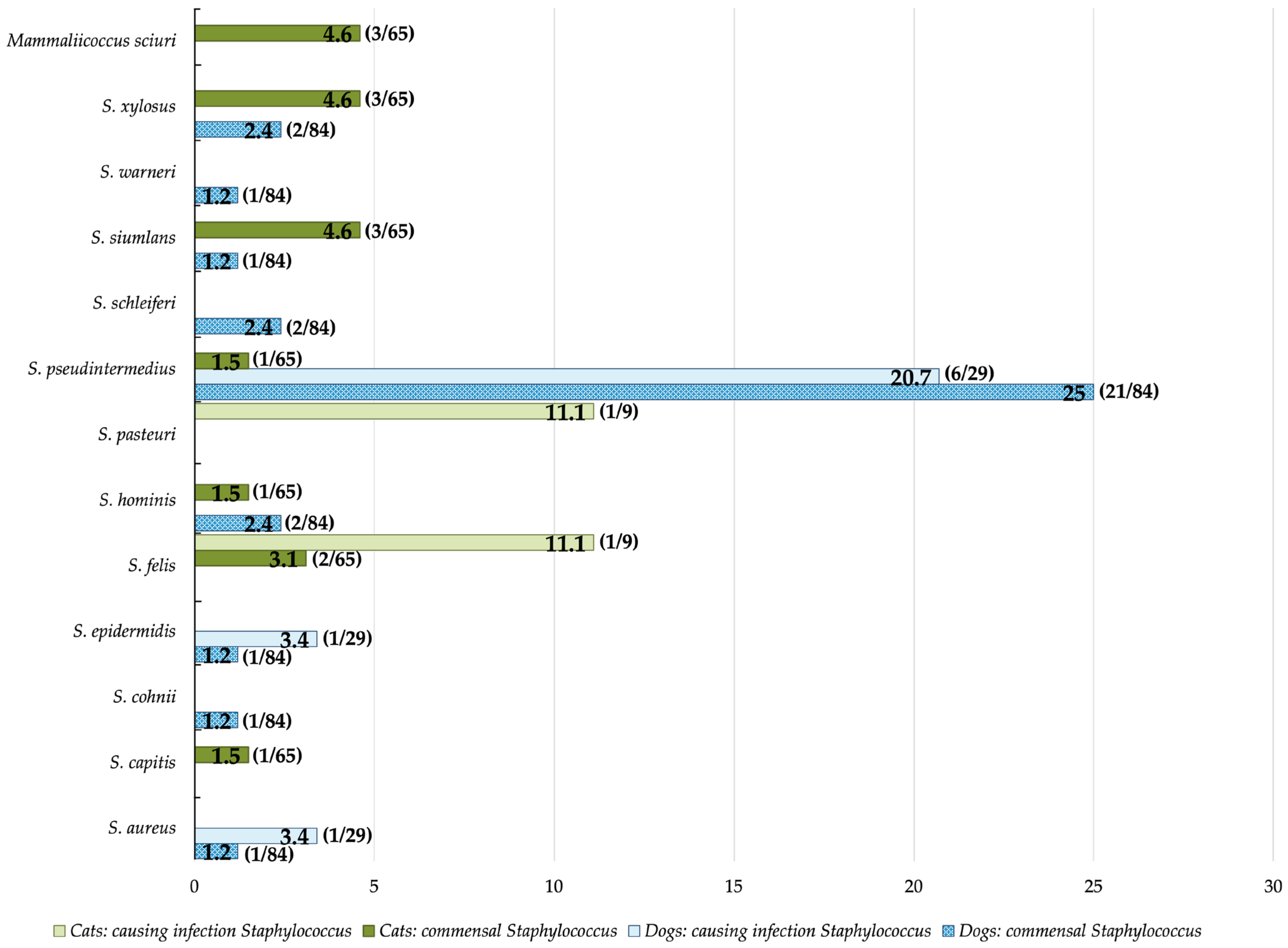
3.2. Staphylococcus Prevalence
3.3. Antimicrobial Susceptibility in Staphylococcus Strains
3.3.1. Methicillin Resistance
3.3.2. Dogs
3.3.3. Cats
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Graham, D.W.; Bergeron, G.; Bourassa, M.W.; Dickson, J.; Gomes, F.; Howe, A.; Kahn, L.H.; Morley, P.S.; Scott, H.M.; Simjee, S.; et al. Complexities in Understanding Antimicrobial Resistance across Domesticated Animal, Human, and Environmental Systems. Ann. N. Y. Acad. Sci. 2019, 1441, 17–30. [Google Scholar] [CrossRef]
- FEDIAF. The European Pet and Food Industry. Annual Report 2023. Available online: https://europeanpetfood.org/about/annual-report/ (accessed on 7 July 2023).
- FEDIAF. The European Pet and Food Industry. Annual Report 2010. Available online: http://www.stray-afp.org/nl/wp-content/uploads/sites/2/2012/08/facts_and_figures_2010.pdf (accessed on 25 September 2023).
- Overgaauw, P.A.M.; Vinke, C.M.; van Hagen, M.A.E.; Lipman, L.J.A. A One Health Perspective on the Human-Companion Animal Relationship with Emphasis on Zoonotic Aspects. Int. J. Environ. Res. Public Health 2020, 17, 3789. [Google Scholar] [CrossRef]
- Marco-Fuertes, A.; Marin, C.; Lorenzo-Rebenaque, L.; Vega, S.; Montoro-Dasi, L. Antimicrobial Resistance in Companion Animals: A New Challenge for the One Health Approach in the European Union. Vet. Sci. 2022, 9, 208. [Google Scholar] [CrossRef]
- World Health Organization. Global Antimicrobial Resistance and Use Surveillance System (GLASS). Report 2022; WHO: Geneva, Switzerland, 2022; Volume 2. [Google Scholar]
- Murray, C.J.; Ikuta, K.S.; Sharara, F.; Swetschinski, L.; Robles Aguilar, G.; Gray, A.; Han, C.; Bisignano, C.; Rao, P.; Wool, E.; et al. Global Burden of Bacterial Antimicrobial Resistance in 2019: A Systematic Analysis. Lancet 2022, 399, 629–655. [Google Scholar] [CrossRef]
- Ye, M.; Su, J.Q.; An, X.L.; Zhu, Y.G. Silencing the Silent Pandemic: Eliminating Antimicrobial Resistance by Using Bacteriophages. Sci. China Life Sci. 2022, 65, 1890. [Google Scholar] [CrossRef] [PubMed]
- Mendelson, M.; Sharland, M.; Mpundu, M. Antibiotic Resistance: Calling Time on the ‘Silent Pandemic’. JAC Antimicrob. Resist. 2022, 4, dlac016. [Google Scholar] [CrossRef] [PubMed]
- EMA Categorisation of Antibiotics Used in Animals Promotes Responsible Use to Protect Public and Animal Health. European Medicines Agency-EMA. Available online: https://www.ema.europa.eu/en/news/categorisation-antibiotics-used-animals-promotes-responsible-use-protect-public-animal-health (accessed on 6 December 2022).
- Mader, R.; Demay, C.; Jouvin-Marche, E.; Ploy, M.C.; Barraud, O.; Bernard, S.; Lacotte, Y.; Pulcini, C.; Weinbach, J.; Berling, C.; et al. Defining the Scope of the European Antimicrobial Resistance Surveillance Network in Veterinary Medicine (EARS-Vet): A Bottom-up and One Health Approach. J. Antimicrob. Chemother. 2022, 77, 816–826. [Google Scholar] [CrossRef]
- Mader, R.; Damborg, P.; Amat, J.P.; Bengtsson, B.; Bourély, C.; Broens, E.M.; Busani, L.; Crespo-Robledo, P.; Filippitzi, M.E.; Fitzgerald, W.; et al. Building the European Antimicrobial Resistance Surveillance Network in Veterinary Medicine (EARS-Vet). Eurosurveillance 2021, 26, 2001359. [Google Scholar] [CrossRef] [PubMed]
- European Centre for Disease Prevention and Control European Antimicrobial Resistance Surveillance Network (EARS-Net). Available online: https://www.ecdc.europa.eu/en/about-us/networks/disease-networks-and-laboratory-networks/ears-net-data (accessed on 19 July 2023).
- Hesp, A.; van Schaik, G.; Wiegel, J.; Heuvelink, A.; Mevius, D.; Veldman, K. Antimicrobial Resistance Monitoring in Commensal and Clinical Escherichia Coli from Broiler Chickens: Differences and Similarities. Prev. Vet. Med. 2022, 204, 105663. [Google Scholar] [CrossRef]
- European Food and Safety Authority. The European Union Summary Report on Antimicrobial Resistance in Zoonotic and Indicator Bacteria from Humans, Animals and Food in 2020/2021. EFSA J. 2023, 21, e07867. [Google Scholar] [CrossRef]
- Kowalewicz, C.; Timmermans, M.; Fretin, D.; Wattiau, P.; Boland, C. An In-House 45-Plex Array for the Detection of Antimicrobial Resistance Genes in Gram-Positive Bacteria. Microbiologyopen 2023, 12, e1341. [Google Scholar] [CrossRef] [PubMed]
- Abusleme, F.; Galarce, N.; Quezada-Aguiluz, M.; Iragüen, D.; González-Rocha, G. Characterization and Antimicrobial Susceptibility of Coagulase-Positive Staphylococcus Isolated in a Veterinary Teaching Hospital in Chile. Rev. Argent. Microbiol. 2022, 54, 192–202. [Google Scholar] [CrossRef] [PubMed]
- Citron, L.E.; Cain, C.L.; Dietrich, J.; Cole, S.D. Genomic and Clinical Case Characterisation of Staphylococcus haemolyticus Isolated from Dogs and Cats in the United States, Including Strains with High-Level Mupirocin Tolerance. Vet. Dermatol. 2023, 34, 298–309. [Google Scholar] [CrossRef] [PubMed]
- Platenik, M.O.; Archer, L.; Kher, L.; Santoro, D. Prevalence of MecA, MecC and Panton-Valentine-Leukocidin Genes in Clinical Isolates of Coagulase Positive Staphylococci from Dermatological Canine Patients. Microorganisms 2022, 10, 2239. [Google Scholar] [CrossRef] [PubMed]
- Wilkinson, D.A.; Rogers, L.E.; Bell, A.; Benschop, J.; Midwinter, A.C. Carriage of Staphylococcus pseudintermedius by Clinically Normal Dogs in Canterbury, New Zealand. N. Z. Vet. J. 2022, 71, 33–36. [Google Scholar] [CrossRef] [PubMed]
- Tabatabaei, S.; Najafifar, A.; Askari Badouei, M.; Zahraei Salehi, T.; Ashrafi Tamai, I.; Khaksar, E.; Abbassi, M.S.; Ghazisaeedi, F. Genetic Characterisation of Methicillin-Resistant Staphylococcus aureus and Staphylococcus pseudintermedius in Pets and Veterinary Personnel in Iran: New Insights into Emerging Methicillin-Resistant S. pseudintermedius (MRSP). J. Glob. Antimicrob. Resist. 2019, 16, 6–10. [Google Scholar] [CrossRef] [PubMed]
- Tamakan, H.; Gocmen, H. Genetic Characterization of Methicillin Resistant Staphylococcus pseudintermedius in Dogs and Cats in Cyprus: Comparison of MRSP and MRSA Results. Pak. J. Zool 2022, 54, 1511–1519. [Google Scholar] [CrossRef]
- Becker, K.; Heilmann, C.; Peters, G. Coagulase-Negative Staphylococci. Clin. Microbiol. Rev. 2014, 27, 870–926. [Google Scholar] [CrossRef]
- Marco-Fuertes, A.; Jordá, J.; Marin, C.; Lorenzo-Rebenaque, L.; Montoro-Dasi, L.; Vega, S. Multidrug-Resistant Escherichia Coli Strains to Last Resort Human Antibiotics Isolated from Healthy Companion Animals in Valencia Region. Antibiotics 2023, 12, 1638. [Google Scholar] [CrossRef]
- Frosini, S.M.; Bond, R.; King, R.H.; Loeffler, A. The Nose Is Not Enough: Multi-Site Sampling Is Best for MRSP Detection in Dogs and Households. Vet. Dermatol. 2022, 33, 576–580. [Google Scholar] [CrossRef]
- Nocera, F.P.; Iovane, G.; DE MARTINO, L.D.; Holbein, B.E. Antimicrobial Activity of the Iron-Chelator, DIBI, against Multidrug-Resistant Canine Methicillin-Susceptible Staphylococcus pseudintermedius: A Preliminary Study of Four Clinical Strains. Pathogens 2022, 11, 656. [Google Scholar] [CrossRef]
- Ardanuy, C.; María, E.C.; Morosini, I.; Torres, C. Detección Fenotípica de mecanismos de Resistencia Grampositivos. Recomendaciones de La Sociedad Española de Enfermedades Infecciosas y Microbiología Clínica (SEIMC). Available online: https://www.seimc.org/contenidos/documentoscientificos/procedimientosmicrobiologia/seimc-procedimientomicrobiologia39.pdf (accessed on 19 July 2023).
- WHO. WHO Medically Important List. A Risk Management Tool for Mitigating Antimicrobial Resistance Due to Non-Human Use. 7 th Revision 2023; WHO: Geneva, Switzerland, 2023. [Google Scholar]
- The European Committee on Antimicrobial Susceptibility Testing. Breakpoint Tables for Interpretation of MICs and Zone Diameters. Version 14.0, 2024. Available online: https://www.eucast.org/fileadmin/src/media/PDFs/EUCAST_files/Breakpoint_tables/v_14.0_Breakpoint_Tables.pdf (accessed on 5 January 2024).
- Clinical and Laboratory Standards Institute. Performance Standards for Antimicrobial Susceptibility Testing, 33rd ed.; CLSI Supplement M100; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2023. [Google Scholar]
- Clinical and Laboratory Standards Institute. Performance Standards for Antimicrobial Disk and Dilution Susceptibility Tests for Bacteria Isolated from Animals, 6th ed.; CLSI Supplement VET01S; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2023. [Google Scholar]
- Magiorakos, A.P.; Srinivasan, A.; Carey, R.B.; Carmeli, Y.; Falagas, M.E.; Giske, C.G.; Harbarth, S.; Hindler, J.F.; Kahlmeter, G.; Olsson-Liljequist, B.; et al. Multidrug-Resistant, Extensively Drug-Resistant and Pandrug-Resistant Bacteria: An International Expert Proposal for Interim Standard Definitions for Acquired Resistance. Clin. Microbiol. Infect. 2012, 18, 268–281. [Google Scholar] [CrossRef] [PubMed]
- Searle, S.R.; Speed, F.M.; Milliken, G.A. Population Marginal Means in the Linear Model: An Alternative to Least Squares Means. Am. Stat. 1980, 34, 216–221. [Google Scholar] [CrossRef]
- Fox, J.; Weisberg, S. An R Companion to Applied Regression, 3rd ed.; Sage: Thousand Oaks, CA, USA, 2019. [Google Scholar]
- Graves, S.; Piepho, H.; Selzer, L.; Dorai-Raj, S. MultcompView: Visualizations of Paired Comparisons. R Package Version 0.1-9. Available online: https://cran.r-project.org/web/packages/multcompView/index.html (accessed on 27 July 2023).
- Velazquez-Meza, M.E.; Galarde-López, M.; Carrillo-Quiróz, B.; Alpuche-Aranda, C.M. Antimicrobial Resistance: One Health Approach. Vet. World 2022, 15, 743. [Google Scholar] [CrossRef] [PubMed]
- Pomba, C.; Rantala, M.; Greko, C.; Baptiste, K.E.; Catry, B.; van Duijkeren, E.; Mateus, A.; Moreno, M.A.; Pyörälä, S.; Ružauskas, M.; et al. Public Health Risk of Antimicrobial Resistance Transfer from Companion Animals. J. Antimicrob. Chemother. 2017, 72, 957–968. [Google Scholar] [CrossRef] [PubMed]
- Despotovic, M.; de Nies, L.; Busi, S.B.; Wilmes, P. Reservoirs of Antimicrobial Resistance in the Context of One Health. Curr. Opin. Microbiol. 2023, 73, 102291. [Google Scholar] [CrossRef] [PubMed]
- Madhaiyan, M.; Wirth, J.S.; Saravanan, V.S. Phylogenomic Analyses of the Staphylococcaceae Family Suggest the Reclassification of Five Species within the Genus Staphylococcus as Heterotypic Synonyms, the Promotion of Five Subspecies to Novel Species, the Taxonomic Reassignment of Five Staphylococcus Species to Mammaliicoccus Gen. Nov., and the Formal Assignment of Nosocomiicoccus to the Family Staphylococcaceae. Int. J. Syst. Evol. Microbiol. 2020, 70, 5926–5936. [Google Scholar] [CrossRef] [PubMed]
- Taxonomy Browser (Mammaliicoccus Sciuri). Available online: https://www.ncbi.nlm.nih.gov/Taxonomy/Browser/wwwtax.cgi?id=1296 (accessed on 7 January 2024).
- de Moura, G.S.; de Carvalho, E.; Ramos Sanchez, E.M.; Sellera, F.P.; Marques, M.F.S.; Heinemann, M.B.; De Vliegher, S.; Souza, F.N.; Mota, R.A. Emergence of Livestock-Associated Mammaliicoccus Sciuri ST71 Co-Harbouring MecA and MecC Genes in Brazil. Vet. Microbiol. 2023, 283, 109792. [Google Scholar] [CrossRef] [PubMed]
- Gandolfi-Decristophoris, P.; Regula, G.; Petrini, O.; Zinsstag, J.; Schelling, E. Prevalence and Risk Factors for Carriage of Multi-Drug Resistant Staphylococci in Healthy Cats and Dogs. J. Vet. Sci. 2013, 14, 449–456. [Google Scholar] [CrossRef]
- Miszczak, M.; Korzeniowska-Kowal, A.; Wzorek, A.; Gamian, A.; Rypuła, K.; Bierowiec, K. Colonization of Methicillin-Resistant Staphylococcus Species in Healthy and Sick Pets: Prevalence and Risk Factors. BMC Vet. Res. 2023, 19, 1–12. [Google Scholar] [CrossRef]
- Bierowiec, K.; Korzeniowska-Kowal, A.; Wzorek, A.; Rypuła, K.; Gamian, A. Prevalence of Staphylococcus Species Colonization in Healthy and Sick Cats. Biomed. Res. Int. 2019, 2019, 4360525. [Google Scholar] [CrossRef]
- Sukur, H.; Esendal, O.M. Presence and Antimicrobial Resistance of Coagulase-Negative Staphylococci Isolated from Animals in a Veterinary Teaching Hospital in Cyprus. Vet. Med. 2020, 65, 191–198. [Google Scholar] [CrossRef]
- Cengiz, S.; Okur, S.; Oz, C.; Turgut, F.; Gumurcinler, B.; Sevuk, N.S.; Kekec, A.I.; Cepoglu, H.; Sevimli, U.; Adiguzel, M.C. Prevalence and Clonal Diversity of Methicillin-Resistant Staphylococcus aureus and Methicillin-Resistant Staphylococcus pseudintermedius Isolated from Dogs and Cats with Eye Discharge. Acta. Microbiol. Immunol. Hung. 2023, 70, 134–141. [Google Scholar] [CrossRef]
- França, A.; Gaio, V.; Lopes, N.; Melo, L.D.R. Virulence Factors in Coagulase-Negative Staphylococci. Pathogens 2021, 10, 170. [Google Scholar] [CrossRef] [PubMed]
- Foster, T.J. Coagulase Negative Staphylococci. In Molecular Medical Microbiology, 3rd ed.; Academic Press: Cambridge, MA, USA, 2024; pp. 681–704. [Google Scholar]
- Abdullahi, I.N.; Lozano, C.; Höfle, Ú.; Cardona-Cabrera, T.; Zarazaga, M.; Torres, C. Antimicrobial Resistome of Coagulase-Negative Staphylococci from Nasotracheal Cavities of Nestlings of Ciconia ciconia in Southern Spain: Detection of MecC-SCCmec Type-XI-Carrying S. Lentus. Comp. Immunol. Microbiol. Infect. Dis. 2023, 99, 102012. [Google Scholar] [CrossRef] [PubMed]
- Sales, I.; Vieira-da-Motta, O.; Tavares, A.; Ruiz-Miranda, C.R.; de Lencastre, H.; Miragaia, M. Impact of Human Created Environments in the Pathogenic Potential and Antimicrobial Resistance of Staphylococci from Wild Neotropical Primates in Brazil. Comp. Immunol. Microbiol. Infect. Dis. 2024, 104, 102094. [Google Scholar] [CrossRef]
- Li, Y.; Fernández, R.; Durán, I.; Molina-López, R.A.; Darwich, L. Antimicrobial Resistance in Bacteria Isolated From Cats and Dogs From the Iberian Peninsula. Front. Microbiol. 2021, 11, 621597. [Google Scholar] [CrossRef]
- Lord, J.; Millis, N.; Jones, R.D.; Johnson, B.; Kania, S.A.; Odoi, A. An Epidemiological Study of the Predictors of Multidrug Resistance and Methicillin Resistance among Staphylococcus spp. Isolated from Canine Specimens Submitted to a Diagnostic Laboratory in Tennessee, USA. PeerJ 2023, 11, e15012. [Google Scholar] [CrossRef]
- Awosile, B.B.; Mcclure, J.T.; Saab, M.E.; Heider, L.C. Antimicrobial Resistance in Bacteria Isolated from Cats and Dogs from the Atlantic Provinces, Canada from 1994–2013. Can. Vet. J. 2018, 59, 885. [Google Scholar] [PubMed]
- DANMAP. Use of Antimicrobial Agents and Occurrence of Antimicrobial Resistance in Bacteria from Food Animals, Food and Humans in Denmark 2022; DANMAP: Copenhagen, Denmark, 2023. [Google Scholar]
- FINRES-Vet. Finnish Veterinary Antimicrobial Resistance Monitoring and Consumption of Antimicrobial Agents 2022; FINRES-Vet: Helsinki, Finland, 2023. [Google Scholar]
- NORM/NORM-Vet. Usage of Antimicrobial Agents and Occurrence of Antimicrobial Resistance in Norway; NORM/NORM-Vet: Tromsø/Oslo, Norway, 2023. [Google Scholar]
- Swedres-Svarm. Sales of Antibiotics and Occurrence of Resistance in Sweden 2022; Swedres-Svarm: Solna/Uppsala, Sweden, 2023. [Google Scholar]
- Tong, Y.C.; Zhang, Y.N.; Li, P.C.; Cao, Y.L.; Ding, D.Z.; Yang, Y.; Lin, Q.Y.; Gao, Y.N.; Sun, S.Q.; Fan, Y.P.; et al. Detection of Antibiotic-Resistant Canine Origin Escherichia coli and the Synergistic Effect of Magnolol in Reducing the Resistance of Multidrug-Resistant Escherichia coli. Front. Vet. Sci. 2023, 10, 1104812. [Google Scholar] [CrossRef]
- Lord, J.; Millis, N.; Jones, R.D.; Johnson, B.; Kania, S.A.; Odoi, A. Patterns of Antimicrobial, Multidrug and Methicillin Resistance among Staphylococcus spp. Isolated from Canine Specimens Submitted to a Diagnostic Laboratory in Tennessee, USA: A Descriptive Study. BMC Vet. Res. 2022, 18, 91. [Google Scholar] [CrossRef]
- Thapa, D.; Pyakurel, S.; Thapa, S.; Lamsal, S.; Chaudhari, M.; Adhikari, N.; Shrestha, D. Staphylococcus aureus with Inducible Clindamycin Resistance and Methicillin Resistance in a Tertiary Hospital in Nepal. Trop. Med. Health 2021, 49, 99. [Google Scholar] [CrossRef]
- Nocera, F.P.; Pizzano, F.; Masullo, A.; Cortese, L.; De Martino, L. Antimicrobial Resistant Staphylococcus Species Colonization in Dogs, Their Owners, and Veterinary Staff of the Veterinary Teaching Hospital of Naples, Italy. Pathogens 2023, 12, 1016. [Google Scholar] [CrossRef]
- Sudhakara Reddy, B.; Sivajothi, S. Methicillin-Resistant Staphylococcus aureus (MRSA) Isolated from Dogs with Recurrent Pyoderma. Artic. J. Dairy Vet. Anim. Res. 2016, 3, 62–65. [Google Scholar] [CrossRef]
- Das, S.; Kabir, A.; Chouhan, C.S.; Shahid, M.A.H.; Habib, T.; Kobir, M.A.; Hossain, M.Z.; Rahman, M.; Nazir, K.H.M.N.H. Antimicrobial Resistance Patterns of Staphylococcus aureus Isolated from Apparently Healthy Pet Cats of Bangladesh. J. Adv. Vet. Anim. Res. 2023, 10, 545–553. [Google Scholar] [CrossRef]
- Morris, D.O.; Rookt, K.A.; Shofer, F.S.; Rankin, S.C. Screening of Staphylococcus aureus, Staphylococcus intermedius, and Staphylococcus schleiferi Isolates Obtained from Small Companion Animals for Antimicrobial Resistance: A Retrospective Review of 749 Isolates (2003–2004). Vet. Dermatol. 2006, 17, 332–337. [Google Scholar] [CrossRef]
- Werth, B.J. Macrolides-Infectious Diseases-MSD. Manual Professional Edition. Available online: https://www.msdmanuals.com/professional/infectious-diseases/bacteria-and-antibacterial-drugs/macrolides (accessed on 9 November 2023).
- Werth, B.J. Clindamycin-Infectious Diseases-MSD. Manual Professional Edition. Available online: https://www.msdmanuals.com/professional/infectious-diseases/bacteria-and-antibacterial-drugs/clindamycin (accessed on 9 November 2023).
- Shrestha, B. D Test: A Simple Test with Big Implication for Staphylococcus aureus Macrolide-Lincosamide-Streptogramin B Resistance Pattern. Rev. Artic. Nepal. Med. Coll J. 2014, 16, 88–94. [Google Scholar] [CrossRef]
- Almuhayawi, M.S.; Alruhaili, M.H.; Gattan, H.S.; Alharbi, M.T.; Nagshabandi, M.; Al Jaouni, S.; Selim, S.; Alanazi, A.; Alruwaili, Y.; Faried, O.A.; et al. Staphylococcus aureus Induced Wound Infections Which Antimicrobial Resistance, Methicillin-and Vancomycin-Resistant: Assessment of Emergence and Cross Sectional Study. Infect. Drug Resist. 2023, 16, 5335–5346. [Google Scholar] [CrossRef] [PubMed]
- Burke, M.; Santoro, D. Prevalence of Multidrug-Resistant Coagulase-Positive Staphylococci in Canine and Feline Dermatological Patients over a 10-Year Period: A Retrospective Study. Microbiol. N. Y. 2023, 169, 001300. [Google Scholar] [CrossRef] [PubMed]
- Srednik, M.E.; Perea, C.A.; Giacoboni, G.I.; Hicks, J.A.; Foxx, C.L.; Harris, B.; Schlater, L.K. Genomic Features of Antimicrobial Resistance in Staphylococcus pseudintermedius Isolated from Dogs with Pyoderma in Argentina and the United States: A Comparative Study. Int. J. Mol. Sci. 2023, 24, 11361. [Google Scholar] [CrossRef] [PubMed]
- Sitovs, A.; Sartini, I.; Giorgi, M. Levofloxacin in Veterinary Medicine: A Literature Review. Res. Vet. Sci. 2021, 137, 111–126. [Google Scholar] [CrossRef]
- Kanafani, Z.A.; Sleiman, A.; Frem, J.A.; Doumat, G.; Gharamti, A.; El Hafi, B.; Doumith, M.; AlGhoribi, M.F.; Kanj, S.S.; Araj, G.F.; et al. Molecular Characterization and Differential Effects of Levofloxacin and Ciprofloxacin on the Potential for Developing Quinolone Resistance among Clinical Pseudomonas aeruginosa Isolates. Front. Microbiol. 2023, 14, 1209224. [Google Scholar] [CrossRef]
- González, J.; Hernandez, L.; Tabera, A.; Bustamante, A.V.; Sanso, A.M. Methicillin-Resistant Staphylococcus aureus and Coagulase-Negative Staphylococcus from School Dining Rooms in Argentina. Foodborne Pathog. Dis. 2023, 21, 44–51. [Google Scholar] [CrossRef] [PubMed]
- Kang, M.H.; Chae, M.J.; Yoon, J.W.; Lee, S.Y.; Yoo, J.H.; Park, H.M. Resistance to Fluoroquinolones and Methicillin in Ophthalmic Isolates of Staphylococcus pseudintermedius from Companion Animals. Can. Vet. J. 2014, 55, 678. [Google Scholar]
- Hamzah, A.M.C.; Yeo, C.C.; Puah, S.M.; Chua, K.H.; Rahman, N.I.A.; Abdullah, F.H.; Othman, N.; Chew, C.H. Tigecycline and Inducible Clindamycin Resistance in Clinical Isolates of Methicillin-Resistant Staphylococcus aureus from Terengganu, Malaysia. J. Med. Microbiol. 2019, 68, 1299–1305. [Google Scholar] [CrossRef]
- Yousefi, M.; Fallah, F.; Arshadi, M.; Pourmand, M.R.; Hashemi, A.; Pourmand, G. Identification of Tigecycline- and Vancomycin-Resistant Staphylococcus aureus Strains among Patients with Urinary Tract Infection in Iran. New Microbes New Infect. 2017, 19, 8–12. [Google Scholar] [CrossRef] [PubMed]
- Dabul, A.N.G.; Camargo, I.L.B.C. Molecular Characterization of Methicillin-Resistant Staphylococcus aureus Resistant to Tigecycline and Daptomycin Isolated in a Hospital in Brazil. Epidemiol. Infect. 2014, 142, 479–483. [Google Scholar] [CrossRef]
- Moawad, A.A.; Hotzel, H.; Awad, O.; Roesler, U.; Hafez, H.M.; Tomaso, H.; Neubauer, H.; El-Adawy, H. Evolution of Antibiotic Resistance of Coagulase-Negative Staphylococci Isolated from Healthy Turkeys in Egypt: First Report of Linezolid Resistance. Microorganisms 2019, 7, 476. [Google Scholar] [CrossRef]
- Papich, M.G. Selection of Antibiotics for Meticillin-Resistant Staphylococcus pseudintermedius: Time to Revisit Some Old Drugs? Vet. Dermatol. 2012, 23, 352-e64. [Google Scholar] [CrossRef] [PubMed]
- Heidary, M.; Sholeh, M.; Koupaei, M.; Asadi, A.; Khah, S.M.; Kheirabadi, F.; Saeidi, P.; Darbandi, A.; Taheri, B.; Ghanavati, R. Prevalence of Tigecycline Resistance in Methicillin-Resistant Staphylococcus aureus: A Systematic Review and Meta-Analysis. Diagn. Microbiol. Infect. Dis. 2024, 108, 116088. [Google Scholar] [CrossRef]
- Costa, S.S.; Ribeiro, R.; Serrano, M.; Oliveira, K.; Ferreira, C.; Leal, M.; Pomba, C.; Couto, I. Staphylococcus aureus Causing Skin and Soft Tissue Infections in Companion Animals: Antimicrobial Resistance Profiles and Clonal Lineages. Antibiotics 2022, 11, 599. [Google Scholar] [CrossRef]
- Bellato, A.; Robino, P.; Stella, M.C.; Scarrone, L.; Scalas, D.; Nebbia, P. Resistance to Critical Important Antibacterials in Staphylococcus pseudintermedius Strains of Veterinary Origin. Antibiotics 2022, 11, 1758. [Google Scholar] [CrossRef]
- Glajzner, P.; Szewczyk, E.M.; Szemraj, M. Pathogenic Potential and Antimicrobial Resistance of Staphylococcus pseudintermedius Isolated from Human and Animals. Folia Microbiol. 2023, 68, 231–243. [Google Scholar] [CrossRef]
- Ayodo, C.; Mugoh, R.; Ita, T.; Ouma, C.; Jepleting, M.; Oduor, B.; Guyah, B.; Omulo, S. Nasal Carriage of Methicillin-Resistant Staphylococcus sciuri Group by Residents of an Urban Informal Settlement in Kenya. East Afr. Health Res. J. 2023, 7, 76. [Google Scholar] [CrossRef]
- Balandin, B.; Lobo, B.; Orden, B.; Román, F.; García, E.; Martínez, R.; Valdivia, M.; Ortega, A.; Fernández, I.; Galdos, P. Emergence of Linezolid-Resistant Coagulase-Negative Staphylococci in an Intensive Care Unit. Infect. Dis. 2016, 48, 343–349. [Google Scholar] [CrossRef]
- Kosowska-Shick, K.; Julian, K.G.; McGhee, P.L.; Appelbaum, P.C.; Whitener, C.J. Molecular and Epidemiologic Characteristics of Linezolid-Resistant Coagulase-Negative Staphylococci at a Tertiary Care Hospital. Diagn. Microbiol. Infect. Dis. 2010, 68, 34–39. [Google Scholar] [CrossRef]
- Werth, B.J. Vancomycin-Infectious Diseases-MSD Manual Professional Edition. Available online: https://www.msdmanuals.com/professional/infectious-diseases/bacteria-and-antibacterial-drugs/vancomycin (accessed on 9 November 2023).
- Ma, S.; Chen, S.; Lyu, Y.; Huang, W.; Liu, Y.; Dang, X.; An, Q.; Song, Y.; Jiao, Y.; Gong, X.; et al. China Antimicrobial Resistance Surveillance Network for Pets (CARPet), 2018 to 2021. One Health Adv. 2023, 1, 7. [Google Scholar] [CrossRef]
- Muzammil, I.; Ijaz, M.; Saleem, M.H.; Ali, M.M. Molecular Characterization of Vancomycin-Resistant Staphylococcus aureus Isolated from Bovine Milk. Zoonoses Public Health 2023, 70, 420–433. [Google Scholar] [CrossRef] [PubMed]
- Werth, B.J. Quinupristin and Dalfopristin-Infectious Diseases-MSD Manual Professional Edition. Available online: https://www.msdmanuals.com/professional/infectious-diseases/bacteria-and-antibacterial-drugs/quinupristin-and-dalfopristin (accessed on 9 November 2023).
- Werth, B.J. Rifamycins-Infectious Diseases-MSD Manual Professional Edition. Available online: https://www.msdmanuals.com/professional/infectious-diseases/bacteria-and-antibacterial-drugs/rifamycins (accessed on 9 November 2023).
- Werth, B.J. Daptomycin-Infectious Diseases-MSD Manual Professional Edition. Available online: https://www.msdmanuals.com/professional/infectious-diseases/bacteria-and-antibacterial-drugs/daptomycin (accessed on 9 November 2023).
- Zhou, Y.; Ji, X.; Liang, B.; Jiang, B.; Li, Y.; Yuan, T.; Zhu, L.; Liu, J.; Guo, X.; Sun, Y. Antimicrobial Resistance and Prevalence of Extended Spectrum β-Lactamase-Producing Escherichia Coli from Dogs and Cats in Northeastern China from 2012 to 2021. Antibiotics 2022, 11, 1506. [Google Scholar] [CrossRef] [PubMed]
- Shariati, A.; Dadashi, M.; Chegini, Z.; Van Belkum, A.; Mirzaii, M.; Khoramrooz, S.S.; Darban-Sarokhalil, D. The Global Prevalence of Daptomycin, Tigecycline, Quinupristin/Dalfopristin, and Linezolid-Resistant Staphylococcus aureus and Coagulase–Negative Staphylococci Strains: A Systematic Review and Meta-Analysis. Antimicrob. Resist. Infect. Control 2020, 9, 56. [Google Scholar] [CrossRef] [PubMed]
- Vercelli, C.; Amadori, M.; Gambino, G.; Re, G. Does Nitrofurantoin Improve the Portfolio of Vets against Resistant Bacteria in Companion Animals? Antibiotics 2023, 12, 911. [Google Scholar] [CrossRef] [PubMed]
| Antibiotic Group | Antibiotic | Abbreviation | WHO | Concentration |
|---|---|---|---|---|
| Aminoglycosides | Gentamicin | GEN | CIA | 2–16 μg/mL |
| Amphenicols | Chloramphenicol | CHL | HIA | 2–16 μg/mL |
| Cephalosporins | Cefoxitin 1 | CXI | HIA | 6 μg/mL |
| Folate inhibitor pathway | Trimethoprim/ sulfamethoxazole | TRS | HIA | 1/19–8/152 μg/mL |
| Glycopeptides | Vancomycin | VAN | NA | 0.25–32 μg/mL |
| Glycylcyclines | Tigecycline | TIG | NA | 0.03–0.5 μg/mL |
| Lincosamides | Clindamycin | CLI | HIA | 0.5–2 μg/mL |
| Lipopeptides | Daptomycin | DAP | NA | 0.5–4 μg/mL |
| Macrolides | Erythromycin | ERY | CIA | 0.25–4 μg/mL |
| Nitrofurans | Nitrofurantoin | NIT | NA | 32–64 μg/mL |
| Oxazolidinones | Linezolid | LIN | NA | 1–8 μg/mL |
| Penicillins | Ampicillin | AMP | HIA | 0.25–8 μg/mL |
| Oxacillin + 2% NaCl 1 | OXA+ | HIA | 0.25–4 μg/mL | |
| Penicillin | PEN | HIA | 0.06–8 μg/mL | |
| Quinolones | Levofloxacin (FQ) | LEV | HPCIA | 0,25–4 μg/mL |
| Ciprofloxacin (FQ) | CIP | HPCIA | 1–2 μg/mL | |
| Moxifloxacin (FQ) | MOX | HPCIA | 0.25–4 μg/mL | |
| Tetracyclines | Tetracycline | TET | HIA | 2–16 μg/mL |
| Ansamycins | Rifampicin | RIF | CIA | 0.5–4 μg/mL |
| Streptogramins | Quinupristin/ dalfopristin | QUD | HIA | 0.5–4 μg/mL |
| D-test | Erythromycin (E) + clindamycin (C) | DT | 4 μg/mL (E) + 0.5 μg/mL (C) |
| Type of Sample | Prevalence of Staphylococcus by Class | Staphylococcus Species | n and (%) Prevalence of Each Species | |
|---|---|---|---|---|
| Dog | Commensal mucosa | CoPS—79.8% | S. aureus | 6 (7.1) |
| S. pseudintermedius | 54 (64.2) | |||
| S. schleiferi | 7 (8.3) | |||
| CoNS—20.2% | S. cohnii | 1 (1.2) | ||
| S. epidermidis | 4 (4.8) | |||
| S. haemolyticus | 1 (1.2) | |||
| S. hominis | 3 (3.6) | |||
| S. sciuri 1 | 2 (2.4) | |||
| S. simulans | 2 (2.4) | |||
| S. warneri | 2 (2.4) | |||
| S. xylosus | 2 (2.4) | |||
| Active skin infection | CoPS—82.8% | S. aureus | 4 (13.8) | |
| S. pseudintermedius | 18 (62.2) | |||
| S. schleiferi | 2 (6.9) | |||
| CoNS—17.2% | S. canis | 1 (3.4) | ||
| S. chromogenes | 1 (3.4) | |||
| S. epidermidis | 2 (6.9) | |||
| S. felis | 1 (3.4) | |||
| Cat | Commensal mucosa | CoPS—16.9% | S. aureus | 6 (9.2) |
| S. pseudintermedius | 3 (4.6) | |||
| S. schleiferi | 2 (3.1) | |||
| CoNS—83.1% | S. capitis | 2 (3.1) | ||
| S. epidermidis | 2 (3.1) | |||
| S. felis | 32 (49.2) | |||
| S. hominis | 1 (1.5) | |||
| S. pettenkoferi | 2 (3.1) | |||
| S. saprophyticus | 1 (1.5) | |||
| S. sciuri 1 | 4 (6.2) | |||
| S. simulans | 6 (9.2) | |||
| S. xylosus | 4 (6.2) | |||
| Active skin infection | CoPS—11.1% | S. aureus | 1 (11.1) | |
| CoNS—88.9% | S. epidermidis | 1 (11.1) | ||
| S. felis | 5 (55.6) | |||
| S. hominis | 1 (11.1) | |||
| S. pasteuri | 1 (11.1) |
| AB Group | AB | % AMR/AB in Commensal S. aureus | % AMR/AB in Infection-Causing S. aureus |
|---|---|---|---|
| Aminoglycosides | GEN | 16.7 a,b (1/6) ± 15.2 | 0 a (0/4) ± 0 |
| Amphenicols | CHL | 83.3 c (5/6) ± 15.2 | 75 b (3/4) ± 21.7 |
| Cephalosporins | CXI | 16.7 a,b (1/6) ± 15.2 | 25 a,b (1/4) ± 21.7 |
| Folate inhibitor pathway | TRS | 0 b (0/6) ± 0 | 0 a (0/4) ± 0 |
| Glycopeptides | VAN | 50 a,c (3/6) ± 20.4 | 0 a (0/4) ± 0 |
| Glycylcyclines | TIG | 16.7 a,b (1/6) ± 15.2 | 25 a,b (1/4) ± 21.7 |
| Lincosamides | CLI | 50 a,c (3/6) ± 20.4 | 75 b (3/4) ± 21.7 |
| Lipopeptides | DAP | 0 b (0/6) ± 0 | 0 a (0/4) ± 0 |
| Macrolides | ERY | 83.3 c (5/6) ± 15.2 | 25 a,b (1/4) ± 25 |
| Nitrofurans | NIT | 0 b (0/6) ± 0 | 0 a (0/4) ± 0 |
| Oxazolidinones | LIN | 83.3 c (5/6) ± 15.2 | 25 a,b (1/4) ± 21.7 |
| Penicillins | AMP | 50 a,c (3/6) ± 20.4 | 50 a,b (2/4) ± 25 |
| PEN | 83.3 c (5/6) ± 15.2 | 75 b (3/4) ± 21.7 | |
| Quinolones | LEV | 16.7 a,b (1/6) ± 15.2 | 0 a (0/4) ± 0 |
| CIP | 0 b (0/6) ± 0 | 0 a (0/4) ± 0 | |
| MOX | 16.7 a,b (1/6) ± 15.2 | 0 a (0/4) ± 0 | |
| Tetracyclines | TET | 33.3 a,b (2/6) ± 19.2 | 25 a,b (1/4) ± 21.7 |
| Ansamycins | RIF | 16.7 a,b (1/6) ± 15.2 | 0 a (0/4) ± 0 |
| Streptogramins | QUD | 0 b (0/6) ± 0 | 25 a,b (1/4) ± 21.7 |
| AB Group | AB | % AMR/AB in Commensal S. pseudintermedius | % AMR/AB in Infection-Causing S. pseudintermedius |
|---|---|---|---|
| Aminoglycosides | GEN | 11.1 a (10/54) ± 4.3 | 27.8 a,b,c,d (5/18) ± 10.6 |
| Amphenicols | CHL | 68.5 e,f (37/54) ± 6.3 | 50 a,c,e (9/18) ± 11.8 |
| Folate inhibitor pathway | TRS | 13 a,i (7/54) ± 4.6 | 22.2 b,c,d (4/18) ± 9.8 |
| Glycopeptides | VAN | 0 g (0/54) ± 0 | 5.6 b,g (1/18) ± 5.4 |
| Glycylcyclines | TIG | 46.3 c,d (25/54) ± 6.8 | 5.6 b,g (1/18) ± 5.4 |
| Lincosamides | CLI | 46.3 c,d (25/54) ± 6.8 | 50 a,c,e (9/18) ± 11.8 |
| Lipopeptides | DAP | 1.9 g,h (1/54) ± 1.8 | 0 g (0/18) ± 0 |
| Macrolides | ERY | 57.4 c,e (31/54) ± 6.7 | 55.6 a,e,f (10/18) ± 11.7 |
| Nitrofurans | NIT | 3.7 a,g,h (2/54) ± 2.6 | 0 g (0/18) ± 0 |
| Oxazolidinones | LIN | 25.9 i,j (14/54) ± 6 | 11.1 b,d,g (2/18) ± 7.4 |
| Penicillins | AMP | 44.4 b,c,d (27/54) ± 6.8 | 66.6 e,f (12/18) ± 11.1 |
| OXA+ | 37 b,d,j (21/54) ± 6.6 | 33.3 a,c,d (6/18) ± 11.1 | |
| PEN | 77.8 f (41/54) ± 5.7 | 83.3 f (15/18) ± 8.8 | |
| Quinolones | LEV | 42.6 b,c,d,j (23/54) ± 6.7 | 44.4 a,c,e (8/18) ± 11.7 |
| CIP | 0 g (0/54) ± 0 | 38.9 a,c,e (7/18) ± 11.5 | |
| MOX | 42.6 b,c,d,j (23/54) ± 6.7 | 33.3 a,c,d (6/18) ± 11.1 | |
| Tetracyclines | TET | 51.9 c,d,e (28/54) ± 6.8 | 50 a,c,e (9/18) ± 11.8 |
| Ansamycins | RIF | 3.7 a,g,h (2/54) ± 2.6 | 0 g (0/18) ± 0 |
| Streptogramins | QUD | 7.4 a,h (3/54) ± 3.6 | 5.6 b,g (1/18) ± 5.4 |
| AB Group | AB | % AMR/AB in Commensal S. aureus | % AMR/AB in Infection-Causing S. aureus |
|---|---|---|---|
| Aminoglycosides | GEN | 0 a (0/6) ± 0 | 0 a (0/1) ± 0 |
| Amphenicols | CHL | 16.7 a,b (1/6) ± 15.2 | 0 a (0/1) ± 0 |
| Cephalosporins | CXI | 0 a (0/6) ± 0 | 0 a (0/1) ± 0 |
| Folate Inhibitor Pathway | TRS | 0 a (0/6) ± 0 | 0 a (0/1) ± 0 |
| Glycopeptides | VAN | 0 a (0/6) ± 0 | 100 b (1/1) ± 0 |
| Glycylcyclines | TIG | 0 a (0/6) ± 0 | 0 a (0/1) ± 0 |
| Lincosamides | CLI | 0 a (0/6) ± 0 | 0 a (0/1) ± 0 |
| Lipopeptides | DAP | 0 a (0/6) ± 0 | 0 a (0/1) ± 0 |
| Macrolides | ERY | 50 b (3/6) ± 20.4 | 100 b (1/1) ± 0 |
| Nitrofurans | NIT | 0 a (0/6) ± 0 | 100 b (1/1) ± 0 |
| Oxazolidinones | LIN | 0 a (0/6) ± 0 | 0 a (0/1) ± 0 |
| Penicillins | AMP | 16.7 a,b (1/6) ± 15.2 | 100 b (1/1) ± 0 |
| PEN | 16.7 a,b (1/6) ± 15.2 | 100 b (1/1) ± 0 | |
| Quinolones | LEV | 0 a (0/6) ± 0 | 100 b (1/1) ± 0 |
| CIP | 0 a (0/6) ± 0 | 100 b (1/1) ± 0 | |
| MOX | 0 a (0/6) ± 0 | 100 b (1/1) ± 0 | |
| Tetracyclines | TET | 0 a (0/6) ± 0 | 100 b (1/1) ± 0 |
| Ansamycins | RIF | 0 a (0/6) ± 0 | 100 b (1/1) ± 0 |
| Streptogramins | QUD | 0 a (0/6) ± 0 | 0 a (0/1) ± 0 |
| AB Group | AB | % AMR/AB in Commensal S. pseudintermedius |
|---|---|---|
| Aminoglycosides | GEN | 0 a (0/3) ± 0 |
| Amphenicols | CHL | 33.3 a,b,c (1/3) ± 27.2 |
| Folate inhibitor pathway | TRS | 0 a (0/3) ± 0 |
| Glycopeptides | VAN | 0 a (0/3) ± 0 |
| Glycylcyclines | TIG | 0 a (0/3) ± 0 |
| Lincosamides | CLI | 66.7 b,c (2/3) |
| Lipopeptides | DAP | 33.3 a,b,c (1/3) ± 27.2 |
| Macrolides | ERY | 66.7 b,c (2/3) ± 27.2 |
| Nitrofurans | NIT | 0 a (0/3) ± 0 |
| Oxazolidinones | LIN | 0 a (0/3) ± 0 |
| Penicillins | AMP | 33.3 a,b (1/3) ± 27.2 |
| OXA+ | 33.3 a,b (1/3) ± 27.2 | |
| PEN | 100 c (3/3) ± 0 | |
| Quinolones | LEV | 0 a (0/3) ± 0 |
| CIP | 33.3 a,b (1/3) ± 27.2 | |
| MOX | 0 a (0/3) ± 0 | |
| Tetracyclines | TET | 0 a (0/3) ± 0 |
| Ansamycins | RIF | 0 a (0/3) ± 0 |
| Streptogramins | QUD | 0 a (0/3) ± 0 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Marco-Fuertes, A.; Marin, C.; Gimeno-Cardona, C.; Artal-Muñoz, V.; Vega, S.; Montoro-Dasi, L. Multidrug-Resistant Commensal and Infection-Causing Staphylococcus spp. Isolated from Companion Animals in the Valencia Region. Vet. Sci. 2024, 11, 54. https://doi.org/10.3390/vetsci11020054
Marco-Fuertes A, Marin C, Gimeno-Cardona C, Artal-Muñoz V, Vega S, Montoro-Dasi L. Multidrug-Resistant Commensal and Infection-Causing Staphylococcus spp. Isolated from Companion Animals in the Valencia Region. Veterinary Sciences. 2024; 11(2):54. https://doi.org/10.3390/vetsci11020054
Chicago/Turabian StyleMarco-Fuertes, Ana, Clara Marin, Concepción Gimeno-Cardona, Violeta Artal-Muñoz, Santiago Vega, and Laura Montoro-Dasi. 2024. "Multidrug-Resistant Commensal and Infection-Causing Staphylococcus spp. Isolated from Companion Animals in the Valencia Region" Veterinary Sciences 11, no. 2: 54. https://doi.org/10.3390/vetsci11020054
APA StyleMarco-Fuertes, A., Marin, C., Gimeno-Cardona, C., Artal-Muñoz, V., Vega, S., & Montoro-Dasi, L. (2024). Multidrug-Resistant Commensal and Infection-Causing Staphylococcus spp. Isolated from Companion Animals in the Valencia Region. Veterinary Sciences, 11(2), 54. https://doi.org/10.3390/vetsci11020054







