Loggerhead Sea Turtle as Possible Source of Transmission for Zoonotic Listeriosis in the Marine Environment
Abstract
Simple Summary
Abstract
1. Introduction
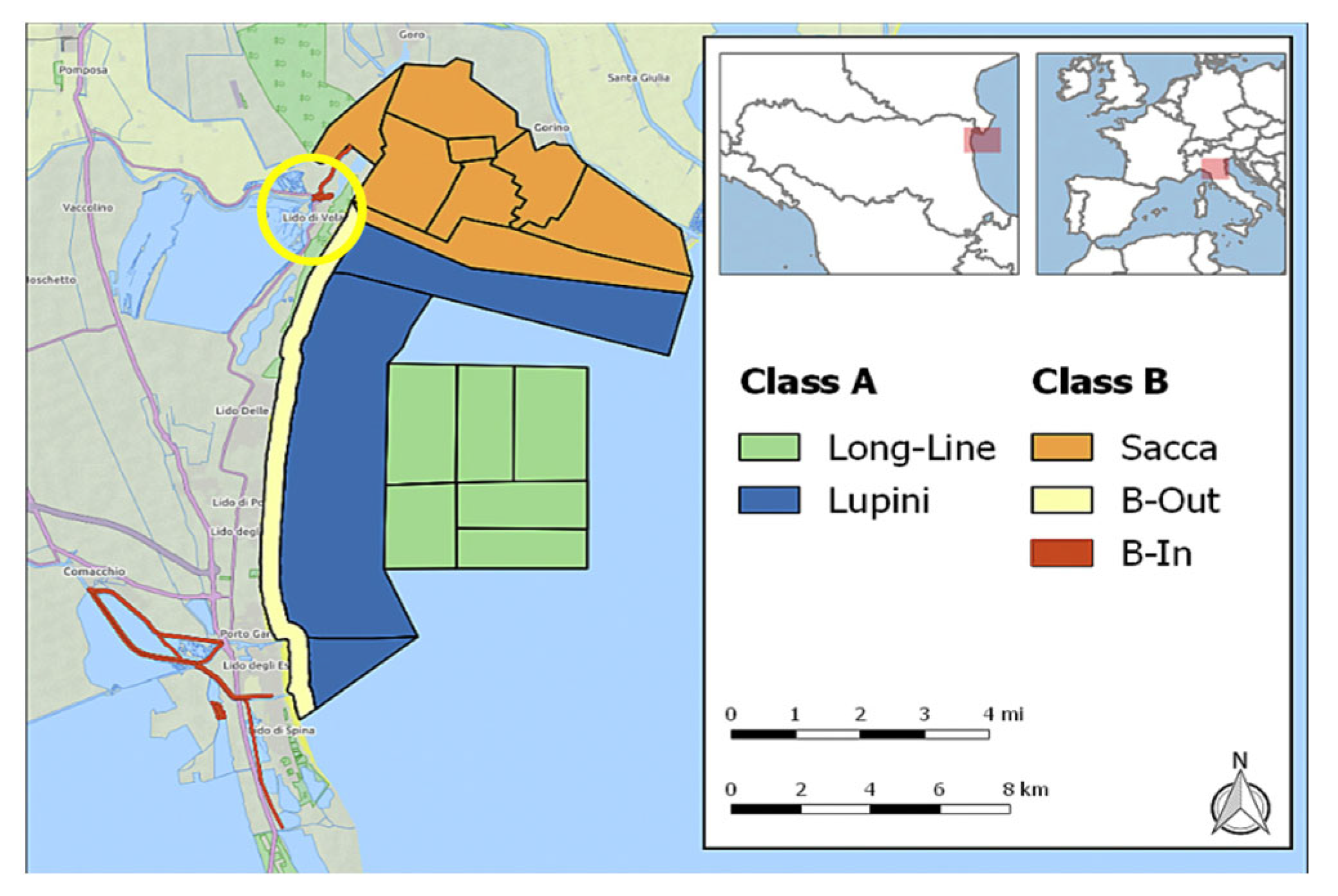
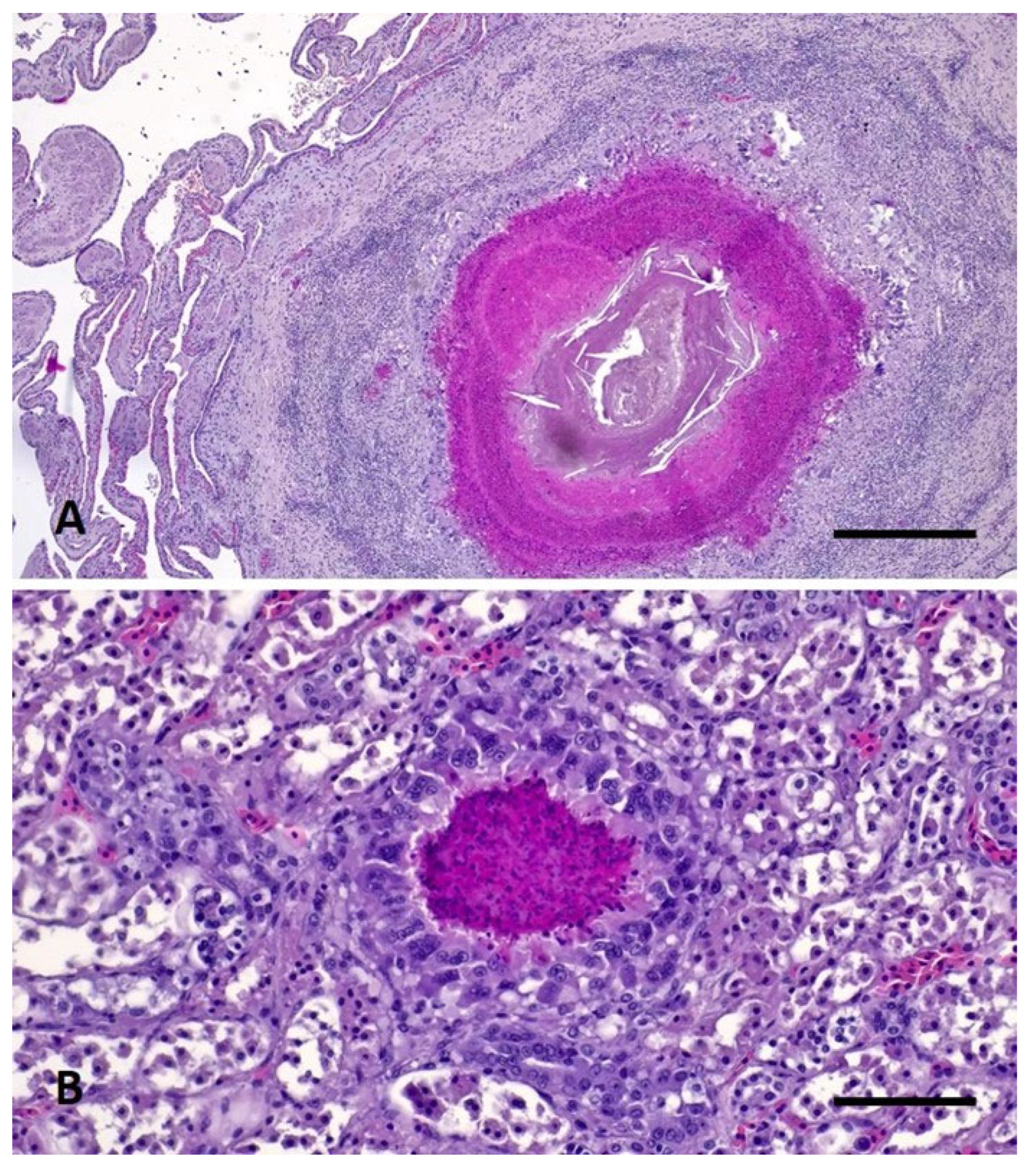
2. Materials, Methods, and Results
3. Discussion
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Buchanan, R.L.; Stahl, H.G.; Whiting, R.C. Effects and Interactions of Temperature, pH, Atmosphere, Sodium Chloride, and Sodium Nitrite on the Growth of Listeria monocytogenes. J. Food Prot. 1989, 52, 844–851. [Google Scholar] [CrossRef]
- Bucur, F.I.; Grigore-Gurgu, L.; Crauwels, P.; Riedel, C.U.; Nicolau, A.I. Resistance of Listeria monocytogenes to Stress Conditions Encountered in Food and Food Processing Environments. Front. Microbiol. 2018, 9, 2700. [Google Scholar] [CrossRef]
- Al-Ghazali, M.R.; Al-Azawi, S.K. Listeria monocytogenes Contamination of Crops Grown on Soil Treated with Sewage Sludge Cake. J. Appl. Bacteriol. 1990, 69, 642–647. [Google Scholar] [CrossRef]
- Odjadjare, E.E.O.; Obi, L.C.; Okoh, A.I. Municipal Wastewater Effluents as a Source of Listerial Pathogens in the Aquatic Milieu of the Eastern Cape Province of South Africa: A Concern of Public Health Importance. Int. J. Environ. Res. Public Heal. 2010, 7, 2376–2394. [Google Scholar] [CrossRef]
- Koopmans, M.M.; Brouwer, M.C.; Vázquez-Boland, J.A.; van de Beek, D. Human Listeriosis. Clin. Microbiol. Rev. 2023, 36, e00060-19. [Google Scholar] [CrossRef]
- Ferreira, V.; Wiedmann, M.; Teixeira, P.; Stasiewicz, M.J. Listeria Monocytogenes Persistence in Food-Associated Environments: Epidemiology, Strain Characteristics, and Implications for Public Health. J. Food Prot. 2014, 77, 150–170. [Google Scholar] [CrossRef]
- Gambarin, P.; Magnabosco, C.; Losio, M.N.; Pavoni, E.; Gattuso, A.; Arcangeli, G.; Favretti, M. Listeria monocytogenes in Ready-to-Eat Seafood and Potential Hazards for the Consumers. Int. J. Microbiol. 2012, 2012, 497635. [Google Scholar] [CrossRef]
- Hellström, S.; Kiviniemi, K.; Autio, T.; Korkeala, H. Listeria monocytogenes is Common in Wild Birds in Helsinki Region and Genotypes Are Frequently Similar with Those Found along the Food Chain. J. Appl. Microbiol. 2008, 104, 883–888. [Google Scholar] [CrossRef]
- Cilia, G.; Turchi, B.; Fratini, F.; Bilei, S.; Bossù, T.; De Marchis, M.L.; Cerri, D.; Pacini, M.I.; Bertelloni, F. Prevalence, Virulence and Antimicrobial Susceptibility of Salmonella spp., Yersinia enterocolitica and Listeria monocytogenes in European Wild Boar (Sus scrofa) Hunted in Tuscany (Central Italy). Pathogens 2021, 10, 93. [Google Scholar] [CrossRef]
- OIE Technical Disease Card. Available online: https://www.woah.org/app/uploads/2021/05/listeria-monocytogenes-infection-with.pdf (accessed on 28 April 2023).
- Di Renzo, L.; De Angelis, M.E.; Torresi, M.; Di Lollo, V.; Di Teodoro, G.; Averaimo, D.; Defourny, S.V.P.; Di Giacinto, F.; Profico, C.; Olivieri, V.; et al. First Report of Septicaemic Listeriosis in a Loggerhead Sea Turtle (Caretta caretta) Stranded along the Adriatic Coast: Strain Detection and Sequencing. Animals 2022, 12, 2364. [Google Scholar] [CrossRef]
- Esposito, E.; Paduano, G.; Iaccarino, D. First Detection of Listeria monocytogenes in Stranded Loggerhead Sea Turtle (Caretta caretta) along the Coast of Campania Region (Southern Italy). In Proceedings of the 7th Mediterranean Conference on Marine Turtles, Tetouan, Morocco, 18–21 October 2022. [Google Scholar]
- Poppi, L.; Marchiori, E. Standard Protocol for Post Mortem Examination on Sea Turtles; Guidelines produced within the Adriatic project “Network for the Conservation of Cetaceans and Sea Turtles in the Adriatic (NETCET) 2013.
- Bacteriological Analytical Manual (BAM). Available online: https://www.fda.gov/food/laboratory-methods-food/bacteriological-analytical-manual-bam (accessed on 20 April 2023).
- Wick, R.R.; Judd, L.M.; Gorrie, C.L.; Holt, K.E. Unicycler: Resolving Bacterial Genome Assemblies from Short and Long Sequencing Reads. PLoS Comput. Biol. 2017, 13, e1005595. [Google Scholar] [CrossRef]
- Van Walle, I.; Björkman, J.T.; Cormican, M.; Dallman, T.; Mossong, J.; Moura, A.; Pietzka, A.; Ruppitsch, W.; Takkinen, J.; European Listeria WGS Typing Group. Retrospective Validation of Whole Genome Sequencing-Enhanced Surveillance of Listeriosis in Europe, 2010 to 2015. Eurosurveillance 2018, 23, 1700798. [Google Scholar] [CrossRef]
- Moura, A.; Criscuolo, A.; Pouseele, H.; Maury, M.M.; Leclercq, A.; Tarr, C.; Björkman, J.T.; Dallman, T.; Reimer, A.; Enouf, V.; et al. Whole Genome-Based Population Biology and Epidemiological Surveillance of Listeria monocytogenes. Nat. Microbiol. 2016, 2, 16185. [Google Scholar] [CrossRef]
- De las Heras, A.; Cain, R.J.; Bielecka, M.K.; Vázquez-Boland, J.A. Regulation of Listeria Virulence: PrfA Master and Commander. Curr. Opin. Microbiol. 2011, 14, 118–127. [Google Scholar] [CrossRef]
- Disson, O.; Moura, A.; Lecuit, M. Making Sense of the Biodiversity and Virulence of Listeria monocytogenes. Trends Microbiol. 2021, 29, 811–822. [Google Scholar] [CrossRef]
- Maury, M.M.; Tsai, Y.-H.; Charlier, C.; Touchon, M.; Chenal-Francisque, V.; Leclercq, A.; Criscuolo, A.; Gaultier, C.; Roussel, S.; Brisabois, A.; et al. Uncovering Listeria monocytogenes Hypervirulence by Harnessing Its Biodiversity. Nat. Genet. 2016, 48, 308–313. [Google Scholar] [CrossRef]
- Plumb, D.C. Veterinary Drug Handbook, 4th ed.; Iowa State University Digital Press: Ames, IA, USA, 2002. [Google Scholar]
- Soldati, G.; Lu, Z.H.; Vaughan, L.; Polkinghorne, A.; Zimmermann, D.R.; Huder, J.B.; Pospischil, A. Detection of Mycobacteria and Chlamydiae in Granulomatous Inflammation of Reptiles: A Retrospective Study. Vet. Pathol. 2004, 41, 388–397. [Google Scholar] [CrossRef]
- Gračan, R.; Buršic, M.; Mladineo, I.; Kučinic, M.; Lazar, B.; Lackovic, G. Gastrointestinal Helminth Community of Loggerhead Sea Turtle Caretta caretta in the Adriatic Sea. Dis. Aquat. Org. 2012, 99, 227–236. [Google Scholar] [CrossRef]
- Girling, S.J.; Fraser, M.A. Listeria monocytogenes Septicaemia in an Inland Bearded Dragon, Pogona vitticeps. J. Herpetol. Med. Surg. 2004, 14, 6–9. [Google Scholar] [CrossRef]
- Matt, C.L.; Ramachandran, A.; Allison, R.W.; Wall, C.R.; Dieterly, A.M.; Brandão, J. Listeria monocytogenes in an inland bearded dragon (Pogona vitticeps). J. Exot. Pet Med. 2019, 30, 76–81. [Google Scholar] [CrossRef]
- Vancraeynest, D.; Pasmans, F.; De Graef, E.; Hermans, K.; Decostere, A. Listeria monocytogenes Associated Myocardial Perforation in a Bearded Dragon (Pogona vitticeps). Vlaams Diergeneeskd. Tijdschr. 2006, 75, 232–234. [Google Scholar]
- Mundt, J.O. Occurrence of Enterococci in Animals in a Wild Environment. Appl. Microbiol. 1963, 11, 136–140. [Google Scholar] [CrossRef]
- Del Turco, E.R.; Bartoletti, M.; Dahl, A.; Cervera, C.; Pericàs, J.M. How Do I Manage a Patient with Enterococcal Bacteraemia? Clin. Microbiol. Infect. 2021, 27, 364–371. [Google Scholar] [CrossRef]
- Parsons, C.; Niedermeyer, J.; Gould, N.; Brown, P.; Strules, J.; Parsons, A.W.; Bernardo Mesa-Cruz, J.; Kelly, M.J.; Hooker, M.J.; Chamberlain, M.J.; et al. Listeria monocytogenes at the Human–Wildlife Interface: Black Bears (Ursus americanus) as Potential Vehicles for Listeria. Microb. Biotechnol. 2020, 13, 706–721. [Google Scholar] [CrossRef]
- Ivanek, R.; Gröhn, Y.T.; Wiedmann, M. Listeria Monocytogenes in Multiple Habitats and Host Populations: Review of Available Data for Mathematical Modeling. Foodborne Pathog. Dis. 2006, 3, 319–336. [Google Scholar] [CrossRef]
- Landrigan, P.J.; Stegeman, J.J.; Fleming, L.E.; Allemand, D.; Anderson, D.M.; Backer, L.C.; Brucker-Davis, F.; Chevalier, N.; Corra, L.; Czerucka, D.; et al. Human Health and Ocean Pollution. Ann. Glob. Health 2020, 86, 151. [Google Scholar] [CrossRef]
- Althaus, D.; Zweifel, C.; Stephan, R. Analysis of a Poultry Slaughter Process: Influence of Process Stages on the Microbiological Contamination of Broiler Carcasses. Ital. J. Food Saf. 2017, 6, 7097. [Google Scholar] [CrossRef]
- Nüesch-Inderbinen, M.; Bloemberg, G.V.; Müller, A.; Stevens, M.J.A.; Cernela, N.; Kollöffel, B.; Stephan, R. Listeriosis Caused by Persistence of Listeria monocytogenes Serotype 4b Sequence Type 6 in Cheese Production Environment. Emerg. Infect. Dis. 2021, 27, 284–288. [Google Scholar] [CrossRef]
- Smith, A.M.; Tau, N.P.; Smouse, S.L.; Allam, M.; Ismail, A.; Ramalwa, N.R.; Disenyeng, B.; Ngomane, M.; Thomas, J. Outbreak of Listeria monocytogenes in South Africa, 2017–2018: Laboratory Activities and Experiences Associated with Whole-Genome Sequencing Analysis of Isolates. Foodborne Pathog. Dis. 2019, 16, 524–530. [Google Scholar] [CrossRef]
- Thomas, J.; Govender, N.; McCarthy, K.M.; Erasmus, L.K.; Doyle, T.J.; Allam, M.; Ismail, A.; Ramalwa, N.; Sekwadi, P.; Ntshoe, G.; et al. Outbreak of Listeriosis in South Africa Associated with Processed Meat. N. Engl. J. Med. 2020, 382, 632–643. [Google Scholar] [CrossRef]
- Hanes, R.M.; Huang, Z. Investigation of Antimicrobial Resistance Genes in Listeria monocytogenes from 2010 through to 2021. Int. J. Environ. Res. Public Heal. 2022, 19, 5506. [Google Scholar] [CrossRef]
- Parra-Flores, J.; Holý, O.; Bustamante, F.; Lepuschitz, S.; Pietzka, A.; Contreras-Fernández, A.; Castillo, C.; Ovalle, C.; Alarcón-Lavín, M.P.; Cruz-Córdova, A.; et al. Virulence and Antibiotic Resistance Genes in Listeria monocytogenes Strains Isolated from Ready-to-Eat Foods in Chile. Front. Microbiol. 2022, 12, 796040. [Google Scholar] [CrossRef]
- Chen, T.; Orsi, R.H.; Chen, R.; Gunderson, M.; Roof, S.; Wiedmann, M.; Childs-Sanford, S.E.; Cummings, K.J. Characterization of Listeria monocytogenes Isolated from Wildlife in Central New York. Vet. Med. Sci. 2022, 8, 1319–1329. [Google Scholar] [CrossRef] [PubMed]
- Schoder, D.; Guldimann, C.; Märtlbauer, E. Asymptomatic Carriage of Listeria monocytogenes by Animals and Humans and Its Impact on the Food Chain. Foods 2022, 11, 3472. [Google Scholar] [CrossRef]
- Casale, P.; Freggi, D.; Cinà, A.; Rocco, M. Spatio-Temporal Distribution and Migration of Adult Male Loggerhead Sea Turtles (Caretta caretta) in the Mediterranean Sea: Further Evidence of the Importance of Neritic Habitats off North Africa. Mar. Biol. 2013, 160, 703–718. [Google Scholar] [CrossRef]
- Arizza, V.; Vecchioni, L.; Caracappa, S.; Sciurba, G.; Berlinghieri, F.; Gentile, A.; Persichetti, M.F.; Arculeo, M.; Alduina, R. New Insights into the Gut Microbiome in Loggerhead Sea Turtles Caretta caretta Stranded on the Mediterranean Coast. PLoS ONE 2019, 14, e0220329. [Google Scholar] [CrossRef]
- Fichi, G.; Cardeti, G.; Cersini, A.; Mancusi, C.; Guarducci, M.; Di Guardo, G.; Terracciano, G. Bacterial and Viral Pathogens Detected in Sea Turtles Stranded along the Coast of Tuscany, Italy. Vet. Microbiol. 2016, 185, 56–61. [Google Scholar] [CrossRef] [PubMed]
- Kuschke, S.G. What Lives on and in the Sea Turtle? A Literature Review of Sea Turtle Bacterial Microbiota. Anim. Microbiome 2022, 4, 52. [Google Scholar] [CrossRef]
- Orós, J.; Torrent, A.; Calabuig, P.; Déniz, S. Diseases and Causes of Mortality among Sea Turtles Stranded in the Canary Islands, Spain (1998–2001). Dis. Aquat. Org. 2005, 63, 13–24. [Google Scholar] [CrossRef]
- Warwick, C.; Arena, P.C.; Steedman, C. Health Implications Associated with Exposure to Farmed and Wild Sea Turtles. JRSM Short Rep. 2013, 4, 1–7. [Google Scholar] [CrossRef]
- Basha, K.A.; Kumar, N.R.; Das, V.; Reshmi, K.; Rao, B.M.; Lalitha, K.V.; Joseph, T.C. Prevalence, Molecular Characterization, Genetic Heterogeneity and Antimicrobial Resistance of Listeria monocytogenes Associated with Fish and Fishery Environment in Kerala, India. Lett. Appl. Microbiol. 2019, 69, 286–293. [Google Scholar] [CrossRef] [PubMed]
- Embarek, P.K.B. Presence, Detection and Growth of Listeria monocytogenes in Seafoods: A Review. Int. J. Food Microbiol. 1994, 23, 17–34. [Google Scholar] [CrossRef] [PubMed]
- Paradiso, R.; Riccardi, M.G.; Cecere, B.; Riccone, N.; Scialla, R.; Anzalone, A.; Cerrone, A.; De Carlo, E.; Borriello, G.; Fusco, G. Whole-Genome Sequencing-Based Characterization of a Listeria monocytogenes Strain from an Aborted Water Buffalo in Southern Italy. Microorganisms 2021, 9, 1875. [Google Scholar] [CrossRef]
- Struthers, J.D.; Kucerova, Z.; Finley, A.; Goe, A.; Huffman, J.; Phair, K. Septicaemic Listeriosis in a White-Faced Saki (Pithecia pithecia). J. Comp. Pathol. 2022, 194, 7–13. [Google Scholar] [CrossRef] [PubMed]
- Farrell, J.A.; Whitmore, L.; Mashkour, N.; Rollinson Ramia, D.R.; Thomas, R.S.; Eastman, C.B.; Burkhalter, B.; Yetsko, K.; Mott, C.; Wood, L.; et al. Detection and Population Genomics of Sea Turtle Species via Noninvasive Environmental DNA Analysis of Nesting Beach Sand Tracks and Oceanic Water. Mol. Ecol. Resour. 2022, 22, 2471–2493. [Google Scholar] [CrossRef]





Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rubini, S.; Baruffaldi, M.; Taddei, R.; D’Annunzio, G.; Scaltriti, E.; Tambassi, M.; Menozzi, I.; Bondesan, G.; Mazzariol, S.; Centelleghe, C.; et al. Loggerhead Sea Turtle as Possible Source of Transmission for Zoonotic Listeriosis in the Marine Environment. Vet. Sci. 2023, 10, 344. https://doi.org/10.3390/vetsci10050344
Rubini S, Baruffaldi M, Taddei R, D’Annunzio G, Scaltriti E, Tambassi M, Menozzi I, Bondesan G, Mazzariol S, Centelleghe C, et al. Loggerhead Sea Turtle as Possible Source of Transmission for Zoonotic Listeriosis in the Marine Environment. Veterinary Sciences. 2023; 10(5):344. https://doi.org/10.3390/vetsci10050344
Chicago/Turabian StyleRubini, Silva, Matilde Baruffaldi, Roberta Taddei, Giulia D’Annunzio, Erika Scaltriti, Martina Tambassi, Ilaria Menozzi, Giulia Bondesan, Sandro Mazzariol, Cinzia Centelleghe, and et al. 2023. "Loggerhead Sea Turtle as Possible Source of Transmission for Zoonotic Listeriosis in the Marine Environment" Veterinary Sciences 10, no. 5: 344. https://doi.org/10.3390/vetsci10050344
APA StyleRubini, S., Baruffaldi, M., Taddei, R., D’Annunzio, G., Scaltriti, E., Tambassi, M., Menozzi, I., Bondesan, G., Mazzariol, S., Centelleghe, C., Corazzola, G., Savini, F., Indio, V., Serraino, A., & Giacometti, F. (2023). Loggerhead Sea Turtle as Possible Source of Transmission for Zoonotic Listeriosis in the Marine Environment. Veterinary Sciences, 10(5), 344. https://doi.org/10.3390/vetsci10050344






