A Multiplex PCR Method for Simultaneous Detection of Infectious Laryngotracheitis Virus and Ornithobacterium rhinotracheale
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Control Sample from Vaccines
2.2. Field Samples
2.3. DNA Extraction and Purification
2.4. Multiplex Primer Design
2.5. Evaluation of Primers
2.6. Optimizing Multiplex PCR
2.7. Determining the Limit of Detection of the Multiplex PCR
2.8. Detection of ILTV and ORT in Field Samples by Multiplex PCR Assay
3. Results
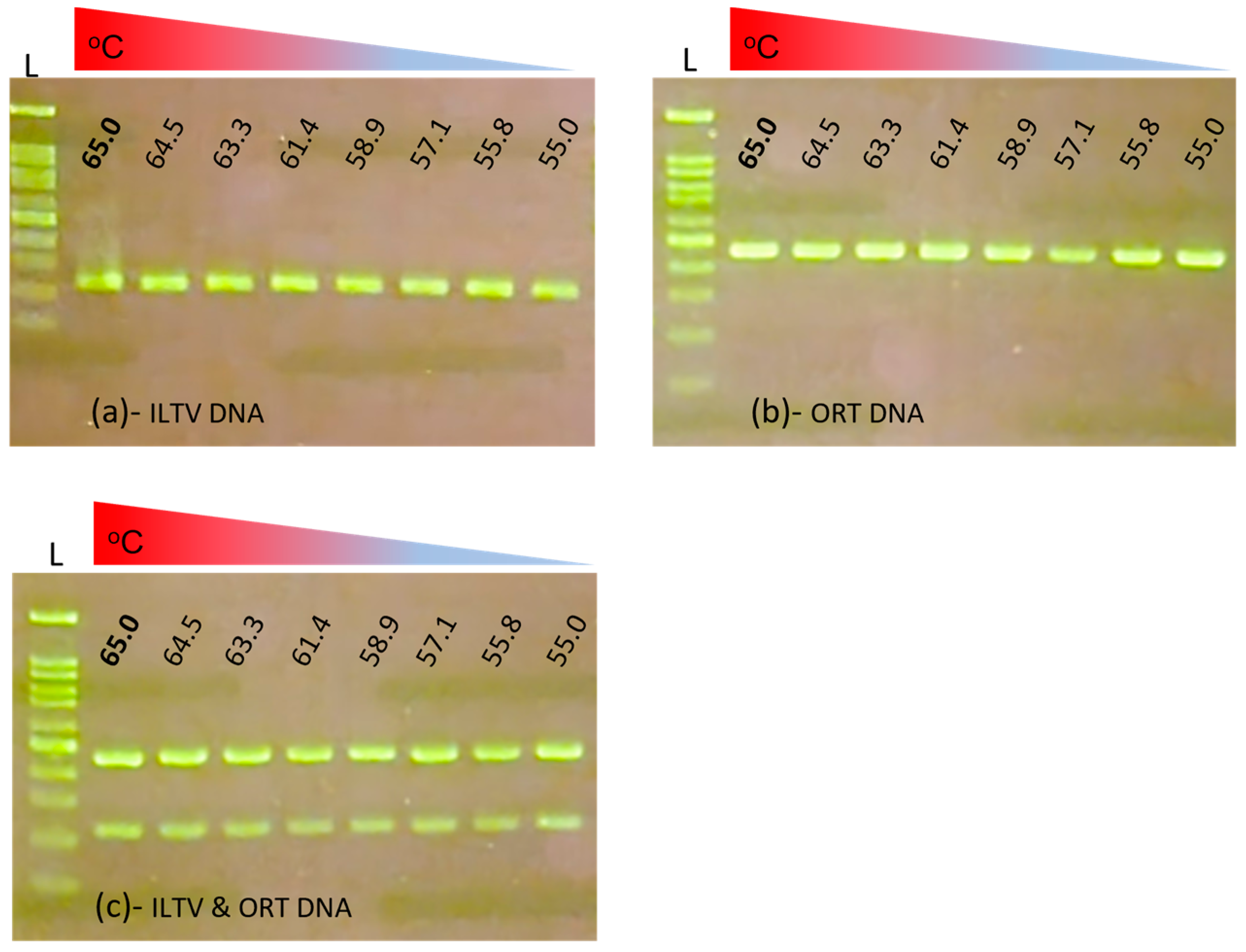
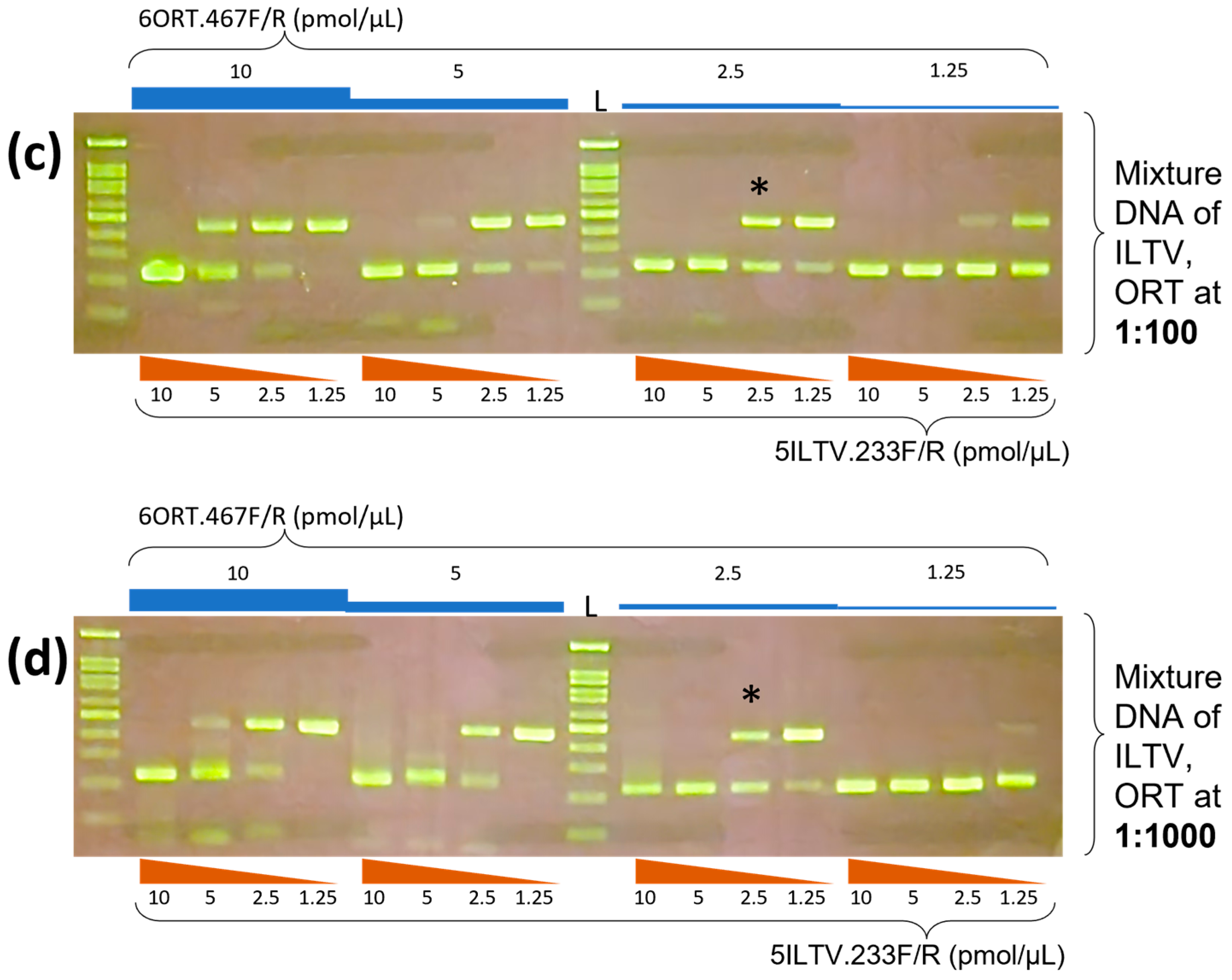
3.1. Optimization of Multiplex PCR
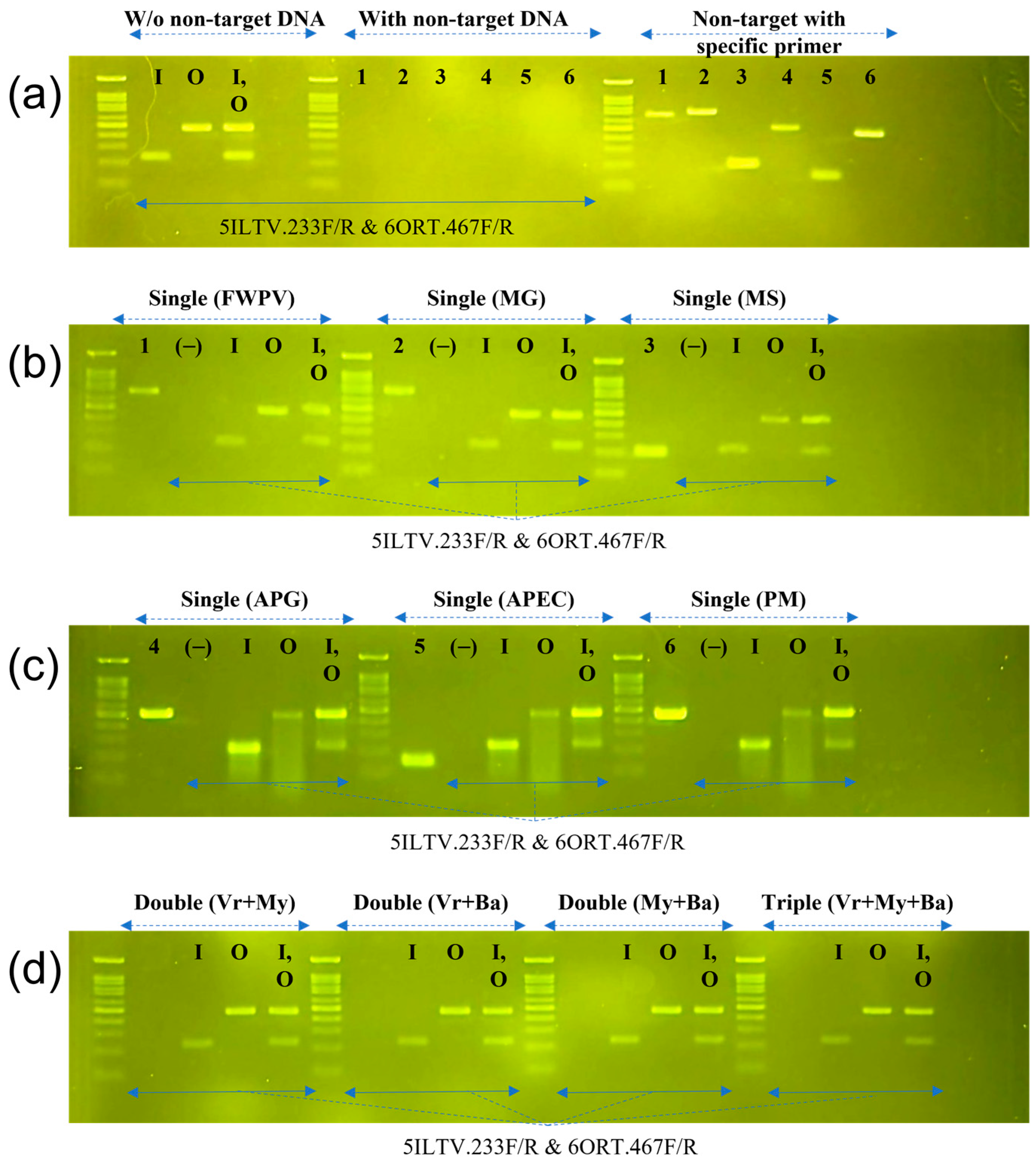
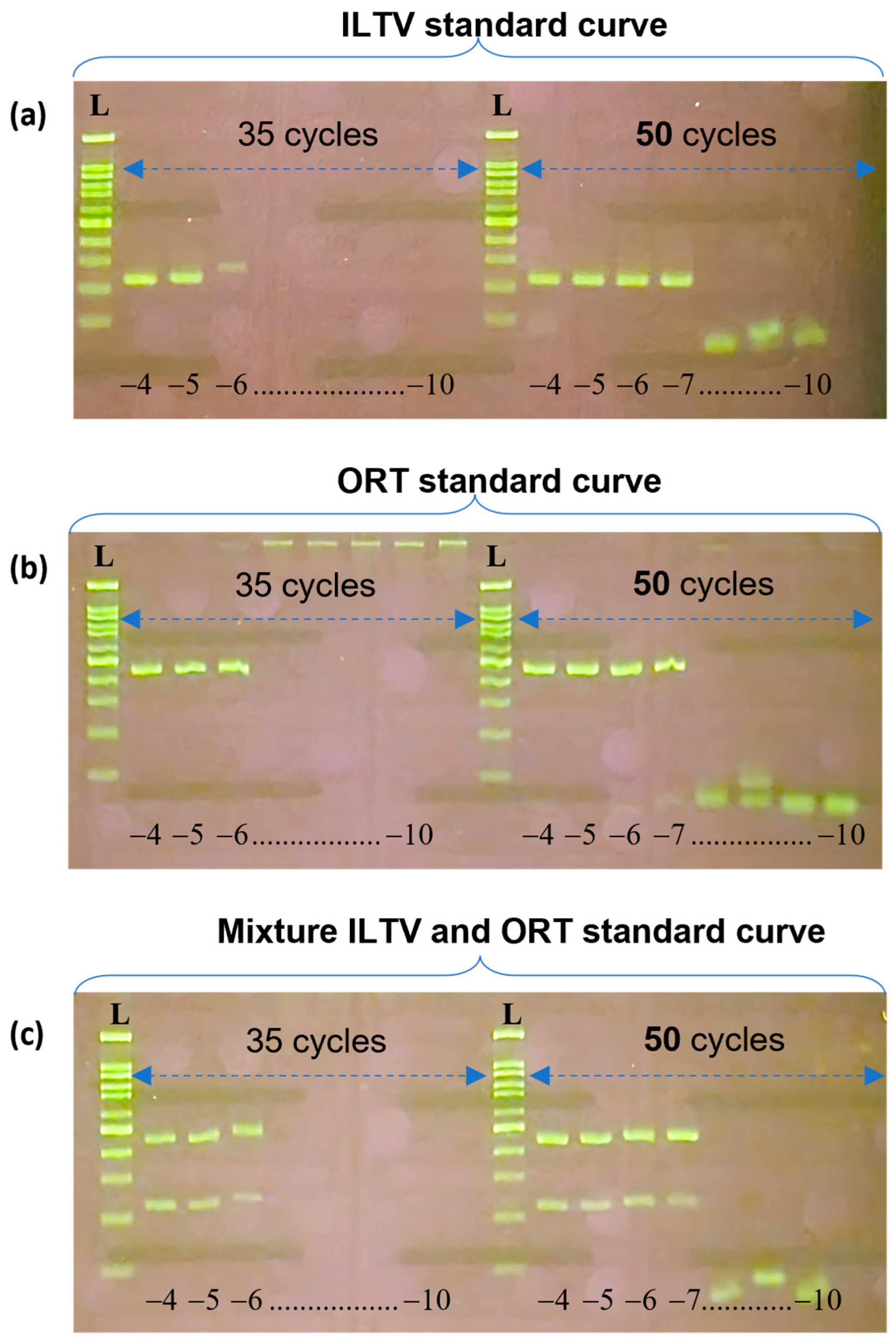
3.2. Specificity and Limit of Detection of Multiplex PCR
3.3. Application of Multiplex PCR on Field Samples
4. Discussion
4.1. Selection of the Target Pathogens and Detection Method
4.2. Multiplex Primer Design and Validation
4.3. Application of the Newly Developed Multiplex PCR
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
| Forward/Reverse 1 | Gene | Sequence (5′-3′) | Reference |
|---|---|---|---|
| TK_792F | TK | TGGCAACGGATTCCGGCA | [29] |
| TK_673R | TTGCAAATAGCGTCTGGTCGATTGAA | [43] | |
| Or01_1607F | Or01 | GCTGAAATTATAAAAATGCC | [17] |
| Or01_1607R | CACAAGCATAGACATTGG | [17] | |
| Or01_1315F | ACATCAGCGGTTTGGTAAGC | This study | |
| Or01_1315R | AGTACTTCCAGTTGCTCCGT | This study |
| Forward/Reverse 1 | Pathogen | Sequence (5′-3′) | Expected Size (bp) | Reference |
|---|---|---|---|---|
| Pox.Po681R | FWPV | CGCCGCATCATCTACTTATC | 681 | [56] |
| Pox.Po681F | CCACACAGCGCCATTCATTA | |||
| TspE4C2.152F | E. coli | CACTATTCGTAAGGTCATCC | 152 | [57] |
| TspE4C2.152R | AGTTTATCGCTGCGGGTCGC | |||
| PM.460F | PM | GCTGTAAACGAACTCGCCAC | 460 | [58] |
| PM.460R | ATCCGCTATTTACCCAGTGG | |||
| MS.207F | MS | GAGAAGCAAAATAGTGATATCA | 202 | [59] |
| MS.207R | CAGTCGTCTCCGAAGTTAACAA | |||
| MG.732F | MG | GGATCCCATCTCGACCACGAGAAAA | 732 | [60] |
| MG.732R | CTTTCAATCAGTGAGTAACTGATGA | |||
| APG.500F | APG | CAATGTCGATCCTGGTACAATGAG | 500 | [61] |
| APG.500R | CAAGGTATCGATCGTCTCTCTACT |
References
- Rajeoni, A.H.; Ghalyanchilangeroudi, A.; Khalesi, B.; Madadi, M.S.; Hosseini, H. The tracheal virome of broiler chickens with respiratory disease complex in Iran: The metagenomics study. Iran. J. Microbiol. 2021, 13, 337–344. [Google Scholar] [CrossRef] [PubMed]
- Sid, H.; Benachour, K.; Rautenschlein, S. Co-infection with multiple respiratory pathogens contributes to increased mortality rates in Algerian poultry flocks. Avian Dis. 2015, 59, 440–446. [Google Scholar] [CrossRef] [PubMed]
- Roussan, D.A.; Haddad, R.; Khawaldeh, G. Molecular survey of avian respiratory pathogens in commercial broiler chicken flocks with respiratory diseases in Jordan. Poult. Sci. 2008, 87, 444–448. [Google Scholar] [CrossRef]
- Hassan, M.S.H.; Abdul-Careem, M.F. Avian viruses that impact table egg production. Animals 2020, 10, 1747. [Google Scholar] [CrossRef] [PubMed]
- Kannaki, T.R.; Priyanka, E.; Haunshi, S.; Subbiah, M. A systematic review and meta-analysis on global prevalence of infectious diseases in backyard chicken in the recent two decades. Indian J. Poult. Sci. 2021, 56, 287–294. [Google Scholar] [CrossRef]
- García, M.; Spatz, S. Infectious Laryngotracheitis. In Diseases of Poultry; John Wiley & Sons: Hoboken, NJ, USA, 2020; pp. 189–209. [Google Scholar] [CrossRef]
- Lee, S.W.; Markham, P.F.; Markham, J.F.; Petermann, I.; Noormohammadi, A.H.; Browning, G.F.; Ficorilli, N.P.; Hartley, C.A.; Devlin, J.M. First complete genome sequence of infectious laryngotracheitis virus. BMC Genom. 2011, 12, 197. [Google Scholar] [CrossRef]
- Blacker, H.P.; Kirkpatrick, N.C.; Rubite, A.; O’Rourke, D.; Noormohammadi, A.H. Epidemiology of recent outbreaks of infectious laryngotracheitis in poultry in Australia. Aust. Vet. J. 2011, 89, 89–94. [Google Scholar] [CrossRef]
- Sabir, A.J.; Olaogun, O.M.; O’Rourke, D.; Fakhri, O.; Coppo, M.J.C.; Devlin, J.M.; Konsak-Ilievski, B.; Noormohammadi, A.H. Full genomic characterisation of an emerging infectious laryngotracheitis virus class 7b from Australia linked to a vaccine strain revealed its identity. Infect. Genet. Evol. J. Mol. Epidemiol. Evol. Genet. Infect. Dis. 2020, 78, 104067. [Google Scholar] [CrossRef]
- Kirkpatrick, N.C.; Mahmoudian, A.; O’Rourke, D.; Noormohammadi, A.H. Differentiation of infectious laryngotracheitis virus isolates by restriction fragment length polymorphic analysis of polymerase chain reaction products amplified from multiple genes. Avian Dis. 2006, 50, 28–34. [Google Scholar] [CrossRef]
- Piccirillo, A.; Lavezzo, E.; Niero, G.; Moreno, A.; Massi, P.; Franchin, E.; Toppo, S.; Salata, C.; Palù, G. Full Genome Sequence-Based Comparative Study of Wild-Type and Vaccine Strains of Infectious Laryngotracheitis Virus from Italy. PLoS ONE 2016, 11, e0149529. [Google Scholar] [CrossRef]
- El-Ghany, W.A.A. An updated comprehensive review on ornithobacteriosis: A worldwide emerging avian respiratory disease. Open Vet. J. 2021, 11, 555–568. [Google Scholar] [CrossRef] [PubMed]
- van Veen, L.; van Empel, P.; Fabri, T. Ornithobacterium rhinotracheale, a Primary Pathogen in Broilers. Avian Dis. 2000, 44, 896–900. [Google Scholar] [CrossRef]
- Boulianne, M.; Blackall, P.J.; Hofacre, C.L.; Ruiz, J.A.; Sandhu, T.S.; Hafez, H.M.; Chin, R.P.; Register, K.B.; Jackwood, M.W. Pasteurellosis and Other Respiratory Bacterial Infections. In Diseases of Poultry; John Wiley & Sons: Hoboken, NJ, USA, 2020; pp. 831–889. [Google Scholar] [CrossRef]
- Van Empel, P.; van den Bosch, H.; Loeffen, P.; Storm, P. Identification and serotyping of Ornithobacterium rhinotracheale. J. Clin. Microbiol. 1997, 35, 418–421. [Google Scholar] [CrossRef] [PubMed]
- Schuijffel, D.F.; Van Empel, P.C.; Segers, R.P.; Van Putten, J.P.; Nuijten, P.J. Vaccine potential of recombinant Ornithobacterium rhinotracheale antigens. Vaccine 2006, 24, 1858–1867. [Google Scholar] [CrossRef] [PubMed]
- Schuijffel, D.F.; van Empel, P.C.; Pennings, A.M.; van Putten, J.P.; Nuijten, P.J. Successful selection of cross-protective vaccine candidates for Ornithobacterium rhinotracheale infection. Infect. Immun. 2005, 73, 6812–6821. [Google Scholar] [CrossRef] [PubMed]
- Abdelaziz, A.M.; Mohamed, M.H.A.; Fayez, M.M.; Al-Marri, T.; Qasim, I.; Al-Amer, A.A. Molecular survey and interaction of common respiratory pathogens in chicken flocks (field perspective). Vet. World 2019, 12, 1975–1986. [Google Scholar] [CrossRef] [PubMed]
- Xie, Z.; Luo, S.; Xie, L.; Liu, J.; Pang, Y.; Deng, X.; Xie, Z.; Fan, Q.; Khan, M.I. Simultaneous typing of nine avian respiratory pathogens using a novel GeXP analyzer-based multiplex PCR assay. J. Virol. Methods 2014, 207, 188–195. [Google Scholar] [CrossRef]
- Croville, G.; Foret, C.; Heuillard, P.; Senet, A.; Delpont, M.; Mouahid, M.; Ducatez, M.F.; Kichou, F.; Guerin, J.-L. Disclosing respiratory co-infections: A broad-range panel assay for avian respiratory pathogens on a nanofluidic PCR platform. Avian Pathol. 2018, 47, 253–260. [Google Scholar] [CrossRef]
- Laamiri, N.; Aouini, R.; Marnissi, B.; Ghram, A.; Hmila, I. A multiplex real-time RT-PCR for simultaneous detection of four most common avian respiratory viruses. Virology 2018, 515, 29–37. [Google Scholar] [CrossRef]
- Saba Shirvan, A.; Mardani, K. Molecular detection of infectious bronchitis and Newcastle disease viruses in broiler chickens with respiratory signs using duplex RT-PCR. J. Vet. Res. Forum 2014, 5, 319–323. [Google Scholar]
- Pang, Y.; Wang, H.; Girshick, T.; Xie, Z.; Khan, M.I. Development and application of a multiplex polymerase chain reaction for avian respiratory agents. Avian Dis. 2002, 46, 691–699. [Google Scholar] [CrossRef]
- Rashid, S.; Naeem, K.; Ahmed, Z.; Saddique, N.; Abbas, M.A.; Malik, S.A. Multiplex polymerase chain reaction for the detection and differentiation of avian influenza viruses and other poultry respiratory pathogens. Poult. Sci. 2009, 88, 2526–2531. [Google Scholar] [CrossRef]
- Nguyen, T.T.; Kwon, H.J.; Kim, I.H.; Hong, S.M.; Seong, W.J.; Jang, J.W.; Kim, J.H. Multiplex nested RT-PCR for detecting avian influenza virus, infectious bronchitis virus and Newcastle disease virus. J. Virol. Methods 2013, 188, 41–46. [Google Scholar] [CrossRef]
- Wang, Z.; Zuo, J.; Gong, J.; Hu, J.; Jiang, W.; Mi, R.; Huang, Y.; Chen, Z.; Phouthapane, V.; Qi, K.; et al. Development of a multiplex PCR assay for the simultaneous and rapid detection of six pathogenic bacteria in poultry. AMB Express 2019, 9, 185. [Google Scholar] [CrossRef] [PubMed]
- Malik, Y.S.; Patnayak, D.P.; Goyal, S.M. Detection of three avian respiratory viruses by single-tube multiplex reverse transcription-polymerase chain reaction assay. J. Vet. Diagn. Investig. 2004, 16, 244–248. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.S.; Song, D.S.; Kim, S.Y.; Lyoo, K.S.; Park, B.K. Detection of porcine circovirus type 2 in feces of pigs with or without enteric disease by polymerase chain reaction. J. Vet. Diagn. Investig. 2003, 15, 369–373. [Google Scholar] [CrossRef] [PubMed]
- Beltrão, N.; Egochega, R.F.; Furian, T.; Rodenbusch, C.; Fallavena, L.C.B.; Pasquali, G.; Canal, C. A sensitive nested-polymerase chain reaction protocol to detect infectious laryngotracheitis virus. Acta Sci. Vet. 2012, 40, 1055. [Google Scholar]
- Numee, S.; Hauck, R.; Hafez, H.M. Detection and typing of Ornithobacterium rhinotracheale from German poultry flocks. Avian Dis. 2012, 56, 654–658. [Google Scholar] [CrossRef]
- Shen, Z.; Qu, W.; Wang, W.; Lu, Y.; Wu, Y.; Li, Z.; Hang, X.; Wang, X.; Zhao, D.; Zhang, C. MPprimer: A program for reliable multiplex PCR primer design. BMC Bioinform. 2010, 11, 143. [Google Scholar] [CrossRef]
- Song, H.; Kim, H.; Kim, S.; Kwon, Y.; Kim, H. Research Note: Simultaneous detection of infectious laryngotracheitis virus, fowlpox virus, and reticuloendotheliosis virus in chicken specimens. Poult. Sci. 2021, 100, 100986. [Google Scholar] [CrossRef]
- Gowthaman, V.; Kumar, S.; Koul, M.; Dave, U.; Murthy, T.; Munuswamy, P.; Tiwari, R.; Karthik, K.; Dhama, K.; Michalak, I.; et al. Infectious laryngotracheitis: Etiology, epidemiology, pathobiology, and advances in diagnosis and control—A comprehensive review. Vet. Q. 2020, 40, 140–161. [Google Scholar] [CrossRef] [PubMed]
- Heikinheimo, A.; Korkeala, H. Multiplex PCR assay for toxinotyping Clostridium perfringens isolates obtained from Finnish broiler chickens. Lett. Appl. Microbiol. 2005, 40, 407–411. [Google Scholar] [CrossRef] [PubMed]
- O’Regan, E.; McCabe, E.; Burgess, C.; McGuinness, S.; Barry, T.; Duffy, G.; Whyte, P.; Fanning, S. Development of a real-time multiplex PCR assay for the detection of multiple Salmonella serotypes in chicken samples. BMC Microbiol. 2008, 8, 156. [Google Scholar] [CrossRef]
- Tahir, M.S.; Mehmood, D.; Sultan, A.U.; Saeed, M.H.; Khan, A.R.; Ansari, F.; Salman, M.M.; Majeed, K.A. A modified strategy of multiplex RT-PCR for simultaneous detection of H5, H7, and H9 subtypes of avian influenza virus based on common forward oligo. J. Appl. Poult. Res. 2016, 25, 322–327. [Google Scholar] [CrossRef]
- Gao, Q.; Yun, B.; Wang, Q.; Jiang, L.; Zhu, H.; Gao, Y.; Qin, L.; Wang, Y.; Qi, X.; Gao, H.; et al. Development and Application of a Multiplex PCR Method for Rapid Differential Detection of Subgroup A, B, and J Avian Leukosis Viruses. J. Clin. Microbiol. 2014, 52, 37–44. [Google Scholar] [CrossRef]
- Caterina, K.M.; Frasca, S.; Girshick, T.; Khan, M.I. Development of a multiplex PCR for detection of avian adenovirus, avian reovirus, infectious bursal disease virus, and chicken anemia virus. Mol. Cell. Probes 2004, 18, 293–298. [Google Scholar] [CrossRef] [PubMed]
- Mettifogo, E.; Buzinhani, M.; Buim, M.; Timenetsky, J.; Ferreira, A. Evaluation of a PCR multiplex for detection and differentiation of Mycoplasma synoviae, M. gallisepticum, and M. gallisepticum strain F-vaccine. Pesqui. Veterinária Bras. 2015, 35, 13–18. [Google Scholar] [CrossRef]
- Kaplinski, L.; Remm, M. MultiPLX: Automatic grouping and evaluation of PCR primers. Methods Mol. Biol. 2015, 1275, 127–142. [Google Scholar] [CrossRef]
- Ou, S.C.; Giambrone, J.J.; Macklin, K.S. Detection of infectious laryngotracheitis virus from darkling beetles and their immature stage (lesser mealworms) by quantitative polymerase chain reaction and virus isolation. J. Appl. Poult. Res. 2012, 21, 33–38. [Google Scholar] [CrossRef]
- Creelan, J.L.; Calvert, V.M.; Graham, D.A.; McCullough, S.J. Rapid detection and characterization from field cases of infectious laryngotracheitis virus by real-time polymerase chain reaction and restriction fragment length polymorphism. Avian Pathol. 2006, 35, 173–179. [Google Scholar] [CrossRef]
- Williams, R.A.; Bennett, M.; Bradbury, J.M.; Gaskell, R.M.; Jones, R.C.; Jordan, F.T.W. Demonstration of sites of latency of infectious laryngotracheitis virus using the polymerase chain reaction. J. Gen. Virol. 1992, 73, 2415–2420. [Google Scholar] [CrossRef]
- Humberd, J.; García, M.; Riblet, S.M.; Resurreccion, R.S.; Brown, T.P. Detection of Infectious Laryngotracheitis Virus in Formalin-Fixed, Paraffin-Embedded Tissues by Nested Polymerase Chain Reaction. Avian Dis. 2002, 46, 64–74. [Google Scholar] [CrossRef]
- Han, M.G.; Kweon, C.H.; Mo, I.P.; Kim, S.J. Pathogenicity and vaccine efficacy of a thymidine kinase gene deleted infectious laryngotracheitis virus expressing the green fluorescent protein gene. Arch. Virol. 2002, 147, 1017–1031. [Google Scholar] [CrossRef] [PubMed]
- van Empel, P.C.; Hafez, H.M. Ornithobacterium rhinotracheale: A review. Avian Pathol. 1999, 28, 217–227. [Google Scholar] [CrossRef] [PubMed]
- Abdelwhab, E.M.; Lüschow, D.; Hafez, H.M. Development of Real-Time Polymerase Chain Reaction Assay for Detection of Ornithobacterium rhinotracheale in Poultry. Avian Dis. 2013, 57, 663–666. [Google Scholar] [CrossRef]
- Hashish, A.; Sinha, A.; Sato, Y.; Macedo, N.R.; El-Gazzar, M. Development and Validation of a New TaqMan Real-Time PCR for the Detection of Ornithobacterium rhinotracheale. Microorganisms 2022, 10, 341. [Google Scholar] [CrossRef] [PubMed]
- Veiga, I.M.B.; Lüschow, D.; Gutzer, S.; Hafez, H.M.; Mühldorfer, K. Phylogenetic relationship of Ornithobacterium rhinotracheale isolated from poultry and diverse avian hosts based on 16S rRNA and rpoB gene analyses. BMC Microbiol. 2019, 19, 31. [Google Scholar] [CrossRef] [PubMed]
- Dieffenbach, C.W.; Lowe, T.M.; Dveksler, G.S. General concepts for PCR primer design. PCR Methods Appl. 1993, 3, S30–S37. [Google Scholar] [CrossRef]
- Werid, G.M.; Zhang, H.; Ibrahim, Y.M.; Pan, Y.; Zhang, L.; Xu, Y.; Zhang, W.; Wang, W.; Chen, H.; Fu, L.; et al. Development of a Multiplex RT-PCR Assay for Simultaneous Detection of Four Potential Zoonotic Swine RNA Viruses. Vet. Sci. 2022, 9, 176. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Liu, F.; Li, Q.; Wu, M.; Lei, L.; Pan, Z. A multiplex RT-PCR assay for rapid and simultaneous detection of four RNA viruses in swine. J. Virol. Methods 2019, 269, 38–42. [Google Scholar] [CrossRef] [PubMed]
- Cao, T.B.P.; Nguyen, V.G.; Nguyen, T.T.; Mai, T.N.; Vu, T.N.; Huynh, T.M.L. Application of Direct PCR for Detection of Several Bacteria in Chicken with Respiratory Disease Complex in Hanoi and Neighbour Vicinity. Vietnam J. Agri. Sci. 2022, 20, 156–165. (In Vietnamese) [Google Scholar]
- Nguyen, V.G.; Chung, H.C.; Do, H.Q.; Nguyen, T.T.; Cao, T.B.; Truong, H.T.; Mai, T.N.; Le, T.T.; Nguyen, T.H.; Le, T.L.; et al. Serological and Molecular Characterization of Avian Metapneumovirus in Chickens in Northern Vietnam. Vet. Sci. 2021, 8, 206. [Google Scholar] [CrossRef] [PubMed]
- Van, N.T.B.; Yen, N.T.P.; Nhung, N.T.; Cuong, N.V.; Kiet, B.T.; Hoang, N.V.; Hien, V.B.; Chansiripornchai, N.; Choisy, M.; Ribas, A.; et al. Characterization of viral, bacterial, and parasitic causes of disease in small-scale chicken flocks in the Mekong Delta of Vietnam. Poult. Sci. 2020, 99, 783–790. [Google Scholar] [CrossRef] [PubMed]
- Gyuranecz, M.; Foster, J.T.; Dan, A.; Ip, H.S.; Egstad, K.F.; Parker, P.G.; Higashiguchi, J.M.; Skinner, M.A.; Hofle, U.; Kreizinger, Z.; et al. Worldwide phylogenetic relationship of avian poxviruses. J. Virol. 2013, 87, 4938–4951. [Google Scholar] [CrossRef]
- Clermont, O.; Christenson, J.K.; Denamur, E.; Gordon, D.M. The Clermont Escherichia coli phylo-typing method revisited: Improvement of specificity and detection of new phylo-groups. Environ. Microbiol. Rep. 2013, 5, 58–65. [Google Scholar] [CrossRef]
- Townsend, K.M.; Frost, A.J.; Lee, C.W.; Papadimitriou, J.M.; Dawkins, H.J. Development of PCR assays for species- and type-specific identification of Pasteurella multocida isolates. J. Clin. Microbiol. 1998, 36, 1096–1100. [Google Scholar] [CrossRef]
- Emam, M.; Hashem, Y.M.; El-Hariri, M.; El-Jakee, J. Detection and antibiotic resistance of Mycoplasma gallisepticum and Mycoplasma synoviae among chicken flocks in Egypt. Vet. World 2020, 13, 1410–1416. [Google Scholar] [CrossRef]
- García, M.; Ikuta, N.; Levisohn, S.; Kleven, S.H. Evaluation and comparison of various PCR methods for detection of Mycoplasma gallisepticum infection in chickens. Avian Dis. 2005, 49, 125–132. [Google Scholar] [CrossRef]
- Chen, X.; Miflin, J.K.; Zhang, P.; Blackall, P.J. Development and application of DNA probes and PCR tests for Haemophilus paragallinarum. Avian Dis. 1996, 40, 398–407. [Google Scholar] [CrossRef]


| Forward/Reverse | Pathogen | Sequence (5′-3′) | Expected Size (bp) |
|---|---|---|---|
| 5ILTV.233F | ILTV | CCGACTTTCGCCGCGTTGTACT | 233 |
| /5ILTV.233R | ACGAGACGCCTCCCGACATTCA | ||
| 6ORT.467F | ORT | AGCGGTGGAGGTGCTAGCCAAT | 467 |
| /6ORT.467R | GCAGCAGGCGCTGGAGTTTCTT |
| Experiment | Template DNA | Primer Mix | Expected Result | ||
|---|---|---|---|---|---|
| Non-Target 1 | Target 2 | ILTV | ORT | ||
| 1 | None | ILTV | 5ILTV.233F /5ILTV.233R and 6ORT.467F /6ORT.467R | + | - |
| 2 | None | ORT | - | + | |
| 3 | None | ILTV; ORT. | + | + | |
| 4 | None | Water | - | - | |
| 5 | Single | None | - | - | |
| 6 | Single | ILTV | + | - | |
| 7 | Single | ORT | - | + | |
| 8 | Single | ILTV; ORT. | + | + | |
| 9 | Double | ILTV | + | - | |
| 10 | Double | ORT | - | + | |
| 11 | Double | ILTV; ORT. | + | + | |
| 12 | Triple | ILTV | + | - | |
| 13 | Triple | ORT | - | + | |
| 14 | Triple | ILTV; ORT. | + | + | |
| 5ILTV.233F/5ILTV.233R Mix (pmol/µL) | |||||
|---|---|---|---|---|---|
| (10) | (5) | (2.5) | (1.25) | ||
| 6ORT.467F/6ORT.467R Mix (pmol/µL) | (10) | 1 | 2 | 3 | 4 |
| (5) | 5 | 6 | 7 | 8 | |
| (2.5) | 9 | 10 | 11 | 12 | |
| (1.25) | 13 | 14 | 15 | 16 | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nguyen, V.-G.; Cao, T.-B.-P.; Le, V.-T.; Truong, H.-T.; Chu, T.-T.-H.; Dang, H.-A.; Nguyen, T.-H.; Le, T.-L.; Huynh, T.-M.-L. A Multiplex PCR Method for Simultaneous Detection of Infectious Laryngotracheitis Virus and Ornithobacterium rhinotracheale. Vet. Sci. 2023, 10, 272. https://doi.org/10.3390/vetsci10040272
Nguyen V-G, Cao T-B-P, Le V-T, Truong H-T, Chu T-T-H, Dang H-A, Nguyen T-H, Le T-L, Huynh T-M-L. A Multiplex PCR Method for Simultaneous Detection of Infectious Laryngotracheitis Virus and Ornithobacterium rhinotracheale. Veterinary Sciences. 2023; 10(4):272. https://doi.org/10.3390/vetsci10040272
Chicago/Turabian StyleNguyen, Van-Giap, Thi-Bich-Phuong Cao, Van-Truong Le, Ha-Thai Truong, Thi-Thanh-Huong Chu, Huu-Anh Dang, Thi-Hoa Nguyen, Thi-Luyen Le, and Thi-My-Le Huynh. 2023. "A Multiplex PCR Method for Simultaneous Detection of Infectious Laryngotracheitis Virus and Ornithobacterium rhinotracheale" Veterinary Sciences 10, no. 4: 272. https://doi.org/10.3390/vetsci10040272
APA StyleNguyen, V.-G., Cao, T.-B.-P., Le, V.-T., Truong, H.-T., Chu, T.-T.-H., Dang, H.-A., Nguyen, T.-H., Le, T.-L., & Huynh, T.-M.-L. (2023). A Multiplex PCR Method for Simultaneous Detection of Infectious Laryngotracheitis Virus and Ornithobacterium rhinotracheale. Veterinary Sciences, 10(4), 272. https://doi.org/10.3390/vetsci10040272






