Some Contributions to the Study of Oenological Lactic Acid Bacteria through Their Interaction with Polyphenols
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials and Reagents
2.2. LAB Strains and E. coli Strain
2.3. Incubations of LAB Strains with Wine Polyphenols
2.4. Growth of LAB Strains in the Presence of Phenolic Compounds
2.5. Growth of Pathogen E. coli in the Presence of Free Supernatants (CFS) from LAB Strains
2.6. Cell Culture Assays: LAB Adhesion and Inhibition of E. coli Adherence to Caco-2 Cells
2.7. Statistical Analysis
3. Results and Discussion
3.1. Metabolism of Wine Polyphenols by LAB Strains
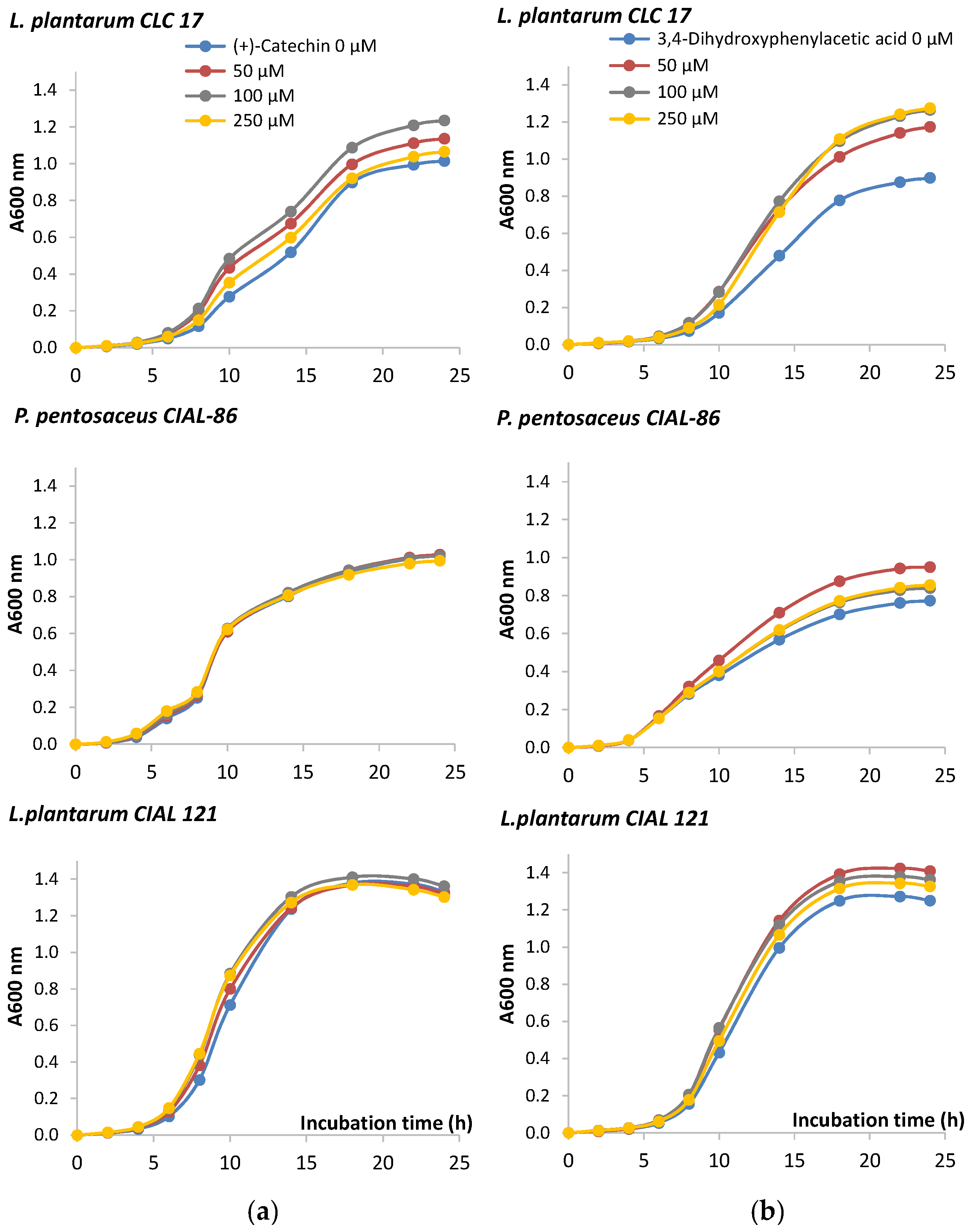
3.2. Effects of Phenolic Compounds on the LAB Growth
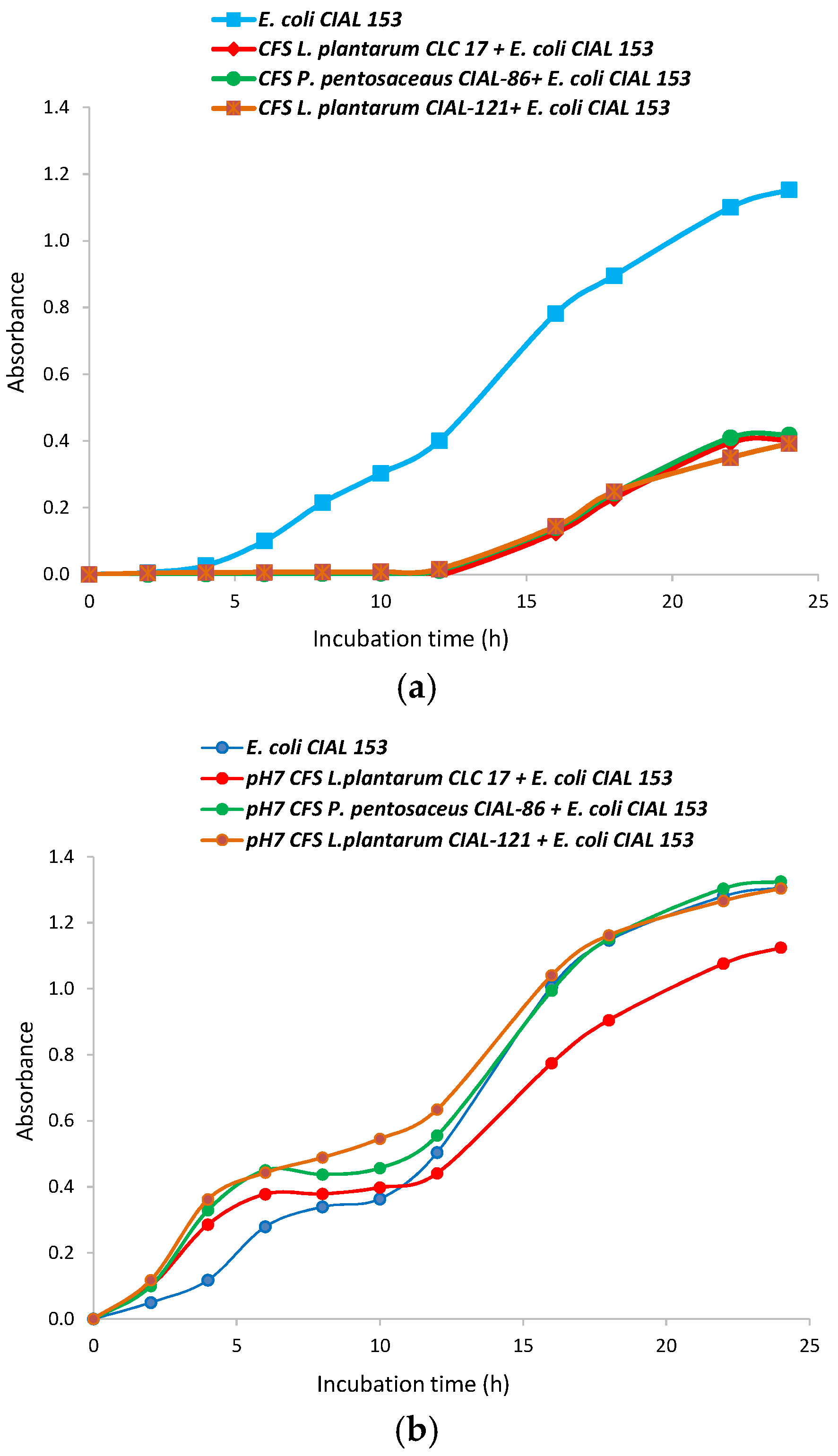
3.3. Effects of the Cell-Free Supernatants (CFS) from the LAB Strains on Growth of Pathogen E. coli
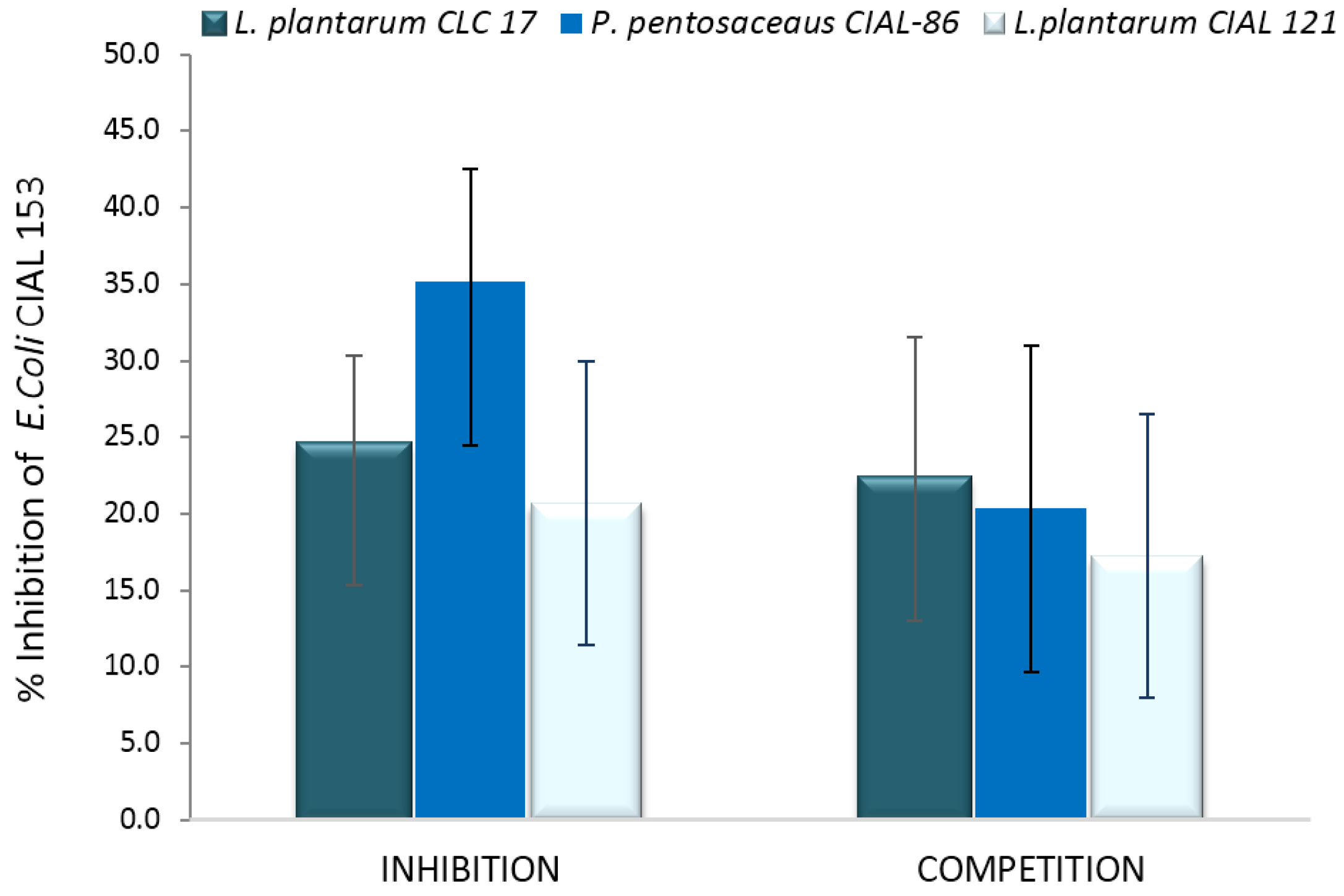
3.4. Effects of LAB on Adherence of Pathogen E. coli to Caco-2 Cells
4. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Sleator, R.D. Designer probiotics: Development and applications in gastrointestinal health. World J. Gastrointest. Pathophysiol. 2015, 6, 73–78. [Google Scholar] [PubMed]
- De Palencia, P.F.; Werning, M.L.; Sierra-Filardi, E.; Duenas, M.T.; Irastorza, A.; Corbi, A.L.; Lopez, P. Probiotic properties of the 2-substituted (1,3)-β-d-glucan-producing bacterium Pediococcus parvulus 2.6. Appl. Environ. Microbiol. 2009, 75, 4887–4891. [Google Scholar] [CrossRef] [PubMed]
- Foligne, B.; Dewulf, J.; Breton, J.; Claisse, O.; Lonvaud-Funel, A.; Pot, B. Probiotic properties of non-conventional lactic acid bacteria: Immunomodulation by Oenococcus oeni. Int. J. Food Microbiol. 2010, 140, 136–145. [Google Scholar] [CrossRef] [PubMed]
- García-Ruiz, A.; González de Llano, D.; Esteban-Fernández, A.; Requena, T.; Bartolomé, B.; Moreno-Arribas, M.V. Assessment of probiotic properties in lactic acid bacteria isolated from wine. Food Microbiol. 2014, 44, 220–225. [Google Scholar] [CrossRef] [PubMed]
- Rossi, M.; Amaretti, A.; Leonardi, A.; Raimondi, S.; Simone, M.; Quartieri, A. Potential impact of probiotic consumption on the bioactivity of dietary phytochemicals. J. Agric. Food Chem. 2013, 61, 9551–9558. [Google Scholar] [CrossRef] [PubMed]
- Pereira-Caro, G.; Oliver, C.M.; Weerakkody, R.; Singh, T.; Conlon, M.; Borges, G.; Sanguansri, L.; Lockett, T.; Roberts, S.A.; Crozier, A.; et al. Chronic administration of a microencapsulated probiotic enhances the bioavailability of orange juice flavanones in humans. Free Radic. Biol. Med. 2015, 84, 206–214. [Google Scholar] [CrossRef] [PubMed]
- Del Rio, D.; Rodriguez-Mateos, A.; Spencer, J.P.E.; Tognolini, M.; Borges, G.; Crozier, A. Dietary (poly)phenolics in human health: Structures, bioavailability, and evidence of protective effects against chronic diseases. Antioxid. Redox Signal. 2013, 18, 1818–1892. [Google Scholar] [CrossRef] [PubMed]
- Muñoz-González, I.; Espinosa-Martos, I.; Rodríguez, J.M.; Jiménez-Girón, A.; Martín-Álvarez, P.J.; Bartolomé, B.; Moreno-Arribas, M.V. Moderate consumption of red wine can modulate human intestinal inflammatory response. J. Agric. Food Chem. 2014, 62, 10567–10575. [Google Scholar] [CrossRef] [PubMed]
- Dueñas, M.; Cueva, C.; Muñoz-González, I.; Jiménez-Girón, A.; Sánchez-Patán, F.; Santos-Buelga, C.; Moreno-Arribas, M.V.; Bartolomé, B. Studies on Modulation of Gut Microbiota by Wine Polyphenols: From Isolated Cultures to Omic Approaches. Antioxidants 2015, 4, 1–21. [Google Scholar] [CrossRef] [PubMed]
- Cardona, F.; Andrés-Lacueva, C.; Tulipani, S.; Tinahones, F.J.; Queipo-Ortuño, M.I. Benefits of polyphenols on gut microbiota and implications in human health. J. Nutr. Biochem. 2013, 24, 1415–1422. [Google Scholar] [CrossRef] [PubMed]
- Boto-Ordoñez, M.; Rothwell, J.A.; Andrés-Lacueva, C.; Manach, C.; Scalbert, A.; Urpi-Sarda, M. Prediction of the wine polyphenol metabolic space: An application of the Phenol-Explorer database. Mol. Nutr. Food Res. 2014, 58, 466–477. [Google Scholar] [CrossRef] [PubMed]
- Muñoz-González, I.; Jiménez-Girón, A.; Martín-Álvarez, P.J.; Bartolomé, B.; Moreno-Arribas, M.V. Profiling of Microbial-Derived Phenolic Metabolites in Human Feces after Moderate Red Wine Intake. J. Agric. Food Chem. 2013, 61, 9470–9479. [Google Scholar] [CrossRef] [PubMed]
- Monagas, M.; Urpi-Sarda, M.; Sánchez-Patán, F.; Llorach, R.; Garrido, I.; Gómez-Cordovés, C.; Andrés-Lacueva, C.; Bartolomé, B. Insights into the metabolism and microbial biotransformation of dietary flavan-3-ols and the bioactivity of their metabolites. Food Funct. 2010, 1, 233–253. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez-Mateos, A.; Vauzour, D.; Krueger, C.G.; Shanmuganayagam, D.; Reed, J.; Calani, L.; Mena, P.; Del Rio, D.; Crozier, A. Bioavailability, bioactivity and impact on health of dietary flavonoids and related compounds: An update. Arch. Toxicol. 2014, 88, 1803–1853. [Google Scholar] [CrossRef] [PubMed]
- Etxeberria, U.; Fernández-Quintela, A.; Milagro, F.I.; Aguirre, L.; Martínez, J.A.; Portillo, M.P. Impact of Polyphenols and Polyphenol-Rich Dietary Sources on Gut Microbiota Composition. J. Agric. Food Chem. 2013, 61, 9517–9533. [Google Scholar] [CrossRef] [PubMed]
- Cueva, C.; Sánchez-Patán, F.; Monagas, M.; Walton, G.E.; Gibson, G.R.; Martín-Álvarez, P.J.; Bartolome, B.; Moreno-Arribas, M.V. In vitro fermentation of grape seed flavan-3-ol fractions by human faecal microbiota: Changes in microbial groups and phenolic metabolites. FEMS Microbiol. Ecol. 2013, 83, 792–805. [Google Scholar] [CrossRef] [PubMed]
- Mosele, J.I.; Macia, A.; Motilva, M.J. Metabolic and microbial modulation of the large intestine ecosystem by non-absorbed diet phenolic compounds. A review. Molecules 2015, 20, 17429–17468. [Google Scholar] [CrossRef] [PubMed]
- Sánchez-Patán, F.; Cueva, C.; Monagas, M.; Walton, G.E.; Gibson, M.G.R.; Quintanilla-López, J.E.; Lebrón-Aguilar, R.; Marín-Álvarez, P.J.; Moreno-Arribas, M.V.; Bartolomé, B. In vitro fermentation of a red wine extract by human gut microbiota: Changes in microbial groups and formation of phenolic metabolites. J. Agric. Food Chem. 2012, 60, 2136–2147. [Google Scholar] [CrossRef] [PubMed]
- Moreno-Arribas, M.V.; Polo, M.C. Occurrence of lactic acid bacteria and biogenic amines in biologically aged wines. Food Microbiol. 2008, 25, 875–881. [Google Scholar] [CrossRef] [PubMed]
- García-Ruiz, A.; Tabasco, R.; Requena, T.; Claisse, O.; Lonvaud-Funel, A.; Bartolome, B.; Moreno-Arribas, M.V. Genetic diversity of Oenoccoccus oeni isolated from wines treated with phenolic extracts as antimicrobial agents. Food Microbiol. 2013, 36, 267–274. [Google Scholar] [CrossRef] [PubMed]
- Martín, R.; Langa, S.; Reviriego, C.; Jimínez, E.; Marín, M.L.; Xaus, J.; Fernández, L.; Rodríguez, J.M. Human milk is a source of lactic acid bacteria for the infant gut. J. Pediatr. 2003, 143, 7540–7548. [Google Scholar] [CrossRef] [PubMed]
- González de Llano, D.; Gil-Sánchez, I.; Esteban-Fernández, A.; Ramos, M.A.; Fernández Díaz, M.; Cueva, C.; Moreno-Arribas, M.V.; Bartolomé, B. Reciprocal beneficial effects between wine polyphenols and probiotics: An exploratory study. Eur. Food Res. Technol. 2016. [Google Scholar] [CrossRef]
- Sánchez-Patán, F.; Monagas, M.; Moreno-Arribas, M.V.; Bartolomé, B. Determination of microbial phenolic acids in human faeces by UPLC-ESI-TQ MS. J. Agric. Food Chem. 2011, 59, 2241–2247. [Google Scholar] [CrossRef] [PubMed]
- Jiménez-Girón, A.; Queipo-Ortuño, M.I.; Boto-Ordoñez, M.; Muñoz-González, I.; Sánchez-Patan, F.; Monagas, M.; Martín-Álvarez, P.J.; Murri, M.; Tinahones, F.J.; Andrés-Lacueva, C.; et al. Comparative study of microbial-derived phenolic metabolites in human feces after intake of gin, red wine, and dealcoholized red wine. J. Agric. Food Chem. 2013, 61, 3909–3915. [Google Scholar] [CrossRef] [PubMed]
- García-Ruiz, A.; Moreno-Arribas, M.V.; Martín-Álvarez, P.J.; Bartolomé, B. Comparative study of the inhibitory effects of wine polyphenols on the growth of enological lactic acid bacteria. Int. J. Food Microbiol. 2011, 145, 426–431. [Google Scholar] [CrossRef] [PubMed]
- Jensen, H.; Grimmer, S.; Naterstad, K.; Axelsson, L. In vitro testing of commercial and potential probiotic lactic acid bacteria. Int. J. Food Microbiol. 2012, 153, 216–222. [Google Scholar] [CrossRef] [PubMed]
- Turchi, B.; Mancini, S.; Fratini, F.; Pedonese, F.; Nuvoloni, R.; Bertelloni, F.; Ebani, V.V.; Cerri, D. Preliminary evaluation of probiotic potential of Lactobacillus plantarum strains isolated from Italian food products. World J. Microbiol. Biotechnol. 2013, 29, 1913–1922. [Google Scholar] [CrossRef] [PubMed]
- Barroso, E.; Van de Wiele, T.; Jiménez-Girón, A.; Muñoz-González, I.; Martín-Alvarez, P.J.; Moreno-Arribas, M.V.; Bartolomé, B.; Peláez, C.; Martínez-Cuesta, M.C.; Requena, T. Lactobacillus plantarum IFPL935 impacts colonic metabolism in a simulator of the human gut microbiota during feeding with red wine polyphenols. Appl. Microbiol. Biotechnol. 2014, 98, 6805–6815. [Google Scholar] [CrossRef] [PubMed]
- Sourabh, A.; Kanwar, S.S.; Sud, R.G.; Ghabru, A.; Sharma, O.P. Influence of phenolic compounds of Kangra tea [Camellia sinensis (L) O Kuntze] on bacterial pathogens and indigenous bacterial probiotics of Western Himalayas. Braz. J. Microbiol. 2013, 44, 709–715. [Google Scholar] [CrossRef] [PubMed]
- Su, P.; Henriksson, A.; Nilsson, C.; Mitchell, H. Synergistic effect of green tea extract and probiotics on the pathogenic bacteria, Staphylococcus aureus and Streptococcus pyogenes. World J. Microbiol. Biotechnol. 2008, 24, 1837–1842. [Google Scholar] [CrossRef]
- Prisciandaro, L.D.; Geier, M.S.; Chua, A.E.; Butler, R.N.; Cummins, A.G.; Sander, G.R.; Howarth, G.S. Probiotic factors partially prevent changes to caspases 3 and 7 activation and transepithelial electrical resistance in a model of 5-fluorouracil-induced epithelial cell damage. Support. Care Cancer 2012, 20, 3205–3210. [Google Scholar] [CrossRef] [PubMed]
- Tejero-Sariñena, S.; Barlow, J.; Costabile, A.; Gibson, G.R.; Rowland, I. In vitro evaluation of the antimicrobial activity of a range of probiotics against pathogens: Evidence for the effects of organic acids. Anaerobe 2012, 18, 530–538. [Google Scholar] [CrossRef] [PubMed]
- Likotrafiti, E.; Valavani, P.; Argiriou, A.; Rhoades, J. In vitro evaluation of potential antimicrobial synbiotics using Lactobacillus kefiri isolated from kefir grains. Int. Dairy J. 2015, 45, 23–30. [Google Scholar] [CrossRef]
- Arena, M.P.; Silvain, A.; Normanno, G.; Grieco, F.; Drider, D.; Spano, G.; Fiocco, D. Lactobacillus plantarum strains as a bio-control strategy against food-borne pathogenic microorganisms. Front. Microbiol. 2016, 7. [Google Scholar] [CrossRef]
- Gueimonde, M.; Jalonen, L.; He, F.; Hiramatsu, M.; Salminen, S. Adhesion and competitive inhibition and displacement of human enteropathogens by selected lactobacilli. Food Res. Int. 2006, 39, 467–471. [Google Scholar] [CrossRef]
- Bustos, I.; Garcia-Cayuela, T.; Hernández-Ledesma, B.; Pelaez, C.; Requena, T.; Martínez-Cuesta, M.C. Effect of flavan-3-ols on the adhesion of potential probiotic lactobacilli to intestinal cells. J. Agric. Food Chem. 2012, 60, 9082–9088. [Google Scholar] [CrossRef] [PubMed]
- Leahy, S.C.; Higgins, D.G.; Fitzgerald, G.F.; Sinderen, D.V. Getting better with bifidobacteria. J. Appl. Microbiol. 2005, 98, 1303–1315. [Google Scholar] [CrossRef] [PubMed]
- Abedi, D.; Feizizadeh, S.; Akbari, V.; Jafarian-Dehkordi, A. In vitro anti-bacterial and anti-adherence effects of Lactobacillus delbrueckii subsp bulgaricus on Escherichia coli. Res Pharm. Sci. 2013, 8, 260–268. [Google Scholar] [PubMed]

| Experimental Conditions | Time (min) | LAB strains | |||
|---|---|---|---|---|---|
| P. pentosaceus CIAL-86 | L. plantarum CIAL-121 | L. plantarum CLC 17 | |||
| Resistance to lysozyme | 30 | 93.9 | 65.1 | 100 | |
| (% survival) | 120 | 88.6 | 50.8 | 86.0 | |
| Tolerance to gastric juice (log CFU/mL) | pH 5.0 | 0 | 8.47 ± 0.05 | 8.08 ± 0.07 | 8.08 ± 0.07 |
| pH 4.1 | 20 | 8.38 ± 0.11 | 8.06 ± 0.08 | 8.19 ± 0.10 | |
| pH 3.0 | 40 | 8.41 ± 0.04 | 8.05 ± 0.13 | 8.08 ± 0.11 | |
| pH 2.1 | 60 | 7.95 ± 0.06 | 6.20 ± 0.28 | 7.95 ± 0.06 | |
| pH 1.8 | 90 | 5.15 ± 0.21 | 5.05 ± 0.38 | 7.31 ± 0.12 | |
| Bile resistance (% growth) | 0.06% | 99.1 | 93.6 | 93.8 | |
| 0.125% | 100.0 | 91.2 | 89.1 | ||
| 0.25% | 88.3 | 89.0 | 77.9 | ||
| 0.5% | 89.5 | 89.5 | 76.8 | ||
| 1.0% | 84.1 | 88.7 | 73.0 | ||
| Samples | Concentrations of phenolic compounds | ||||
|---|---|---|---|---|---|
| t = 0 h | t = 6 h | t = 24 h | |||
| ∑C (ppm) | ∑C (ppm) | % a | ∑C (ppm) | % a | |
| Wine extract | 25.6 | 25.8 | 100.7 | 25.3 | 98.6 |
| L. plantarum CLC 17 + wine extract | 24.5 | 23.0 | 94.2 | 32.5 | 133.2 ** |
| P. pentosaceus CIAL-86 + wine extract | 28.7 | 27.9 | 97.4 | 26.9 | 93.8 |
| L. plantarum CIAL-121 + wine extract | 25.5 | 22.6 | 88.7 | 25.23 | 99.1 |
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
De Llano, D.G.; Gil‐Sánchez, I.; Esteban‐Fernández, A.; Ramos, A.M.; Cueva, C.; Moreno‐Arribas, M.V.; Bartolomé, B. Some Contributions to the Study of Oenological Lactic Acid Bacteria through Their Interaction with Polyphenols. Beverages 2016, 2, 27. https://doi.org/10.3390/beverages2040027
De Llano DG, Gil‐Sánchez I, Esteban‐Fernández A, Ramos AM, Cueva C, Moreno‐Arribas MV, Bartolomé B. Some Contributions to the Study of Oenological Lactic Acid Bacteria through Their Interaction with Polyphenols. Beverages. 2016; 2(4):27. https://doi.org/10.3390/beverages2040027
Chicago/Turabian StyleDe Llano, Dolores González, Irene Gil‐Sánchez, Adelaida Esteban‐Fernández, Alba M. Ramos, Carolina Cueva, M. Victoria Moreno‐Arribas, and Begoña Bartolomé. 2016. "Some Contributions to the Study of Oenological Lactic Acid Bacteria through Their Interaction with Polyphenols" Beverages 2, no. 4: 27. https://doi.org/10.3390/beverages2040027
APA StyleDe Llano, D. G., Gil‐Sánchez, I., Esteban‐Fernández, A., Ramos, A. M., Cueva, C., Moreno‐Arribas, M. V., & Bartolomé, B. (2016). Some Contributions to the Study of Oenological Lactic Acid Bacteria through Their Interaction with Polyphenols. Beverages, 2(4), 27. https://doi.org/10.3390/beverages2040027