Lanthanide-Loaded Nanoparticles as Potential Fluorescent and Mass Probes for High-Content Protein Analysis
Abstract
1. Introduction
2. Materials and Methods
2.1. Reagents and Cell Lines
2.2. Nanoparticle Synthesis and Characterization
2.3. Lanthanide Loading on the Nanoparticles and Loading Characterization
2.4. Imaging and Flow Cytometry
2.5. Western Blot
3. Results
3.1. Nanoparticle Characterization
3.2. Lanthanide Loading Capability of Our Nanoparticles
3.3. Antibody-Conjugated Nanoparticles Bind to Cells Expressing Target Proteins with High Specificity
4. Discussion
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Yao, Y.; Liu, R.; Shin, M.S.; Trentalange, M.; Allore, H.; Nassar, A.; Kang, I.; Pober, J.S.; Montgomery, R.R. CyTOF supports efficient detection of immune cell subsets from small samples. J. Immunol. Methods 2014, 415, 1–5. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez, H.; Pennington, S.R. Revolutionizing Precision Oncology through Collaborative Proteogenomics and Data Sharing. Cell 2018, 173, 535–539. [Google Scholar] [CrossRef] [PubMed]
- Waters, J.C. Accuracy and precision in quantitative fluorescence microscopy. J. Cell Biol. 2009, 185, 1135–1148. [Google Scholar] [CrossRef] [PubMed]
- Giesen, C.; Mairinger, T.; Khoury, L.; Waentig, L.; Jakubowski, N.; Panne, U. Multiplexed immunohistochemical detection of tumor markers in breast cancer tissue using laser ablation inductively coupled plasma mass spectrometry. Anal. Chem. 2011, 83, 8177–8183. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Yan, S.; Lin, B.; Shi, Q.; Lu, Y. Chapter Eight—Single-Cell Proteomics for Cancer Immunotherapy. In Advances in Cancer Research; Broome, A.-M., Ed.; Academic Press: Cambridge, MA, USA, 2018; Volume 139, pp. 185–207. [Google Scholar]
- Bendall, S.C.; Nolan, G.P.; Roederer, M.; Chattopadhyay, P.K. A deep profiler’s guide to cytometry. Trends Immunol. 2012, 33, 323–332. [Google Scholar] [CrossRef] [PubMed]
- Yantasee, W.; Fryxell, G.E.; Addleman, R.S.; Wiacek, R.J.; Koonsiripaiboon, V.; Pattamakomsan, K.; Sukwarotwat, V.; Xu, J.; Raymond, K.N. Selective removal of lanthanides from natural waters, acidic streams and dialysate. J. Hazard. Mater. 2009, 168, 1233–1238. [Google Scholar] [CrossRef] [PubMed]
- Ngamcherdtrakul, W.; Morry, J.; Gu, S.; Castro, D.J.; Goodyear, S.M.; Sangvanich, T.; Reda, M.M.; Lee, R.; Mihelic, S.A.; Beckman, B.L.; et al. Cationic Polymer Modified Mesoporous Silica Nanoparticles for Targeted SiRNA Delivery to HER2+ Breast Cancer. Adv. Funct. Mater. 2015, 25, 2646–2659. [Google Scholar] [CrossRef] [PubMed]
- Duan, C.; Liang, L.; Li, L.; Zhang, R.; Xu, Z.P. Recent progress in upconversion luminescence nanomaterials for biomedical applications. J. Mater. Chem. B 2018, 6, 192–209. [Google Scholar] [CrossRef]
- Liang, L.; Care, A.; Zhang, R.; Lu, Y.; Packer, N.H.; Sunna, A.; Qian, Y.; Zvyagin, A.V. Facile Assembly of Functional Upconversion Nanoparticles for Targeted Cancer Imaging and Photodynamic Therapy. ACS Appl. Mater. Interfaces 2016, 8, 11945–11953. [Google Scholar] [CrossRef] [PubMed]
- Liang, L.; Lu, Y.; Zhang, R.; Care, A.; Ortega, T.A.; Deyev, S.M.; Qian, Y.; Zvyagin, A.V. Deep-penetrating photodynamic therapy with KillerRed mediated by upconversion nanoparticles. Acta Biomater. 2017, 51, 461–470. [Google Scholar] [CrossRef] [PubMed]
- Dong, J.; Xue, M.; Zink, J.I. Functioning of nanovalves on polymer coated mesoporous silica Nanoparticles. Nanoscale 2013, 5, 10300–10306. [Google Scholar] [CrossRef] [PubMed]
- Morry, J.; Ngamcherdtrakul, W.; Gu, S.; Reda, M.; Castro, D.J.; Sangvanich, T.; Gray, J.W.; Yantasee, W. Targeted treatment of metastatic breast cancer by PLK1 siRNA delivered by an antioxidant nanoparticle platform. Mol. Cancer Ther. 2017, 16, 763–772. [Google Scholar] [CrossRef] [PubMed]
- Ngamcherdtrakul, W.; Sangvanich, T.; Reda, M.; Gu, S.; Bejan, D.; Yantasee, W. Lyophilization and stability of antibody-conjugated mesoporous silica nanoparticle with cationic polymer and PEG for siRNA delivery. Int. J. Nanomed. 2018, 13, 4015–4027. [Google Scholar] [CrossRef]
- Lin, W.; Hou, Y.; Lu, Y.; Abdelrahman, A.I.; Cao, P.; Zhao, G.; Tong, L.; Qian, J.; Baranov, V.; Nitz, M.; et al. A high-sensitivity lanthanide nanoparticle reporter for mass cytometry: Tests on microgels as a proxy for cells. Langmuir 2014, 30, 3142–3153. [Google Scholar] [CrossRef] [PubMed]
- Kay, A.W.; Strauss-Albee, D.M.; Blish, C.A. Application of Mass Cytometry (CyTOF) for Functional and Phenotypic Analysis of Natural Killer Cells. Methods Mol. Biol. 2016, 1441, 13–26. [Google Scholar] [CrossRef] [PubMed]
- Levenson, R.M.; Borowsky, A.D.; Angelo, M. Immunohistochemistry and mass spectrometry for highly multiplexed cellular molecular imaging. Lab. Investig. A J. Tech. Methods Pathol. 2015, 95, 397–405. [Google Scholar] [CrossRef]
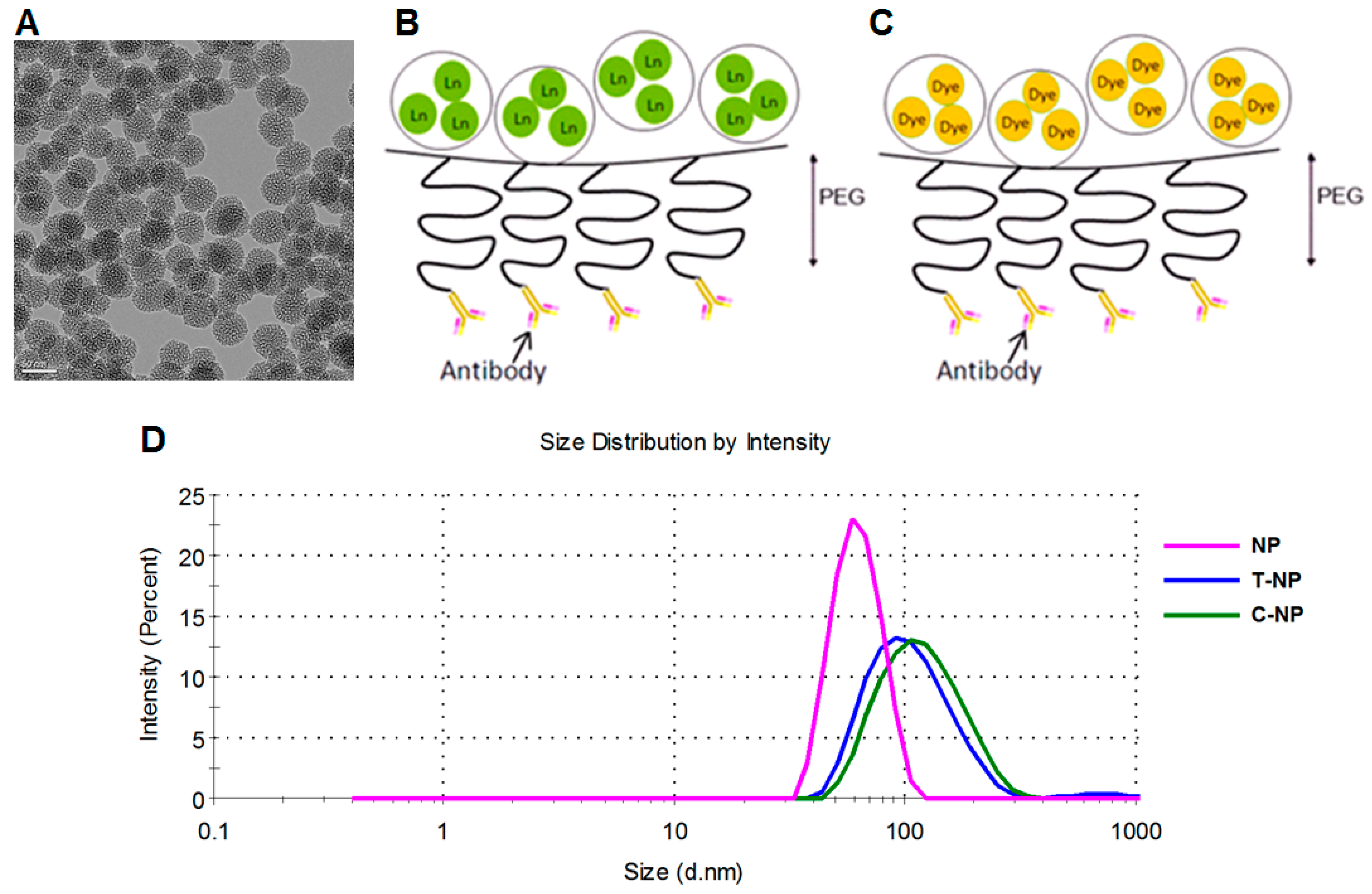
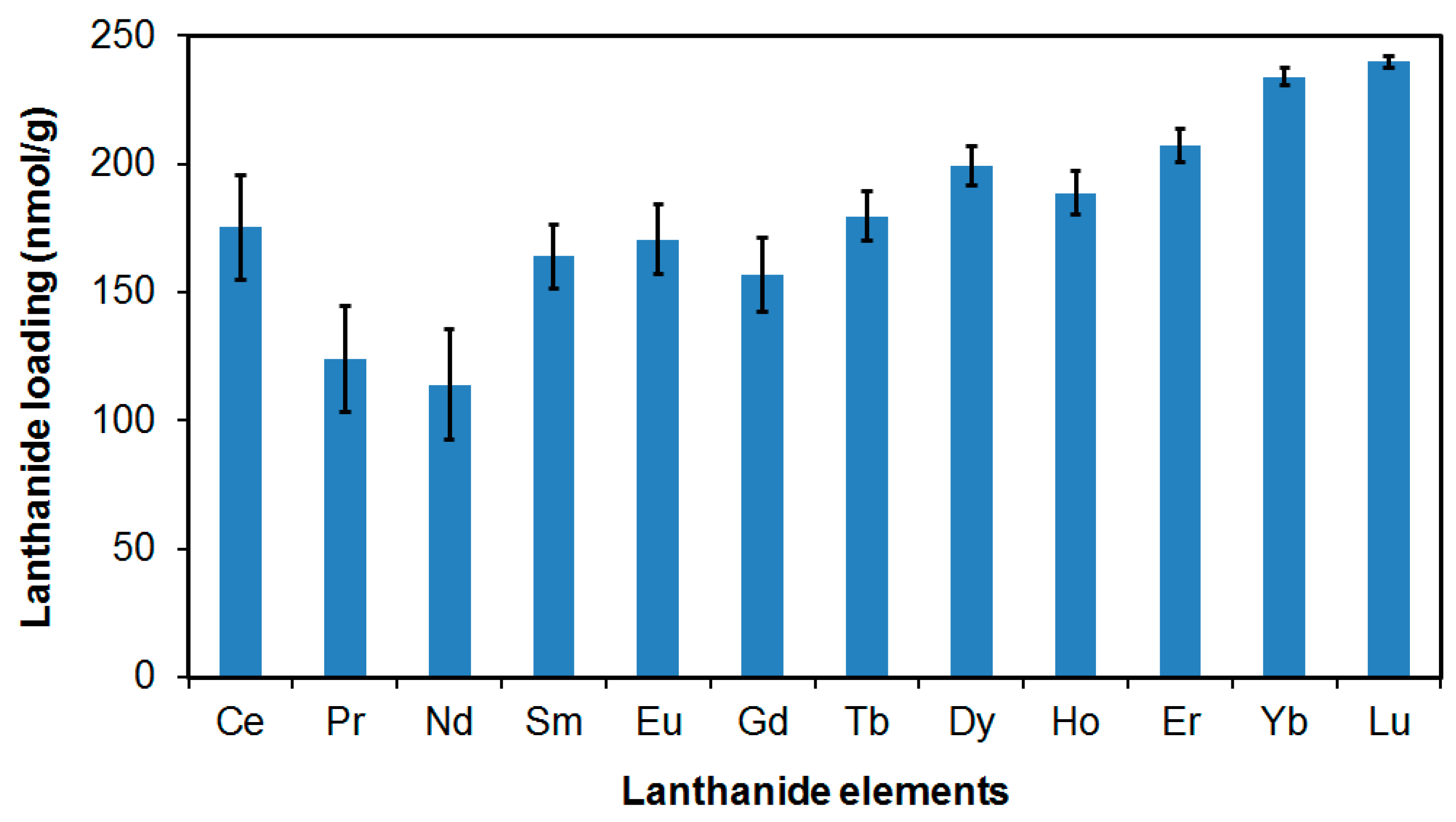
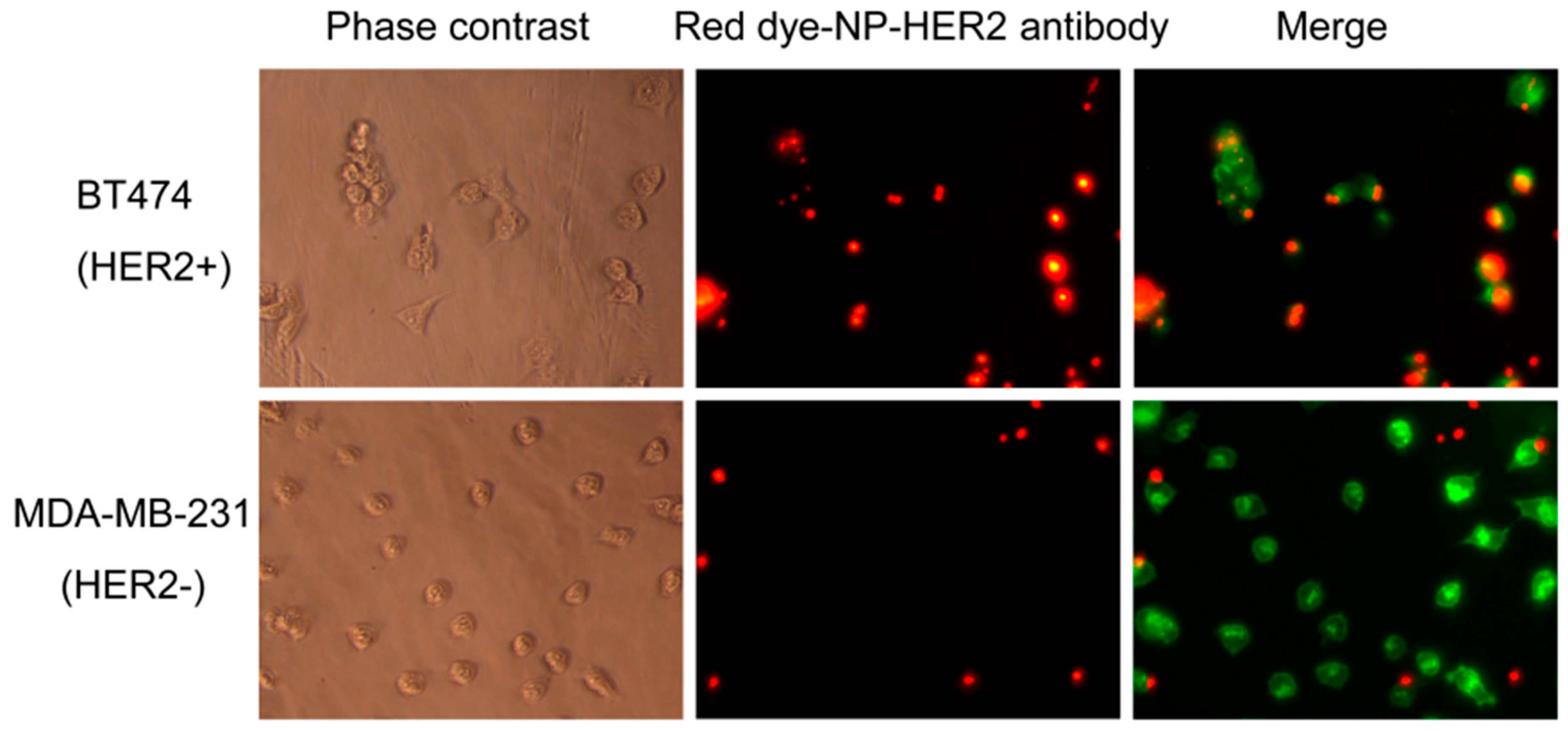
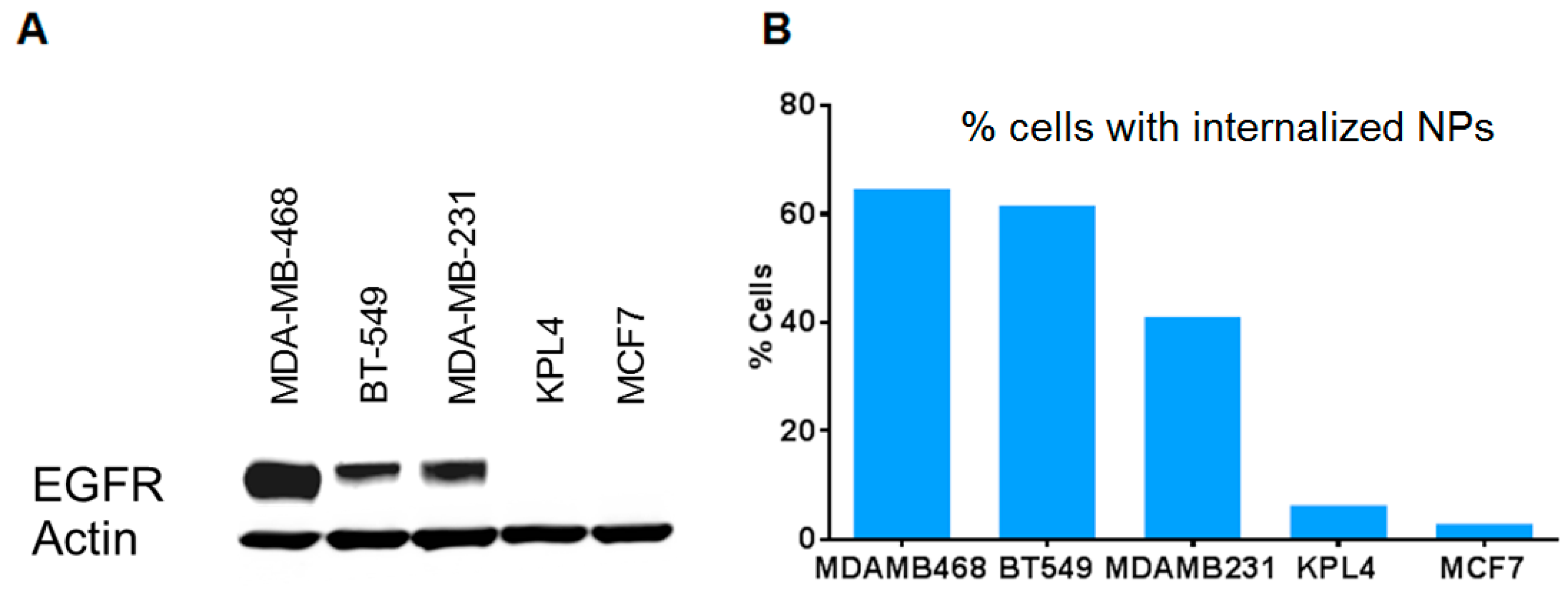
| MSNP Surface Functionality | Tm Loading (mg Tm/g Material) |
|---|---|
| Hydroxyl | 14.6 ± 0.6 |
| Aminated | 25.3 ± 1.0 |
| Phosphonated | 32.4 ± 1.7 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ngamcherdtrakul, W.; Sangvanich, T.; Goodyear, S.; Reda, M.; Gu, S.; Castro, D.J.; Punnakitikashem, P.; Yantasee, W. Lanthanide-Loaded Nanoparticles as Potential Fluorescent and Mass Probes for High-Content Protein Analysis. Bioengineering 2019, 6, 23. https://doi.org/10.3390/bioengineering6010023
Ngamcherdtrakul W, Sangvanich T, Goodyear S, Reda M, Gu S, Castro DJ, Punnakitikashem P, Yantasee W. Lanthanide-Loaded Nanoparticles as Potential Fluorescent and Mass Probes for High-Content Protein Analysis. Bioengineering. 2019; 6(1):23. https://doi.org/10.3390/bioengineering6010023
Chicago/Turabian StyleNgamcherdtrakul, Worapol, Thanapon Sangvanich, Shaun Goodyear, Moataz Reda, Shenda Gu, David J. Castro, Primana Punnakitikashem, and Wassana Yantasee. 2019. "Lanthanide-Loaded Nanoparticles as Potential Fluorescent and Mass Probes for High-Content Protein Analysis" Bioengineering 6, no. 1: 23. https://doi.org/10.3390/bioengineering6010023
APA StyleNgamcherdtrakul, W., Sangvanich, T., Goodyear, S., Reda, M., Gu, S., Castro, D. J., Punnakitikashem, P., & Yantasee, W. (2019). Lanthanide-Loaded Nanoparticles as Potential Fluorescent and Mass Probes for High-Content Protein Analysis. Bioengineering, 6(1), 23. https://doi.org/10.3390/bioengineering6010023






