Nut and Bolt Microfluidics with Helical Minichannel for Counting CD4+ T-Cells
Abstract
1. Introduction
2. Technical Development
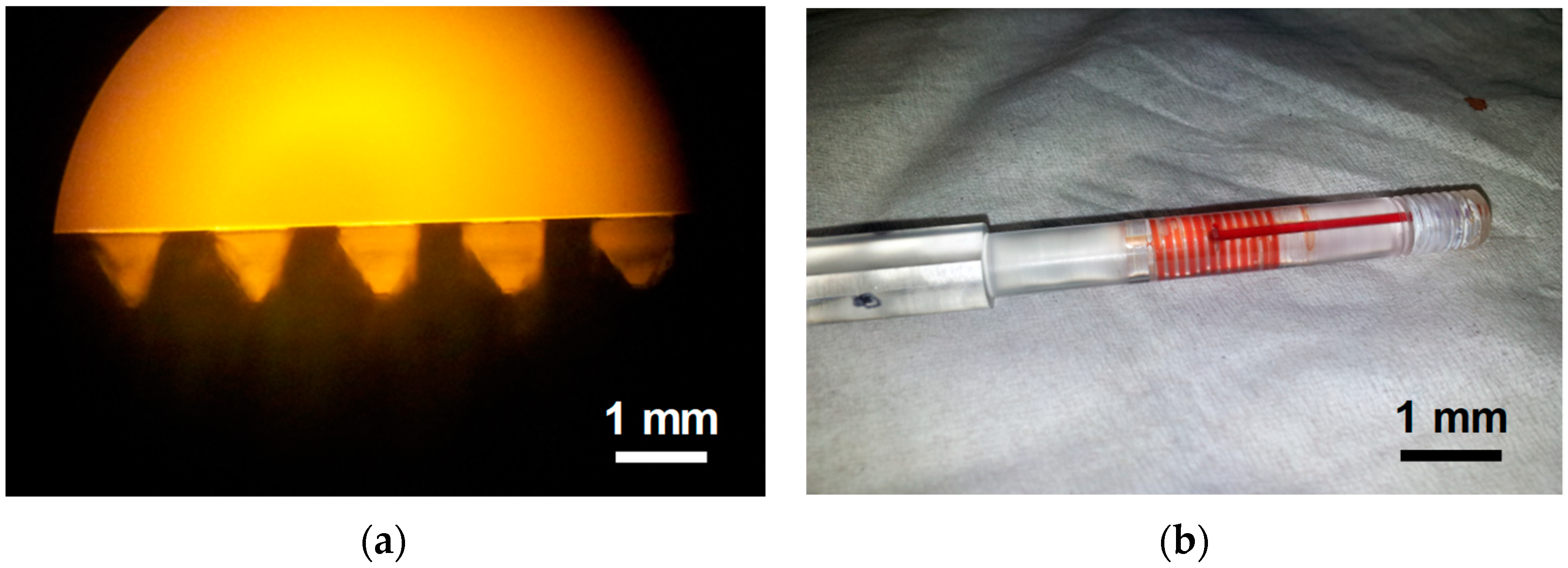
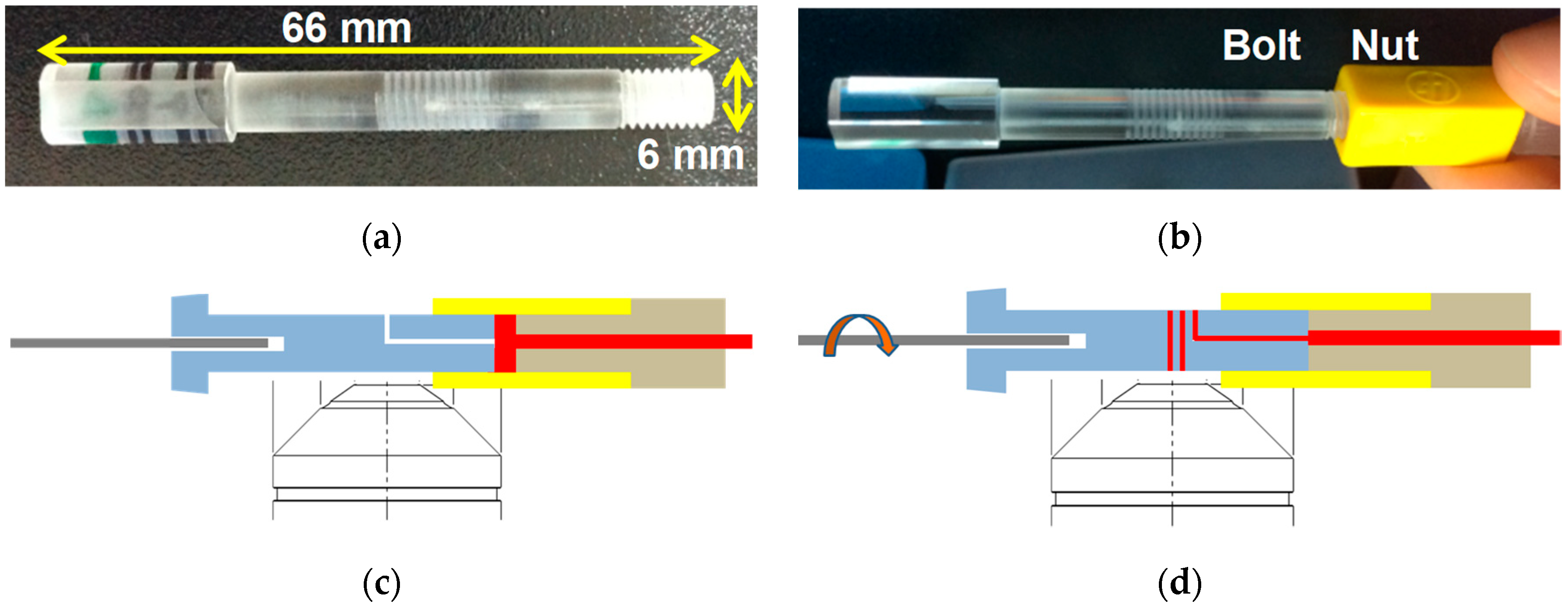
2.1. Fabrication of Sample Cartridge
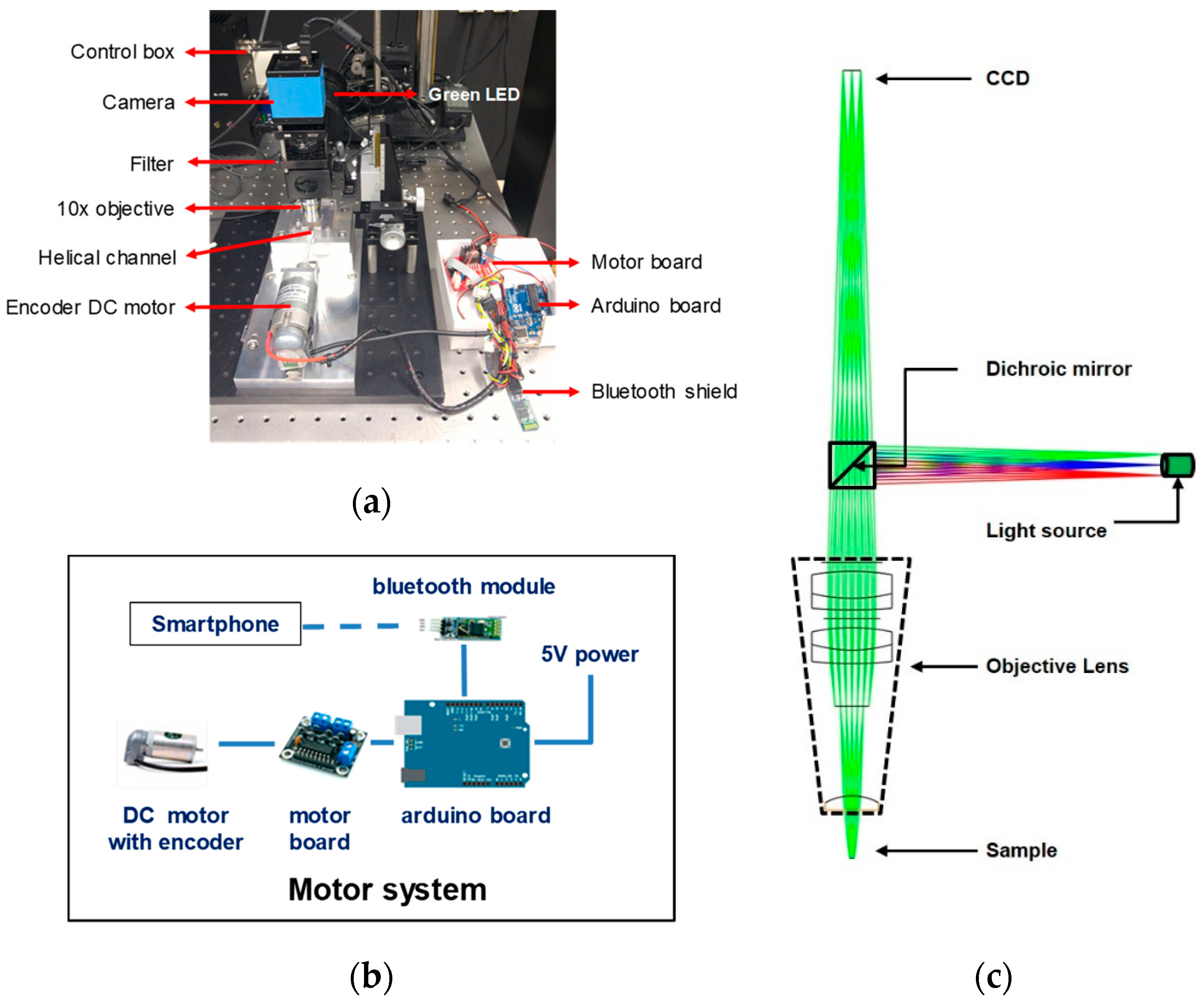
2.2. Fluorescence Imaging Setup
2.3. Optimization of Fluorescence Imaging Parameters
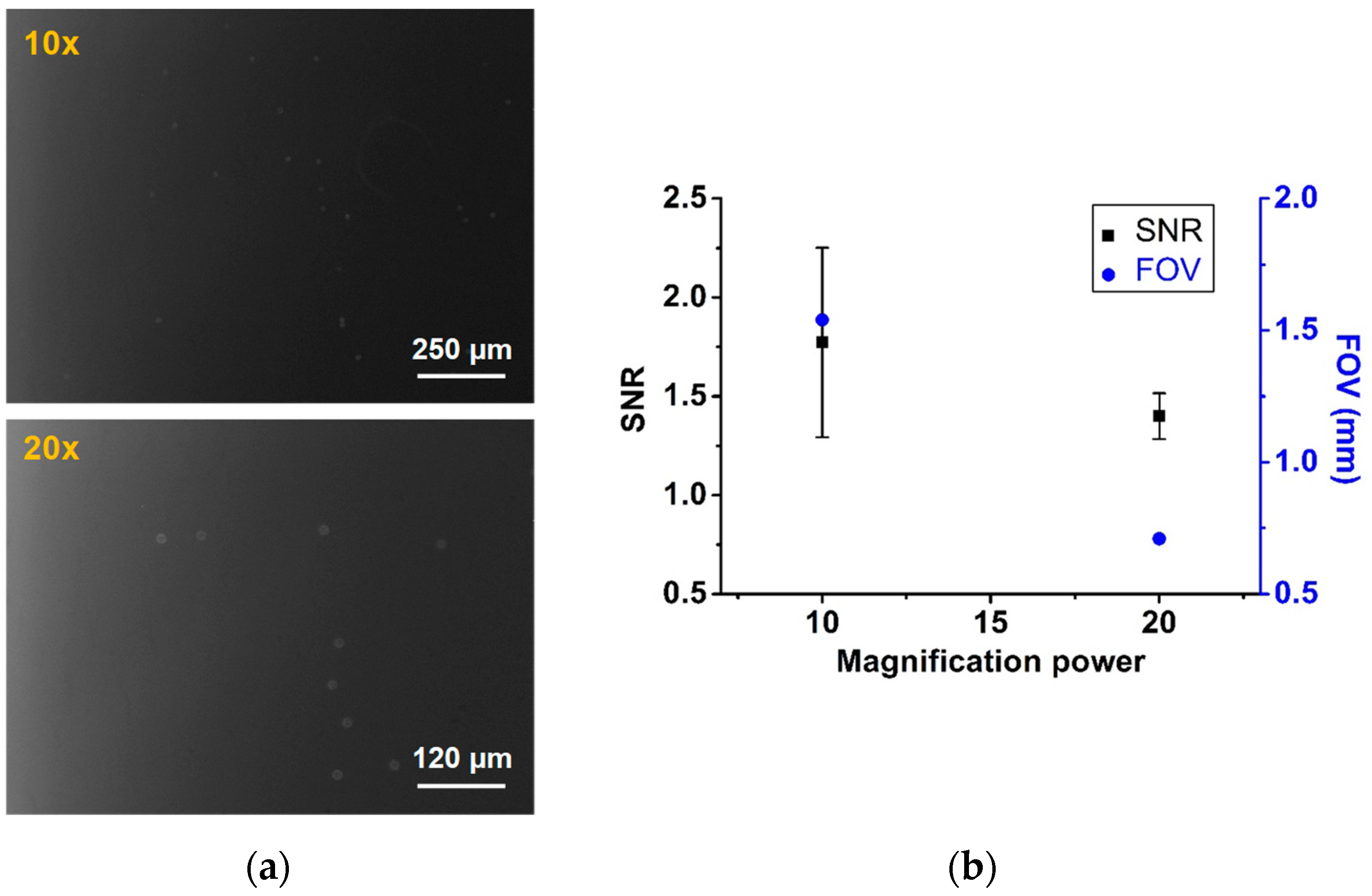
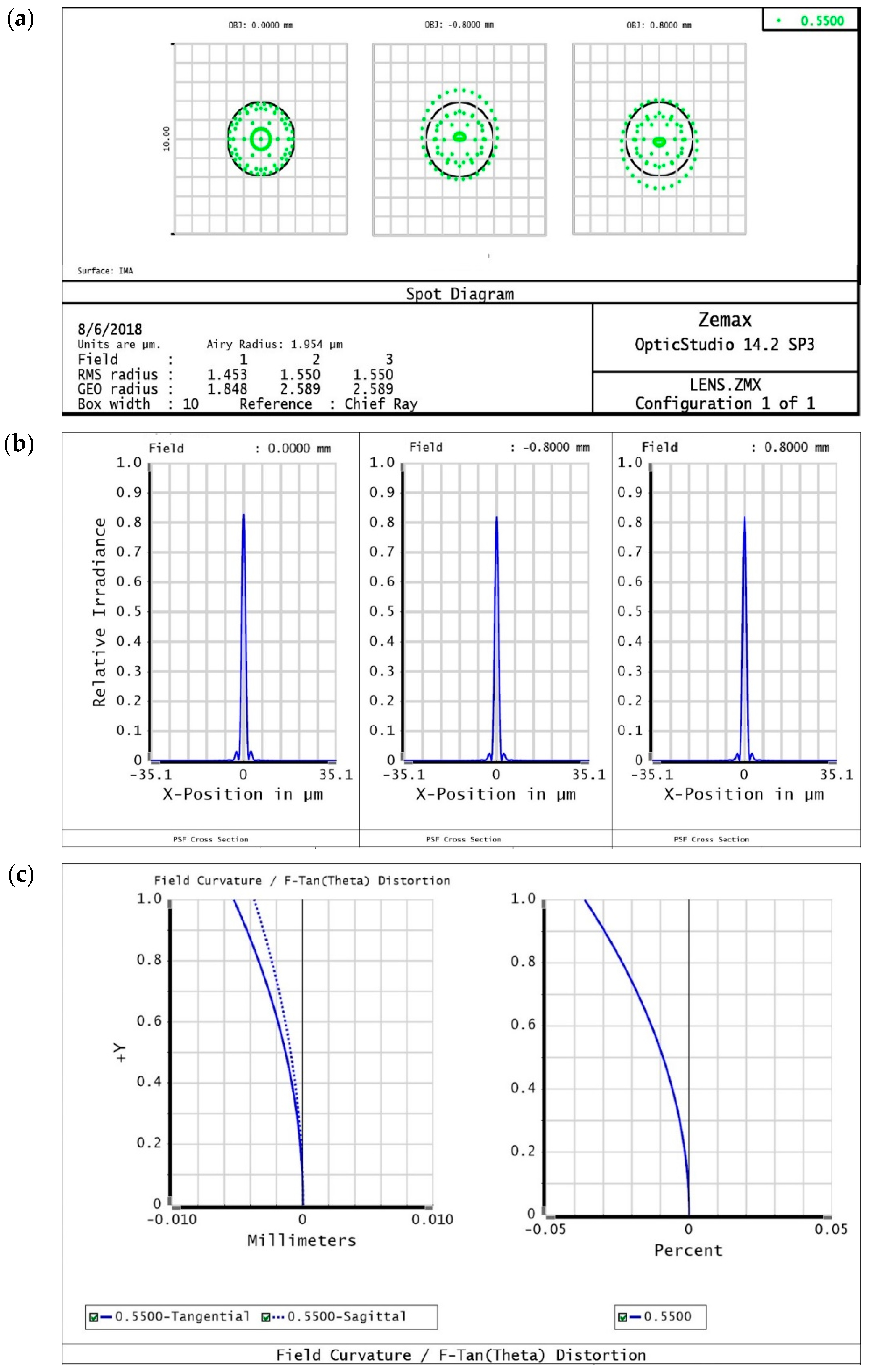
2.3.1. Objective Lens
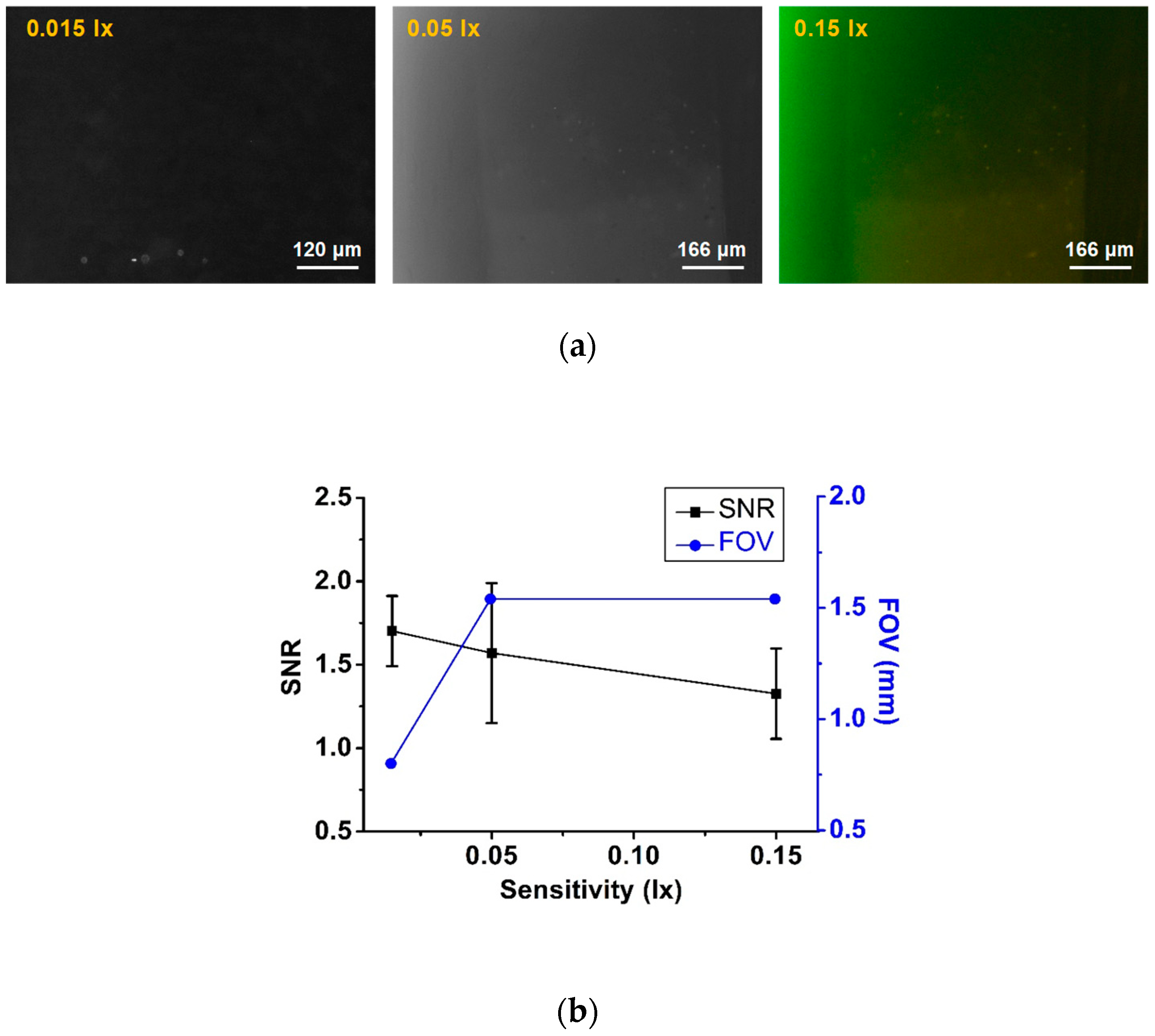
2.3.2. Field-of-view
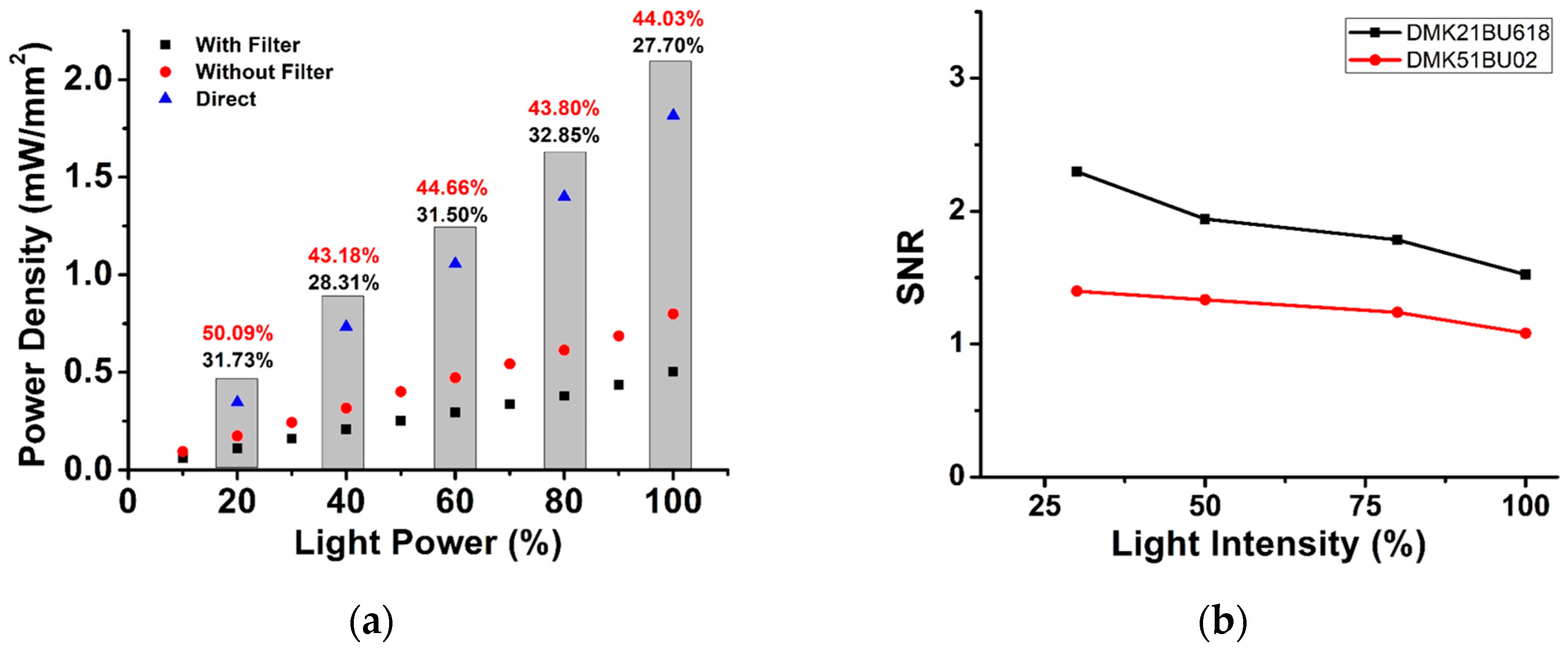
2.3.3. Light Power
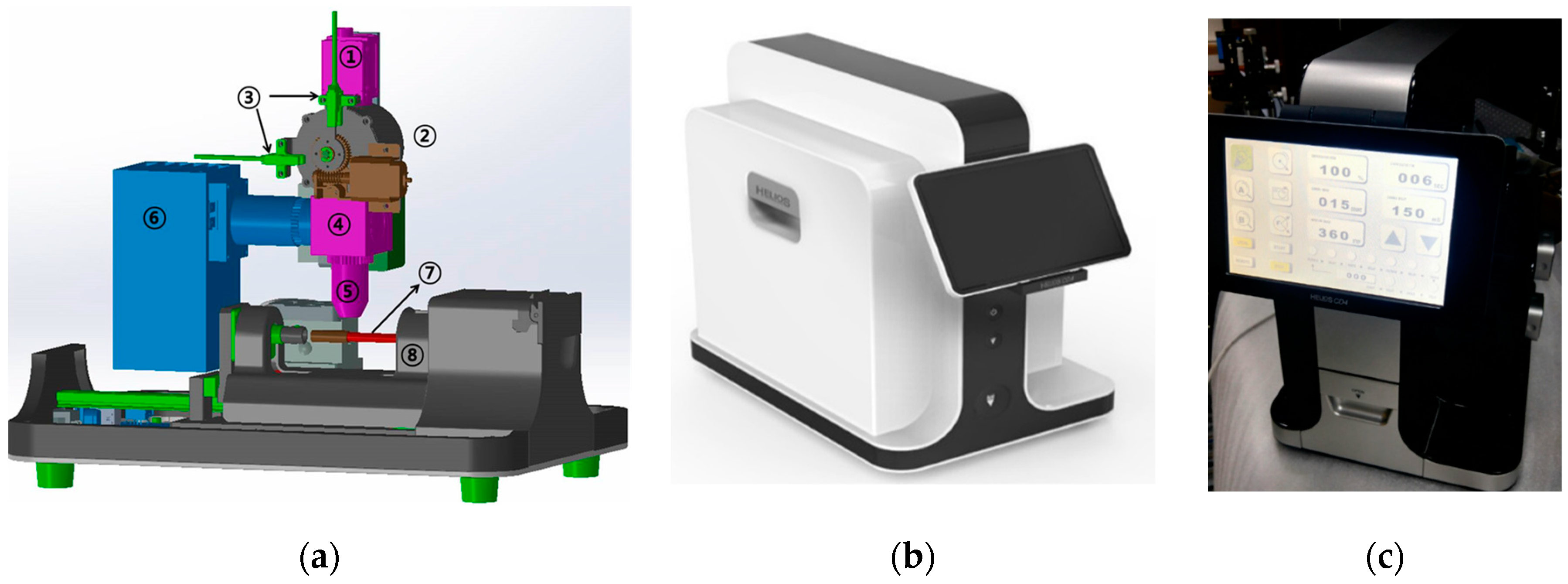
2.4. Prototype Development
3. Materials and Methods
3.1. Optical Simulation
3.2. Sample Preparation
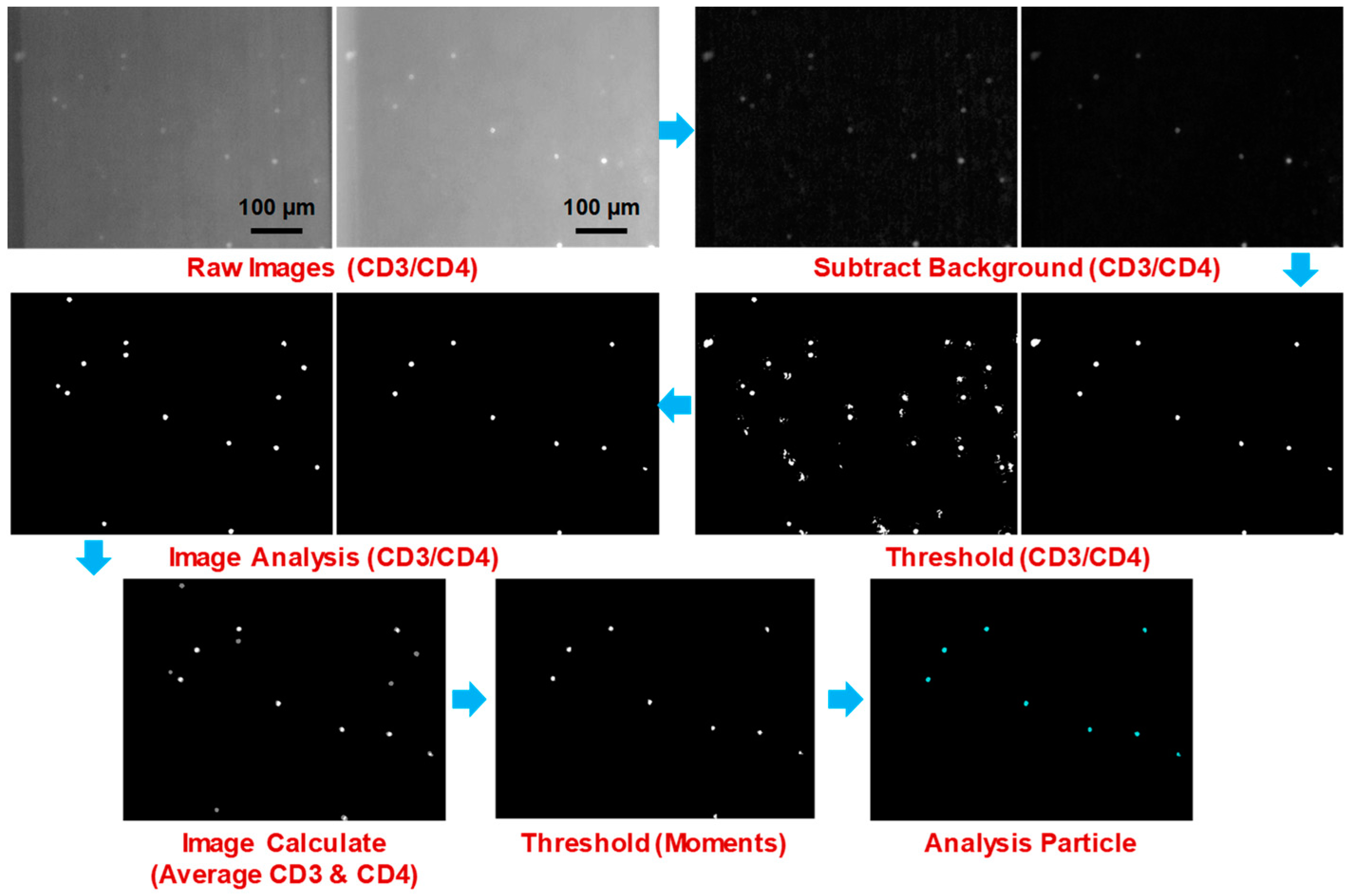
3.3. Image Processing
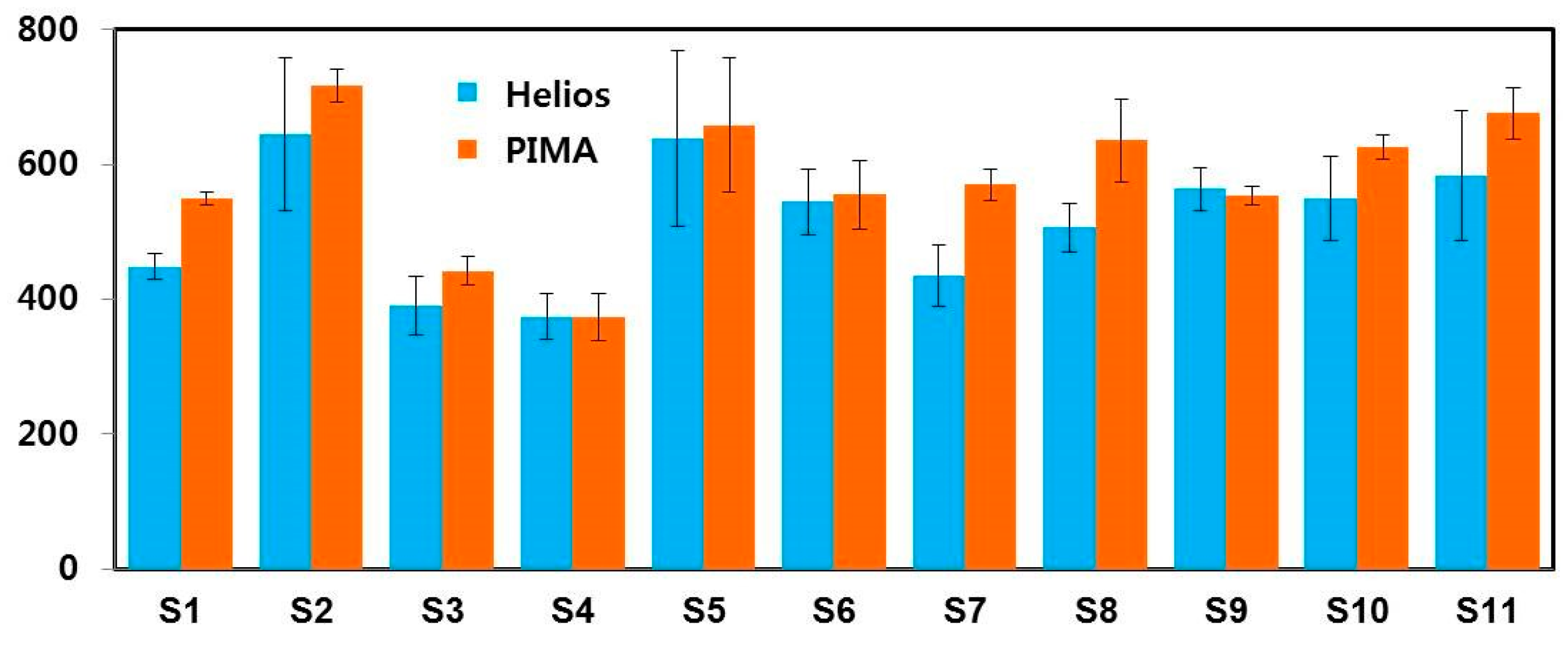
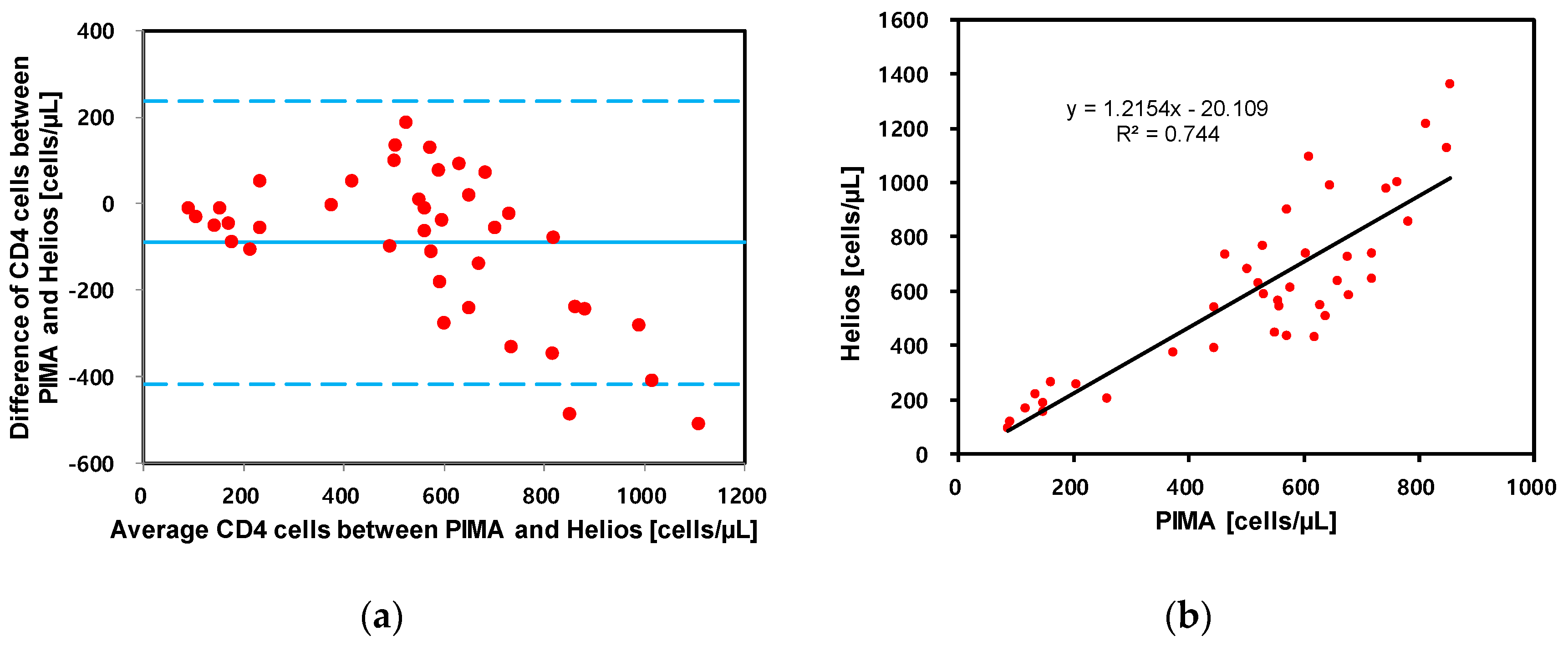
4. Results and Discussion
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Young, P.W.; Kim, A.A.; Wamicwe, J.; Nyagah, L.; Kiama, C.; Stover, J. HIV associated mortality in the era of antiretroviral therapy scale-up. PLoS ONE 2017, 12, e0181837. [Google Scholar]
- Johnson, L.F.; May, M.T.; Dorrington, R.E.; Cornell, M.; Boulle, A.; Egger, M. Estimating the impact of antiretroviral treatment on adult mortality trends in South Africa: A mathematical modelling study. PLoS Med. 2017, 14, e1002468. [Google Scholar] [CrossRef] [PubMed]
- Nasi, M.; Biasi, S.D.; Bianchini, E.; Gibellini, L.; Pinti, M.; Scacchetti, T. Reliable and accurate CD4+ T cell count and percent by the portable flow cytometer CyFlow MiniPOC and “CD4 Easy Count Kit-Dry”, as revealed by the comparison with the gold standard dual platform technology. PLoS ONE 2015, 10, e0116848. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, T.; Zoëga Andreasen, S.; Wolff, A.; Duong Bang, D. From lab on a chip to point of care devices: The role of open source microcontrollers. Micromachines 2018, 9, 403. [Google Scholar] [CrossRef] [PubMed]
- Glynn, M.T.; Kinahan, D.J.; Ducrée, J. CD4 counting technologies for HIV therapy monitoring in resource-poor settings-state-of-the-art and emerging microtechnologies. Lab Chip 2013, 13, 2731–2748. [Google Scholar] [CrossRef] [PubMed]
- Larson, B.; Schnippel, K.; Ndibongo, B.; Long, L.; Fox, M.P.; Rosen, S. How to estimate the cost of point-of-care CD4 testing in program settings: An example using the Alere PimaTM Analyzer in South Africa. PLoS ONE 2012, 7, e35444. [Google Scholar] [CrossRef] [PubMed]
- Cho, M.-O.; Kim, S.; Lee, J.Y.; Oh, J.H.; Kim, J.Y.; Bong, S.R.; Chung, C.; Kim, J.K. Performance evaluation of an automated image-based fluorescence CD4+ cell analyzer. Technol. Health Care 2018, 26, 867–871. [Google Scholar] [CrossRef] [PubMed]
- Bae, S.Y.; Park, H.C.; Oh, J.S.; Yoon, S.Y.; Park, D.W.; Choi, I.K.; Kim, H.J.; Oh, J.H.; Hur, D.S.; Chung, C.; et al. Absolute CD4+ cell count using a plastic microchip and a microscopic cell counter. Cytom. Part B Clin. Cytom. 2009, 76, 345–353. [Google Scholar] [CrossRef] [PubMed]
- Shourav, M.K.; Kim, K.; Kim, S.; Kim, J.K. Wide field-of-view fluorescence imaging with optical-quality curved microfluidic chamber for absolute cell counting. Micromachines 2016, 7, 125. [Google Scholar] [CrossRef] [PubMed]
- Cho, M.-O.; Chang, H.M.; Lee, D.; Yu, Y.G.; Han, H.; Kim, J.K. Selective detection and automated counting of fluorescently-labeled chrysotile asbestos using a dual-mode high-throughput microscopy (DM-HTM) method. Sensors 2013, 13, 5686–5699. [Google Scholar] [CrossRef] [PubMed]
- Park, E.; Kim, S.; Cho, M.-O.; Kim, K.; Shourav, M.K.; Kim, S.; Lee, J.; Kim, J.K. Evaluation of particle counting by smartphone-based fluorescence SmartScope and particle positioning in spinning helical channel. J. Korea Ind. Inf. Syst. Res. 2015, 20, 19–28. [Google Scholar]
- Grunwald, D.; Shenoy, S.M.; Burke, S.S.; Singer, R.H. Calibrating excitation light fluxes for quantitative light microscopy in cell biology. Nat. Protoc. 2008, 3, 1809–1814. [Google Scholar] [CrossRef] [PubMed]
- Cho, M.-O.; Yoon, S.; Han, H.; Kim, J.K. Automated counting of airborne asbestos fibers by a high-throughput microscopy (HTM) method. Sensors 2011, 11, 7231–7242. [Google Scholar] [CrossRef] [PubMed]
- Burger, W.; Burge, M.J. Digital Image Processing: An Algorithmic Introduction Using Java, 2nd ed.; Springer: London, UK, 2016. [Google Scholar]
- Nordenfelt, P.; Cooper, J.; Hochstetter, A. Matrix-masking to balance nonuniform illumination in microscopy. Opt. Express 2018, 26, 17279–17288. [Google Scholar] [CrossRef] [PubMed]
- Kim, B.; Choi, Y.J.; Seo, H.; Shin, E.C.; Choi, S. Deterministic migration-based separation of white blood cells. Small 2016, 12, 5159–5168. [Google Scholar] [CrossRef] [PubMed]
- Lee, W.; Kwon, D.; Choi, W.; Jung, G.Y.; Au, A.K.; Folch, A.; Jeon, S. 3D-printed microfluidic device for the detection of pathogenic bacteria using size-based separation in helical channel with trapezoid cross-section. Sci. Rep. 2015, 5, 7717. [Google Scholar] [CrossRef] [PubMed]
- Wu, L.; Guan, G.; Hou, H.W.; Bhagat, A.A.S.; Han, J. Separation of leukocytes from blood using spiral channel with trapezoid cross-section. Anal. Chem. 2012, 84, 9324–9331. [Google Scholar] [CrossRef] [PubMed]
- Kuntaegowdanahalli, S.S.; Bhagat, A.A.; Kumar, G.; Papautsky, I. Inertial microfluidics for continuous particle separation in spiral microchannels. Lab Chip 2009, 9, 2973–2980. [Google Scholar] [CrossRef] [PubMed]
- Prasad, B.; Kim, J.K. CFD analysis of geometric parameters that affect dean flow in a helical microchannel. J. Korean Soc. Mar. Eng. 2014, 38, 1269–1274. [Google Scholar] [CrossRef]
| Component | Model No. | Company |
|---|---|---|
| Arduino Board | Arduino Uno (R3) | LK EMBEDDED, Seoul, Korea |
| Bluetooth Shield | HC-06 | LK EMBEDDED, Seoul, Korea |
| Camera | DMK21BU618 | Imaging Source, Bremen, Germany |
| DC Encoder Motor | MB-3352E-0521-24V | Robotmart, Seoul, Korea |
| Filters | XF108-2 Cy3/695AF55 CY5M | Omega Optical, Brattleboro, VT, USA |
| Light-Emitting Diode | Custom-made Green LED with Controller | BoardLab, Incheon, Korea |
| Motor board | AM-DC1-3D | NEWTC, Seoul, Korea |
| Motor Power Supply | PS-2425 | Robotmart, Seoul, Korea |
| Objective lens | UPLFLN10 × 2 | Olympus, Tokyo, Japan |
| CD4+ T-Cell Group | Bland–Altman Difference between PIMA and Helios (Limits of Agreement) | T-Test (p-Value) | |
|---|---|---|---|
| <200 | −48.15 (−114.76–18.46) | n = 7 | 0.01 |
| 200–350 | −1.76 (−105.94–102.43) | n = 2 | 0.97 |
| 350–500 | −80.28 (−325.55–162.99) | n = 4 | 0.34 |
| >500 | −109.03 (−485.57–267.51) | n = 27 | 0.00 |
| Total | −90.14 (−416–236.34) | n = 40 | 0.00 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, J.K.; Shourav, M.K.; Cho, M.-O.; Lee, Y. Nut and Bolt Microfluidics with Helical Minichannel for Counting CD4+ T-Cells. Bioengineering 2019, 6, 24. https://doi.org/10.3390/bioengineering6010024
Kim JK, Shourav MK, Cho M-O, Lee Y. Nut and Bolt Microfluidics with Helical Minichannel for Counting CD4+ T-Cells. Bioengineering. 2019; 6(1):24. https://doi.org/10.3390/bioengineering6010024
Chicago/Turabian StyleKim, Jung Kyung, Mohiuddin Khan Shourav, Myoung-Ock Cho, and Yein Lee. 2019. "Nut and Bolt Microfluidics with Helical Minichannel for Counting CD4+ T-Cells" Bioengineering 6, no. 1: 24. https://doi.org/10.3390/bioengineering6010024
APA StyleKim, J. K., Shourav, M. K., Cho, M.-O., & Lee, Y. (2019). Nut and Bolt Microfluidics with Helical Minichannel for Counting CD4+ T-Cells. Bioengineering, 6(1), 24. https://doi.org/10.3390/bioengineering6010024





