Insights in Pharmaceutical Pollution: The Prospective Role of eDNA Metabarcoding
Abstract
1. Introduction
2. Pharmaceuticals and Pollution: Routes and Pathways
2.1. Antibiotics
2.2. Hormones and Endocrine-Disrupting Chemicals
2.3. Analgesics and Nonsteroidal Anti-Inflammatory Drugs
2.4. Psychotropic and Antiepileptic Drugs
2.5. β-Blockers
2.6. Chemotherapy and Anticancer Drugs
3. Treatment Methods for Pharmaceutical Pollution
4. Methods of Analysis, Detecting, and Monitoring of Pharmaceutical Pollution
4.1. Bio-Monitoring and Pharmaceutical Surveillance Methods
4.1.1. Bioindicator Species: Methods and Platforms
4.1.2. eDNA Metabarcoding
5. Environmental Impact of Different Sources of Pharmaceutical Pollution
5.1. Alteration of Microbial Communities Due to Pharmaceutical Contamination
5.2. Effects of Pharmaceuticals on Aquatic Invertebrates, Plants, and Fishes
5.3. Bioaccumulation and Trophic Transfer of Pharmaceuticals in Aquatic Food Webs
6. Future Perspectives
7. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Goel, P.K. Water Pollution: Causes, Effects and Control; New Age International: New Delhi, India, 2006. [Google Scholar]
- Gautam, P.K.; Gautam, R.K.; Banerjee, S.; Chattopadhyaya, M.C.; Pandey, J.D. Heavy Metals in the Environment: Fate, Transport, Toxicity and Remediation Technologies. In Heavy Metals: Sources, Toxicity and Remediation Techniques; Pathania, S., Ed.; Nova Science Publishers: Hauppauge, NY, USA, 2016; ISBN 9781634847407. [Google Scholar]
- Obinnaa, I.B.; Ebere, E.C. A Review: Water Pollution by Heavy Metal and Organic Pollutants: Brief Review of Sources, Effects and Progress on Remediation with Aquatic Plants. Anal. Methods Environ. Chem. J. 2019, 2, 5–38. [Google Scholar] [CrossRef]
- Tongesayi, T.; Fedick, P.; Lechner, L.; Brock, C.; Le Beau, A.; Bray, C. Daily Bioaccessible Levels of Selected Essential but Toxic Heavy Metals from the Consumption of Non-Dietary Food Sources. Food Chem. Toxicol. 2013, 62, 142–147. [Google Scholar] [CrossRef] [PubMed]
- Świacka, K.; Maculewicz, J.; Kowalska, D.; Grace, M.R. Do Pharmaceuticals Affect Microbial Communities in Aquatic Environments? A Review. Front. Environ. Sci. 2023, 10, 1093920. [Google Scholar] [CrossRef]
- Patel, M.; Kumar, R.; Kishor, K.; Mlsna, T.; Pittman, C.U.; Mohan, D. Pharmaceuticals of Emerging Concern in Aquatic Systems: Chemistry, Occurrence, Effects, and Removal Methods. Chem. Rev. 2019, 119, 3510–3673. [Google Scholar] [CrossRef] [PubMed]
- Carlsson, C.; Johansson, A.K.; Alvan, G.; Bergman, K.; Kühler, T. Are Pharmaceuticals Potent Environmental Pollutants? Part I: Environmental Risk Assessments of Selected Active Pharmaceutical Ingredients. Sci. Total Environ. 2006, 364, 67–87. [Google Scholar] [CrossRef]
- Kock, A.; Glanville, H.C.; Law, A.C.; Stanton, T.; Carter, L.J.; Taylor, J.C. Emerging Challenges of the Impacts of Pharmaceuticals on Aquatic Ecosystems: A Diatom Perspective. Sci. Total Environ. 2023, 878, 162939. [Google Scholar] [CrossRef]
- Hignite, C.; Azarnoff, D.L. Drugs and Drug Metabolites as Environmental Contaminants: Chlorophenoxyisobutyrate and Salicyclic Acid in Sewage Water Effluent. Life Sci. 1977, 20, 337–342. [Google Scholar] [CrossRef]
- Cardoso, O.; Porcher, J.M.; Sanchez, W. Factory-Discharged Pharmaceuticals Could Be a Relevant Source of Aquatic Environment Contamination: Review of Evidence and Need for Knowledge. Chemosphere 2014, 115, 20–30. [Google Scholar] [CrossRef]
- Pozzebon, E.A.; Seifert, L. Emerging Environmental Health Risks Associated with the Land Application of Biosolids: A Scoping Review. Environ. Health 2023, 22, 57. [Google Scholar] [CrossRef]
- Nguyen, M.K.; Lin, C.; Nguyen, H.L.; Hung, N.T.Q.; La, D.D.; Nguyen, X.H.; Chang, S.W.; Chung, W.J.; Nguyen, D.D. Occurrence, Fate, and Potential Risk of Pharmaceutical Pollutants in Agriculture: Challenges and Environmentally Friendly Solutions. Sci. Total Environ. 2023, 899, 165323. [Google Scholar] [CrossRef]
- aus der Beek, T.; Weber, F.A.; Bergmann, A.; Hickmann, S.; Ebert, I.; Hein, A.; Küster, A. Pharmaceuticals in the Environment-Global Occurrences and Perspectives. Environ. Toxicol. Chem. 2016, 35, 823–835. [Google Scholar] [CrossRef]
- Kovalakova, P.; Cizmas, L.; McDonald, T.J.; Marsalek, B.; Feng, M.; Sharma, V.K. Occurrence and Toxicity of Antibiotics in the Aquatic Environment: A Review. Chemosphere 2020, 251, 126351. [Google Scholar] [CrossRef]
- Valdez-Carrillo, M.; Abrell, L.; Ramírez-Hernández, J.; Reyes-López, J.A.; Carreón-Diazconti, C. Pharmaceuticals as Emerging Contaminants in the Aquatic Environment of Latin America: A Review. Environ. Sci. Pollut. Res. 2020, 27, 44863–44891. [Google Scholar] [CrossRef]
- Hughes, S.R.; Kay, P.; Brown, L.E. Global Synthesis and Critical Evaluation of Pharmaceutical Data Sets Collected from River Systems. Environ. Sci. Technol. 2013, 47, 661–677. [Google Scholar] [CrossRef]
- Kolpin, D.W.; Furlong, E.T.; Meyer, M.T.; Thurman, E.M.; Zaugg, S.D.; Barber, L.B.; Buxton, H.T. Pharmaceuticals, Hormones, and Other Organic Wastewater Contaminants in U.S. Streams, 1999-2000: A National Reconnaissance. Environ. Sci. Technol. 2002, 36, 1202–1211. [Google Scholar] [CrossRef] [PubMed]
- Fekadu, S.; Alemayehu, E.; Dewil, R.; Van der Bruggen, B. Pharmaceuticals in Freshwater Aquatic Environments: A Comparison of the African and European Challenge. Sci. Total Environ. 2019, 654, 324–337. [Google Scholar] [CrossRef] [PubMed]
- Hussain, A.; Ashique, S.; Zaheen Hassan, M.; Afzal, O.; Asiri, Y.I.; Kumar, P.; Dua, K.; Webster, T.J.; Altamimi, A.S.A.; Altamimi, M.A. Pharmaceutical Contaminants in Aquatic Systems, Conventional and Green Strategies, Recent Updates, Challenges and Policies, and Potential Outcomes. J. Mol. Liq. 2023, 389, 122905. [Google Scholar] [CrossRef]
- Mahapatra, I.; Clark, J.R.A.; Dobson, P.J.; Owen, R.; Lynch, I.; Lead, J.R. Expert Perspectives on Potential Environmental Risks from Nanomedicines and Adequacy of the Current Guideline on Environmental Risk Assessment. Environ. Sci. Nano 2018, 5, 1873–1889. [Google Scholar] [CrossRef]
- Wilkinson, J.L.; Boxall, A.B.A.; Kolpin, D.W.; Leung, K.M.Y.; Lai, R.W.S.; Galbán-Malagón, C.; Adell, A.D.; Mondon, J.; Metian, M.; Marchant, R.A.; et al. Pharmaceutical Pollution of the World’s Rivers. Proc. Natl. Acad. Sci. USA 2022, 119, e2113947119. [Google Scholar] [CrossRef]
- Feckler, A.; Wolfram, J.; Schulz, R.; Bundschuh, M. Reducing Pollution to Levels Not Harming Biodiversity and Ecosystem Functions: A Perspective on the Post-2020 Global Biodiversity Framework. Curr. Opin. Environ. Sci. Health 2023, 35, 100495. [Google Scholar] [CrossRef]
- Fatimazahra, S.; Latifa, M.; Laila, S.; Monsif, K. Review of Hospital Effluents: Special Emphasis on Characterization, Impact, and Treatment of Pollutants and Antibiotic Resistance. Environ. Monit. Assess. 2023, 195, 393. [Google Scholar] [CrossRef]
- Fernandes, J.P.; Almeida, C.M.R.; Salgado, M.A.; Carvalho, M.F.; Mucha, A.P. Pharmaceutical Compounds in Aquatic Environments— Occurrence, Fate and Bioremediation Prospective. Toxics 2021, 9, 257. [Google Scholar] [CrossRef]
- Salimi, M.; Esrafili, A.; Gholami, M.; Jonidi Jafari, A.; Rezaei Kalantary, R.; Farzadkia, M.; Kermani, M.; Sobhi, H.R. Contaminants of Emerging Concern: A Review of New Approach in AOP Technologies. Environ. Monit. Assess. 2017, 189, 414. [Google Scholar] [CrossRef] [PubMed]
- Ortúzar, M.; Esterhuizen, M.; Olicón-Hernández, D.R.; González-López, J.; Aranda, E. Pharmaceutical Pollution in Aquatic Environments: A Concise Review of Environmental Impacts and Bioremediation Systems. Front. Microbiol. 2022, 13, 869332. [Google Scholar] [CrossRef]
- Lin, J.Y.; Zhang, Y.; Bian, Y.; Zhang, Y.X.; Du, R.Z.; Li, M.; Tan, Y.; Feng, X.S. Non-Steroidal Anti-Inflammatory Drugs (NSAIDs) in the Environment: Recent Updates on the Occurrence, Fate, Hazards and Removal Technologies. Sci. Total Environ. 2023, 904, 166897. [Google Scholar] [CrossRef] [PubMed]
- Santos, L.H.M.L.M.; Rodríguez-Mozaz, S.; Barceló, D. Microplastics as Vectors of Pharmaceuticals in Aquatic Organisms—An Overview of Their Environmental Implications. Case Stud. Chem. Environ. Eng. 2021, 3, 100079. [Google Scholar] [CrossRef]
- Prestinaci, F.; Pezzotti, P.; Pantosti, A. Antimicrobial Resistance: A Global Multifaceted Phenomenon. Pathog. Glob. Health 2015, 109, 309–318. [Google Scholar] [CrossRef]
- Świacka, K.; Maculewicz, J.; Kowalska, D.; Caban, M.; Smolarz, K.; Świeżak, J. Presence of Pharmaceuticals and Their Metabolites in Wild-Living Aquatic Organisms—Current State of Knowledge. J. Hazard. Mater. 2022, 424, 127350. [Google Scholar] [CrossRef] [PubMed]
- Rios-Miguel, A.B.; van Bergen, T.J.H.M.; Zillien, C.; Ragas, A.M.J.; van Zelm, R.; Jetten, M.S.M.; Hendriks, A.J.; Welte, C.U. Predicting and Improving the Microbial Removal of Organic Micropollutants during Wastewater Treatment: A Review. Chemosphere 2023, 333, 138908. [Google Scholar] [CrossRef]
- Pinheiro, M.; Martins, I.; Raimundo, J.; Caetano, M.; Neuparth, T.; Santos, M.M. Stressors of Emerging Concern in Deep-Sea Environments: Microplastics, Pharmaceuticals, Personal Care Products and Deep-Sea Mining. Sci. Total Environ. 2023, 876, 162557. [Google Scholar] [CrossRef]
- Bethke, K.; Kropidłowska, K.; Stepnowski, P.; Caban, M. Review of Warming and Acidification Effects to the Ecotoxicity of Pharmaceuticals on Aquatic Organisms in the Era of Climate Change. Sci. Total Environ. 2023, 877, 162829. [Google Scholar] [CrossRef]
- Hejna, M.; Kapuścińska, D.; Aksmann, A. Pharmaceuticals in the Aquatic Environment: A Review on Eco-Toxicology and the Remediation Potential of Algae. Int. J. Environ. Res. Public Health 2022, 19, 7717. [Google Scholar] [CrossRef] [PubMed]
- Carvalho, I.T.; Santos, L. Antibiotics in the Aquatic Environments: A Review of the European Scenario. Environ. Int. 2016, 94, 736–757. [Google Scholar] [CrossRef]
- Nannou, C.; Ofrydopoulou, A.; Evgenidou, E.; Heath, D.; Heath, E.; Lambropoulou, D. Antiviral Drugs in Aquatic Environment and Wastewater Treatment Plants: A Review on Occurrence, Fate, Removal and Ecotoxicity. Sci. Total Environ. 2020, 699, 134322. [Google Scholar] [CrossRef] [PubMed]
- Eguchi, K.; Nagase, H.; Ozawa, M.; Endoh, Y.S.; Goto, K.; Hirata, K.; Miyamoto, K.; Yoshimura, H. Evaluation of Antimicrobial Agents for Veterinary Use in the Ecotoxicity Test Using Microalgae. Chemosphere 2004, 57, 1733–1738. [Google Scholar] [CrossRef] [PubMed]
- González-Pleiter, M.; Gonzalo, S.; Rodea-Palomares, I.; Leganés, F.; Rosal, R.; Boltes, K.; Marco, E.; Fernández-Piñas, F. Toxicity of Five Antibiotics and Their Mixtures towards Photosynthetic Aquatic Organisms: Implications for Environmental Risk Assessment. Water Res. 2013, 47, 2050–2064. [Google Scholar] [CrossRef]
- Świacka, K.; Michnowska, A.; Maculewicz, J.; Caban, M.; Smolarz, K. Toxic Effects of NSAIDs in Non-Target Species: A Review from the Perspective of the Aquatic Environment. Environ. Pollut. 2021, 273, 115891. [Google Scholar] [CrossRef]
- Cleuvers, M. Mixture Toxicity of the Anti-Inflammatory Drugs Diclofenac, Ibuprofen, Naproxen, and Acetylsalicylic Acid. Ecotoxicol. Environ. Saf. 2004, 59, 309–315. [Google Scholar] [CrossRef]
- Martins, N.; Pereira, R.; Abrantes, N.; Pereira, J.; Gonçalves, F.; Marques, C.R. Ecotoxicological Effects of Ciprofloxacin on Freshwater Species: Data Integration and Derivation of Toxicity Thresholds for Risk Assessment. Ecotoxicology 2012, 21, 1167–1176. [Google Scholar] [CrossRef]
- Załęska-Radziwiłł, M.; Łebkowska, M.; Affek, K.; Zarzeczna, A. Environmental Risk Assessment of Selected Pharmaceuticals Present in Surface Waters in Relation to Animals. Arch. Environ. Prot. 2011, 37, 31–42. [Google Scholar]
- Hartwig, C.; Muth-Köhne, E.; Düring, R.A. Screening for Ecotoxicological Effects of Antiepileptic Drugs in Biologically Treated Waste Water Originating from an Epilepsy Ward by Danio Rerio Embryos. Environ. Sci. Eur. 2013, 25, 29. [Google Scholar] [CrossRef]
- Argaluza, J.; Domingo-Echaburu, S.; Orive, G.; Medrano, J.; Hernandez, R.; Lertxundi, U. Environmental Pollution with Psychiatric Drugs. World J. Psychiatry 2021, 11, 791–804. [Google Scholar] [CrossRef] [PubMed]
- Duarte, I.A.; Reis-Santos, P.; Novais, S.C.; Rato, L.D.; Lemos, M.F.L.; Freitas, A.; Pouca, A.S.V.; Barbosa, J.; Cabral, H.N.; Fonseca, V.F. Depressed, Hypertense and Sore: Long-Term Effects of Fluoxetine, Propranolol and Diclofenac Exposure in a Top Predator Fish. Sci. Total Environ. 2020, 712, 136564. [Google Scholar] [CrossRef]
- Aguirre-Martínez, G.V.; Del Valls, T.A.; Martín-Díaz, M.L. Identification of Biomarkers Responsive to Chronic Exposure to Pharmaceuticals in Target Tissues of Carcinus Maenas. Mar. Environ. Res. 2013, 87–88, 1–11. [Google Scholar] [CrossRef]
- Yamamoto, H.; Nakamura, Y.; Moriguchi, S.; Nakamura, Y.; Honda, Y.; Tamura, I.; Hirata, Y.; Hayashi, A.; Sekizawa, J. Persistence and Partitioning of Eight Selected Pharmaceuticals in the Aquatic Environment: Laboratory Photolysis, Biodegradation, and Sorption Experiments. Water Res. 2009, 43, 351–362. [Google Scholar] [CrossRef] [PubMed]
- Pal, A.; Gin, K.Y.H.; Lin, A.Y.C.; Reinhard, M. Impacts of Emerging Organic Contaminants on Freshwater Resources: Review of Recent Occurrences, Sources, Fate and Effects. Sci. Total Environ. 2010, 408, 6062–6069. [Google Scholar] [CrossRef]
- Lapworth, D.J.; Baran, N.; Stuart, M.E.; Ward, R.S. Emerging Organic Contaminants in Groundwater: A Review of Sources, Fate and Occurrence. Environ. Pollut. 2012, 163, 287–303. [Google Scholar] [CrossRef]
- Kummerer, K. Pharmaceuticals in the Environment. Annu. Rev. Environ. Resour. 2010, 35, 57–75. [Google Scholar] [CrossRef]
- Kasprzyk-Hordern, B. Pharmacologically Active Compounds in the Environment and Their Chirality. Chem. Soc. Rev. 2010, 39, 4466–4503. [Google Scholar] [CrossRef]
- Kasprzyk-Hordern, B.; Baker, D.R. Estimation of Community-Wide Drugs Use via Stereoselective Profiling of Sewage. Sci. Total Environ. 2012, 423, 142–150. [Google Scholar] [CrossRef] [PubMed]
- Miller, T.H.; Bury, N.R.; Owen, S.F.; MacRae, J.I.; Barron, L.P. A Review of the Pharmaceutical Exposome in Aquatic Fauna. Environ. Pollut. 2018, 239, 129–146. [Google Scholar] [CrossRef] [PubMed]
- Wilkinson, J.L.; Boxall, A.B.A.; Kolpin, D.W. A Novel Method to Characterise Levels of Pharmaceutical Pollution in Large-Scale Aquatic Monitoring Campaigns. Appl. Sci. 2019, 9, 1368. [Google Scholar] [CrossRef]
- Kontana, A.; Papadimitriou, C.A.; Samaras, P.; Zdragas, A.; Yiangou, M. Bioassays and Biomarkers for Ecotoxicological Assessment of Reclaimed Municipal Wastewater. Water Sci. Technol. 2008, 57, 947–953. [Google Scholar] [CrossRef]
- Maranho, L.A.; Baena-Nogueras, R.M.; Lara-Martín, P.A.; DelValls, T.A.; Martín-Díaz, M.L. Bioavailability, Oxidative Stress, Neurotoxicity and Genotoxicity of Pharmaceuticals Bound to Marine Sediments. The Use of the Polychaete Hediste Diversicolor as Bioindicator Species. Environ. Res. 2014, 134, 353–365. [Google Scholar] [CrossRef]
- Mercado, S.A.S.; Galvis, D.G.V. Paracetamol Ecotoxicological Bioassay Using the Bioindicators Lens Culinaris Med. and Pisum Sativum L. Environ. Sci. Pollut. Res. 2023, 30, 61965–61976. [Google Scholar] [CrossRef] [PubMed]
- Rodrigues, S.; Pinto, I.; Martins, F.; Formigo, N.; Antunes, S.C. Can Biochemical Endpoints Improve the Sensitivity of the Biomonitoring Strategy Using Bioassays with Standard Species, for Water Quality Evaluation? Ecotoxicol. Environ. Saf. 2021, 215, 112151. [Google Scholar] [CrossRef]
- Gavrilescu, M.; Demnerová, K.; Aamand, J.; Agathos, S.; Fava, F. Emerging Pollutants in the Environment: Present and Future Challenges in Biomonitoring, Ecological Risks and Bioremediation. New Biotechnol. 2015, 32, 147–156. [Google Scholar] [CrossRef] [PubMed]
- Chauhan, B.; Dodamani, S.; Malik, S.; Almalki, W.H.; Haque, S.; Sayyed, R.Z. Microbial Approaches for Pharmaceutical Wastewater Recycling and Management for Sustainable Development: A Multicomponent Approach. Environ. Res. 2023, 237, 116983. [Google Scholar] [CrossRef]
- Mishra, S.; Singh, A.K.; Cheng, L.; Hussain, A.; Maiti, A. Occurrence of Antibiotics in Wastewater: Potential Ecological Risk and Removal through Anaerobic–Aerobic Systems. Environ. Res. 2023, 226, 115678. [Google Scholar] [CrossRef]
- Lin, Y.; Zhong, W.; Zhang, X.; Zhou, X.; He, L.; Lv, J.; Zhao, Z. Environmental DNA Metabarcoding Revealed the Impacts of Anthropogenic Activities on Phytoplankton Diversity in Dianchi Lake and Its Three Inflow Rivers. Ecol. Evol. 2023, 13, e10088. [Google Scholar] [CrossRef]
- Cardinale, B.J.; Duffy, J.E.; Gonzalez, A.; Hooper, D.U.; Perrings, C.; Venail, P.; Narwani, A.; MacE, G.M.; Tilman, D.; Wardle, D.A.; et al. Biodiversity Loss and Its Impact on Humanity. Nature 2012, 486, 59–67. [Google Scholar] [CrossRef]
- Shim, K.Y.; Shin, H.; Yeo, I.C.; Kim, K.R.; Kwak, I.S.; Jeong, C.B. Environmental DNA Surveillance of Biocontamination in a Drinking Water Treatment Plant. J. Hazard. Mater. 2023, 456, 131656. [Google Scholar] [CrossRef] [PubMed]
- Bernhard, A.E.; Field, K.G. Identification of Nonpoint Sources of Fecal Pollution in Coastal Waters by Using Host-Specific 16S Ribosomal DNA Genetic Markers from Fecal Anaerobes. Appl. Environ. Microbiol. 2000, 66, 1587–1594. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Peng, Y.; Fang, W.; Altermatt, F.; Xie, Y.; Yang, J.; Zhang, X. Application of Environmental DNA Metabarcoding for Predicting Anthropogenic Pollution in Rivers. Environ. Sci. Technol. 2018, 52, 11708–11719. [Google Scholar] [CrossRef]
- Wang, S.; Zhang, P.; Zhang, D.; Chang, J. Evaluation and Comparison of the Benthic and Microbial Indices of Biotic Integrity for Urban Lakes Based on Environmental DNA and Its Management Implications. J. Environ. Manag. 2023, 341, 118026. [Google Scholar] [CrossRef] [PubMed]
- Zan, R.; Blackburn, A.; Plaimart, J.; Acharya, K.; Walsh, C.; Stirling, R.; Kilsby, C.G.; Werner, D. Environmental DNA Clarifies Impacts of Combined Sewer Overflows on the Bacteriology of an Urban River and Resulting Risks to Public Health. Sci. Total Environ. 2023, 889, 164282. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Yang, J.; Zhang, Y.; Shi, J.; Yu, H.; Zhang, X. eDNAeDNA Biomonitoring Revealed the Ecological Effects of Water Diversion Projects between Yangtze River and Tai Lake. Water Res. 2022, 210, 117994. [Google Scholar] [CrossRef]
- Aulsebrook, L.C.; Wong, B.B.M.; Hall, M.D. Can Pharmaceutical Pollution Alter the Spread of Infectious Disease? A Case Study Using Fluoxetine. Philos. Trans. R. Soc. B Biol. Sci. 2023, 378, 20220010. [Google Scholar] [CrossRef]
- Barnes, M.A.; Turner, C.R.; Jerde, C.L.; Renshaw, M.A.; Chadderton, W.L.; Lodge, D.M. Environmental Conditions Influence eDNA Persistence in Aquatic Systems. Environ. Sci. Technol. 2014, 48, 1819–1827. [Google Scholar] [CrossRef]
- Greco, M.; Lejzerowicz, F.; Reo, E.; Caruso, A.; Maccotta, A.; Coccioni, R.; Pawlowski, J.; Frontalini, F. Environmental RNA Outperforms eDNA Metabarcoding in Assessing Impact of Marine Pollution: A Chromium-Spiked Mesocosm Test. Chemosphere 2022, 298, 134239. [Google Scholar] [CrossRef] [PubMed]
- Jerde, C.L.; Olds, B.P.; Shogren, A.J.; Andruszkiewicz, E.A.; Mahon, A.R.; Bolster, D.; Tank, J.L. Influence of Stream Bottom Substrate on Retention and Transport of Vertebrate Environmental DNA. Environ. Sci. Technol. 2016, 50, 8770–8779. [Google Scholar] [CrossRef]
- Suarez-Menendez, M.; Planes, S.; Garcia-Vazquez, E.; Ardura, A. Early Alert of Biological Risk in a Coastal Lagoon Through eDNA Metabarcoding. Front. Ecol. Evol. 2020, 8, 9. [Google Scholar] [CrossRef]
- Moher, D.; Liberati, A.; Tetzlaff, J.; Altman, D.G.; Antes, G.; Atkins, D.; Barbour, V.; Barrowman, N.; Berlin, J.A.; Clark, J.; et al. Preferred Reporting Items for Systematic Reviews and Meta-Analyses: The PRISMA Statement. PLoS Med. 2009, 6, e1000097. [Google Scholar] [CrossRef]
- Barra Caracciolo, A.; Topp, E.; Grenni, P. Pharmaceuticals in the Environment: Biodegradation and Effects on Natural Microbial Communities. A Review. J. Pharm. Biomed. Anal. 2015, 106, 25–36. [Google Scholar] [CrossRef] [PubMed]
- Arnold, K.E.; Brown, A.R.; Brown, A.R.; Ankley, G.T.; Sumpter, J.P. Medicating the Environment: Assessing Risks of Pharmaceuticals to Wildlife and Ecosystems. Philos. Trans. R. Soc. B Biol. Sci. 2014, 369, 20130569. [Google Scholar] [CrossRef] [PubMed]
- Grenni, P.; Ancona, V.; Barra Caracciolo, A. Ecological Effects of Antibiotics on Natural Ecosystems: A Review. Microchem. J. 2018, 136, 25–39. [Google Scholar] [CrossRef]
- King, K.C.; Hall, M.D.; Wolinska, J. Infectious Disease Ecology and Evolution in a Changing World. Philos. Trans. R. Soc. B Biol. Sci. 2023, 378, 20220002. [Google Scholar] [CrossRef]
- Caban, M.; Stepnowski, P. How to Decrease Pharmaceuticals in the Environment? A Review. Environ. Chem. Lett. 2021, 19, 3115–3138. [Google Scholar] [CrossRef]
- Bilal, M.; Mehmood, S.; Rasheed, T.; Iqbal, H.M.N. Antibiotics Traces in the Aquatic Environment: Persistence and Adverse Environmental Impact. Curr. Opin. Environ. Sci. Health 2020, 13, 68–74. [Google Scholar] [CrossRef]
- Bondarczuk, K.; Piotrowska-Seget, Z. Microbial Diversity and Antibiotic Resistance in a Final Effluent-Receiving Lake. Sci. Total Environ. 2019, 650, 2951–2961. [Google Scholar] [CrossRef]
- Lai, C.C.; Chen, S.Y.; Ko, W.C.; Hsueh, P.R. Increased Antimicrobial Resistance during the COVID-19 Pandemic. Int. J. Antimicrob. Agents 2021, 57, 106324. [Google Scholar] [CrossRef]
- López-Serna, R.; Jurado, A.; Vázquez-Suñé, E.; Carrera, J.; Petrović, M.; Barceló, D. Occurrence of 95 Pharmaceuticals and Transformation Products in Urban Groundwaters Underlying the Metropolis of Barcelona, Spain. Environ. Pollut. 2013, 174, 305–315. [Google Scholar] [CrossRef] [PubMed]
- Michaelis, C.; Grohmann, E. Horizontal Gene Transfer of Antibiotic Resistance Genes in Biofilms. Antibiotics 2023, 12, 328. [Google Scholar] [CrossRef]
- Mukhtar, A.; Manzoor, M.; Gul, I.; Zafar, R.; Jamil, H.I.; Niazi, A.K.; Ali, M.A.; Park, T.J.; Arshad, M. Phytotoxicity of Different Antibiotics to Rice and Stress Alleviation upon Application of Organic Amendments. Chemosphere 2020, 258, 127353. [Google Scholar] [CrossRef] [PubMed]
- Peng, X.; Yu, Y.; Tang, C.; Tan, J.; Huang, Q.; Wang, Z. Occurrence of Steroid Estrogens, Endocrine-Disrupting Phenols, and Acid Pharmaceutical Residues in Urban Riverine Water of the Pearl River Delta, South China. Sci. Total Environ. 2008, 397, 158–166. [Google Scholar] [CrossRef]
- Peng, X.; Zhang, K.; Tang, C.; Huang, Q.; Yu, Y.; Cui, J. Distribution Pattern, Behavior, and Fate of Antibacterials in Urban Aquatic Environments in South China. J. Environ. Monit. 2011, 13, 446–454. [Google Scholar] [CrossRef]
- Samal, K.; Mahapatra, S.; Hibzur Ali, M. Pharmaceutical Wastewater as Emerging Contaminants (EC): Treatment Technologies, Impact on Environment and Human Health. Energy Nexus 2022, 6, 100076. [Google Scholar] [CrossRef]
- Serwecińska, L. Antimicrobials and Antibiotic-Resistant Bacteria: A Risk to the Environment and to Public Health. Water 2020, 12, 3313. [Google Scholar] [CrossRef]
- Valcárcel, Y.; González Alonso, S.; Rodríguez-Gil, J.L.; Gil, A.; Catalá, M. Detection of Pharmaceutically Active Compounds in the Rivers and Tap Water of the Madrid Region (Spain) and Potential Ecotoxicological Risk. Chemosphere 2011, 84, 1336–1348. [Google Scholar] [CrossRef] [PubMed]
- Andrade-Eiroa, A.; Canle, M.; Leroy-Cancellieri, V.; Cerdà, V. Solid-Phase Extraction of Organic Compounds: A Critical Review (Part I). Trends Anal. Chem. 2016, 80, 641–654. [Google Scholar] [CrossRef]
- Braun, J.M. Early-Life Exposure to EDCs: Role in Childhood Obesity and Neurodevelopment. Nat. Rev. Endocrinol. 2017, 13, 161–173. [Google Scholar] [CrossRef]
- Forte, M.; Di Lorenzo, M.; Carrizzo, A.; Valiante, S.; Vecchione, C.; Laforgia, V.; De Falco, M. Nonylphenol Effects on Human Prostate Non Tumorigenic Cells. Toxicology 2016, 357–358, 21–32. [Google Scholar] [CrossRef] [PubMed]
- Forte, M.; Di Lorenzo, M.; Iachetta, G.; Mita, D.G.; Laforgia, V.; De Falco, M. Nonylphenol Acts on Prostate Adenocarcinoma Cells via Estrogen Molecular Pathways. Ecotoxicol. Environ. Saf. 2019, 180, 412–419. [Google Scholar] [CrossRef] [PubMed]
- Gröger, T.M.; Käfer, U.; Zimmermann, R. Gas Chromatography in Combination with Fast High-Resolution Time-of-Flight Mass Spectrometry: Technical Overview and Perspectives for Data Visualization. Trends Anal. Chem. 2020, 122, 115677. [Google Scholar] [CrossRef]
- Heindel, J.J.; Newbold, R.; Schug, T.T. Endocrine Disruptors and Obesity. Nat. Rev. Endocrinol. 2015, 11, 653–661. [Google Scholar] [CrossRef]
- Li, C.; Wei, Y.; Zhang, S.; Tan, W. Advanced Methods to Analyze Steroid Estrogens in Environmental Samples. Environ. Chem. Lett. 2020, 18, 543–559. [Google Scholar] [CrossRef]
- Marotta, V.; Russo, G.; Gambardella, C.; Grasso, M.; La Sala, D.; Chiofalo, M.G.; D’Anna, R.; Puzziello, A.; Docimo, G.; Masone, S.; et al. Human Exposure to Bisphenol AF and Diethylhexylphthalate Increases Susceptibility to Develop Differentiated Thyroid Cancer in Patients with Thyroid Nodules. Chemosphere 2019, 218, 885–894. [Google Scholar] [CrossRef]
- Nadal, A.; Quesada, I.; Tudurí, E.; Nogueiras, R.; Alonso-Magdalena, P. Endocrine-Disrupting Chemicals and the Regulation of Energy Balance. Nat. Rev. Endocrinol. 2017, 13, 536–546. [Google Scholar] [CrossRef]
- Izadi, P.; Izadi, P.; Salem, R.; Papry, S.A.; Magdouli, S.; Pulicharla, R.; Brar, S.K. Non-Steroidal Anti-Inflammatory Drugs in the Environment: Where Were We and How Far We Have Come? Environ. Pollut. 2020, 267, 115370. [Google Scholar] [CrossRef] [PubMed]
- Jan-Roblero, J.; Cruz-Maya, J.A. Ibuprofen: Toxicology and Biodegradation of an Emerging Contaminant. Molecules 2023, 28, 2097. [Google Scholar] [CrossRef]
- Parolini, M. Toxicity of the Non-Steroidal Anti-Inflammatory Drugs (NSAIDs) Acetylsalicylic Acid, Paracetamol, Diclofenac, Ibuprofen and Naproxen towards Freshwater Invertebrates: A Review. Sci. Total Environ. 2020, 740, 140043. [Google Scholar] [CrossRef] [PubMed]
- Tyumina, E.A.; Bazhutin, G.A.; Cartagena Gómez, A.d.P.; Ivshina, I.B. Nonsteroidal Anti-Inflammatory Drugs as Emerging Contaminants. Microbiology 2020, 89, 148–163. [Google Scholar] [CrossRef]
- Huynh, N.C.; Nguyen, T.T.T.; Nguyen, D.T.C.; Tran, T. Van Occurrence, Toxicity, Impact and Removal of Selected Non-Steroidal Anti-Inflammatory Drugs (NSAIDs): A Review. Sci. Total Environ. 2023, 898, 165317. [Google Scholar] [CrossRef] [PubMed]
- Gao, S.; Yang, F. Behavioral Changes and Neurochemical Responses in Chinese Rare Minnow Exposed to Four Psychoactive Substances. Sci. Total Environ. 2022, 808, 152100. [Google Scholar] [CrossRef]
- Gupta, I.; Clauder-Münster, S.; Klaus, B.; Järvelin, A.I.; Aiyar, R.S.; Benes, V.; Wilkening, S.; Huber, W.; Pelechano, V.; Steinmetz, L.M. Alternative Polyadenylation Diversifies Post-Transcriptional Regulation by Selective RNA-Protein Interactions. Mol. Syst. Biol. 2014, 10, 719. [Google Scholar] [CrossRef] [PubMed]
- Mousel, D.; Bastian, D.; Firk, J.; Palmowski, L.; Pinnekamp, J. Removal of Pharmaceuticals from Wastewater of Health Care Facilities. Sci. Total Environ. 2021, 751, 141310. [Google Scholar] [CrossRef]
- Huggett, D.B.; Brooks, B.W.; Peterson, B.; Foran, C.M.; Schlenk, D. Toxicity of Select Beta Adrenergic Receptor-Blocking Pharmaceuticals (B-Blockers) on Aquatic Organisms. Arch. Environ. Contam. Toxicol. 2002, 43, 229–235. [Google Scholar] [CrossRef] [PubMed]
- Maszkowska, J.; Stolte, S.; Kumirska, J.; Łukaszewicz, P.; Mioduszewska, K.; Puckowski, A.; Caban, M.; Wagil, M.; Stepnowski, P.; Białk-Bielińska, A. Beta-Blockers in the Environment: Part II. Ecotoxicity Study. Sci. Total Environ. 2014, 493, 1122–1126. [Google Scholar] [CrossRef]
- De Oliveira, L.L.D.; Antunes, S.C.; Gonçalves, F.; Rocha, O.; Nunes, B. Acute and Chronic Ecotoxicological Effects of Four Pharmaceuticals Drugs on Cladoceran Daphnia Magna. Drug Chem. Toxicol. 2016, 39, 13–21. [Google Scholar] [CrossRef]
- Rutere, C.; Posselt, M.; Ho, A.; Horn, M.A. Biodegradation of Metoprolol in Oxic and Anoxic Hyporheic Zone Sediments: Unexpected Effects on Microbial Communities. Appl. Microbiol. Biotechnol. 2021, 105, 6103–6115. [Google Scholar] [CrossRef]
- Li, D.; Chen, H.; Liu, H.; Schlenk, D.; Mu, J.; Lacorte, S.; Ying, G.G.; Xie, L. Anticancer Drugs in the Aquatic Ecosystem: Environmental Occurrence, Ecotoxicological Effect and Risk Assessment. Environ. Int. 2021, 153, 106543. [Google Scholar] [CrossRef]
- Nassour, C.; Barton, S.J.; Nabhani-Gebara, S.; Saab, Y.; Barker, J. Occurrence of Anticancer Drugs in the Aquatic Environment: A Systematic Review. Environ. Sci. Pollut. Res. 2020, 27, 1339–1347. [Google Scholar] [CrossRef]
- Rowney, N.C.; Johnson, A.C.; Williams, R.J. Cytotoxic Drugs in Drinking Water: A Prediction and Risk Assessment Exercise for the Thames Catchment in the United Kingdom. Environ. Toxicol. Chem. 2009, 28, 2733–2743. [Google Scholar] [CrossRef] [PubMed]
- Kümmerer, K. Antibiotics in the Aquatic Environment—A Review—Part I. Chemosphere 2009, 75, 417–434. [Google Scholar] [CrossRef]
- Gore, A.C.; Chappell, V.A.; Fenton, S.E.; Flaws, J.A.; Nadal, A.; Prins, G.S.; Toppari, J.; Zoeller, R.T. Executive Summary to EDC-2: The Endocrine Society’s Second Scientific Statement on Endocrine-Disrupting Chemicals. Endocr. Rev. 2015, 36, 593–602. [Google Scholar] [CrossRef] [PubMed]
- Kabir, E.R.; Rahman, M.S.; Rahman, I. A Review on Endocrine Disruptors and Their Possible Impacts on Human Health. Environ. Toxicol. Pharmacol. 2015, 40, 241–258. [Google Scholar] [CrossRef]
- Kiyama, R.; Wada-Kiyama, Y. Estrogenic Endocrine Disruptors: Molecular Mechanisms of Action. Environ. Int. 2015, 83, 11–40. [Google Scholar] [CrossRef]
- Gore, A.C.; Crews, D.; Doan, L.L.; Merrill, M.L.; Patisaul, H.; Zota, A. Introduction to Endocrine Disrupting Chemicals (EDCs): A Guide for Public Interest Organizations and Policymakers; IPEN: Göteborg, Sweden, 2014; pp. 21–22. [Google Scholar]
- Tijani, J.O.; Fatoba, O.O.; Babajide, O.O.; Petrik, L.F. Pharmaceuticals, Endocrine Disruptors, Personal Care Products, Nanomaterials and Perfluorinated Pollutants: A Review. Environ. Chem. Lett. 2016, 14, 27–49. [Google Scholar] [CrossRef]
- Huang, S.; Qi, Z.; Ma, S.; Li, G.; Long, C.; Yu, Y. A Critical Review on Human Internal Exposure of Phthalate Metabolites and the Associated Health Risks. Environ. Pollut. 2021, 279, 116941. [Google Scholar] [CrossRef]
- De Coster, S.; Van Larebeke, N. Endocrine-Disrupting Chemicals: Associated Disorders and Mechanisms of Action. J. Environ. Public Health 2012, 2012, 713696. [Google Scholar] [CrossRef]
- Monneret, C. What Is an Endocrine Disruptor? Comptes Rendus. Biol. 2017, 340, 403–405. [Google Scholar] [CrossRef] [PubMed]
- Akhbarizadeh, R.; Russo, G.; Rossi, S.; Golianova, K.; Moore, F.; Guida, M.; De Falco, M.; Grumetto, L. Emerging Endocrine Disruptors in Two Edible Fish from the Persian Gulf: Occurrence, Congener Profile, and Human Health Risk Assessment. Mar. Pollut. Bull. 2021, 166, 112241. [Google Scholar] [CrossRef]
- Vieira, W.T.; de Farias, M.B.; Spaolonzi, M.P.; da Silva, M.G.C.; Vieira, M.G.A. Removal of Endocrine Disruptors in Waters by Adsorption, Membrane Filtration and Biodegradation. A Review. Environ. Chem. Lett. 2020, 18, 1113–1143. [Google Scholar] [CrossRef]
- Gonsioroski, A.; Mourikes, V.E.; Flaws, J.A. Endocrine Disruptors in Water and Their Effects on the Reproductive System. Int. J. Mol. Sci. 2020, 21, 1929. [Google Scholar] [CrossRef]
- Feng, L.; van Hullebusch, E.D.; Rodrigo, M.A.; Esposito, G.; Oturan, M.A. Removal of Residual Anti-Inflammatory and Analgesic Pharmaceuticals from Aqueous Systems by Electrochemical Advanced Oxidation Processes. A Review. Chem. Eng. J. 2013, 228, 944–964. [Google Scholar] [CrossRef]
- Azmi Hassali, M.; Shakeel, S. Unused and Expired Medications Disposal Practices among the General Public in Selangor, Malaysia. Pharmacy 2020, 8, 196. [Google Scholar] [CrossRef]
- Vieno, N.; Tuhkanen, T.; Kronberg, L. Elimination of Pharmaceuticals in Sewage Treatment Plants in Finland. Water Res. 2007, 41, 1001–1012. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Sun, H.; Zhang, Y.; Alder, A.C. Occurrence and Enantiomer Profiles of β-Blockers in Wastewater and a Receiving Water Body and Adjacent Soil in Tianjin, China. Sci. Total Environ. 2019, 650, 1122–1130. [Google Scholar] [CrossRef] [PubMed]
- Godoy, A.A.; Kummrow, F.; Pamplin, P.A.Z. Occurrence, Ecotoxicological Effects and Risk Assessment of Antihypertensive Pharmaceutical Residues in the Aquatic Environment—A Review. Chemosphere 2015, 138, 281–291. [Google Scholar] [CrossRef] [PubMed]
- Maszkowska, J.; Stolte, S.; Kumirska, J.; Łukaszewicz, P.; Mioduszewska, K.; Puckowski, A.; Caban, M.; Wagil, M.; Stepnowski, P.; Białk-Bielińska, A. Beta-Blockers in the Environment: Part I. Mobility and Hydrolysis Study. Sci. Total Environ. 2014, 493, 1112–1121. [Google Scholar] [CrossRef]
- Heath, E.; Filipič, M.; Kosjek, T.; Isidori, M. Fate and Effects of the Residues of Anticancer Drugs in the Environment. Environ. Sci. Pollut. Res. 2016, 23, 14687–14691. [Google Scholar] [CrossRef] [PubMed]
- Wormington, A.M.; De María, M.; Kurita, H.G.; Bisesi, J.H.; Denslow, N.D.; Martyniuk, C.J. Antineoplastic Agents: Environmental Prevalence and Adverse Outcomes in Aquatic Organisms. Environ. Toxicol. Chem. 2020, 39, 967–985. [Google Scholar] [CrossRef]
- Grassi, M.; Kaykioglu, G.; Belgiorno, V.; Lofrano, G. Removal of Emerging Contaminants from Water and Wastewater by Adsorption Process. In Emerging Compounds Removal from Wastewater Natural and Solar Based Treatments; Lofrano, G., Ed.; Springer: Salerno, Italy, 2012; pp. 15–37. [Google Scholar]
- Falisse, E.; Voisin, A.S.; Silvestre, F. Impacts of Triclosan Exposure on Zebrafish Early-Life Stage: Toxicity and Acclimation Mechanisms. Aquat. Toxicol. 2017, 189, 97–107. [Google Scholar] [CrossRef]
- Khan, F.I.; Husain, T.; Hejazi, R. An Overview and Analysis of Site Remediation Technologies. J. Environ. Manag. 2004, 71, 95–122. [Google Scholar] [CrossRef]
- Kuppusamy, S.; Palanisami, T.; Megharaj, M.; Venkateswarlu, K.; Naidu, R. In-Situ Remediation Approaches for the Management of Contaminated Sites: A Comprehensive Overview. In Reviews of Environmental Contamination and Toxicology; de Voogt, P., Ed.; Springer: New York, NY, USA, 2016; Volume 236, pp. 1–115. [Google Scholar]
- Chang, C.Y.; Chang, J.S.; Vigneswaran, S.; Kandasamy, J. Pharmaceutical Wastewater Treatment by Membrane Bioreactor Process—A Case Study in Southern Taiwan. Desalination 2008, 234, 393–401. [Google Scholar] [CrossRef]
- Madukasi, E.I.; Dai, X.; He, C.; Zhou, J. Potentials of Phototrophic Bacteria in Treating Pharmaceutical Wastewater. Int. J. Environ. Sci. Technol. 2010, 7, 165–174. [Google Scholar] [CrossRef]
- Spina, F.; Anastasi, A.E.; Prigione, V.P.; Tigini, V.; Varese, G. Biological Treatment of Industrial Wastewaters: A Fungal Approach. Chem. Eng. Trans. 2012, 27, 175–180. [Google Scholar]
- Haq, I.; Raj, A. Endocrine-Disrupting Pollutants in Industrial Wastewater and Their and Detoxification. In Emerging and Eco-Friendly Approaches for Waste Management; Bharagava, R.N., Chowdhary, P., Eds.; Springer: Singapore, 2019; pp. 121–142. ISBN 9789811086694. [Google Scholar]
- Leusch, F.D.L.; Neale, P.A.; Busetti, F.; Card, M.; Humpage, A.; Orbell, J.D.; Ridgway, H.F.; Stewart, M.B.; van de Merwe, J.P.; Escher, B.I. Transformation of Endocrine Disrupting Chemicals, Pharmaceutical and Personal Care Products during Drinking Water Disinfection. Sci. Total Environ. 2019, 657, 1480–1490. [Google Scholar] [CrossRef]
- Abd El-Gawad, S.A.; Abd ElAziz, H.M. Effective Removal of Chemical Oxygen Demand and Phosphates from Aqueous Medium Using Entrapped Activated Carbon in Alginate. MOJ Biol. Med. 2018, 3, 227–236. [Google Scholar] [CrossRef]
- Martín de Vidales, M.J.; Rua, J.; de Juan, J.L.M.; Fernández-Martínez, F.; Dos Santos-García, A.J. Degradation of Contaminants of Emerging Concern by Electrochemical Oxidation: Coupling of Ultraviolet and Ultrasound Radiations. Materials 2020, 13, 5551. [Google Scholar] [CrossRef]
- Qin, K.; Zhao, Q.; Yu, H.; Xia, X.; Li, J.; He, S.; Wei, L.; An, T. A Review of Bismuth-Based Photocatalysts for Antibiotic Degradation: Insight into the Photocatalytic Degradation Performance, Pathways and Relevant Mechanisms. Environ. Res. 2021, 199, 111360. [Google Scholar] [CrossRef]
- An, T.; Yang, H.; Li, G.; Song, W.; Cooper, W.J.; Nie, X. Kinetics and Mechanism of Advanced Oxidation Processes (AOPs) in Degradation of Ciprofloxacin in Water. Appl. Catal. B 2010, 94, 288–294. [Google Scholar] [CrossRef]
- Chen, J.; Luo, H.; Shi, H.; Li, G.; An, T. Anatase TiO2 Nanoparticles-Carbon Nanotubes Composite: Optimization Synthesis and the Relationship of Photocatalytic Degradation Activity of Acyclovir in Water. Appl. Catal. A Gen. 2014, 485, 188–195. [Google Scholar] [CrossRef]
- Li, G.; Nie, X.; Chen, J.; Wong, P.K.; An, T.; Yamashita, H.; Zhao, H. Enhanced Simultaneous PEC Eradication of Bacteria and Antibiotics by Facilely Fabricated High-Activity {001} Facets TiO2 Mounted onto TiO2 Nanotubular Photoanode. Water Res. 2016, 101, 597–605. [Google Scholar] [CrossRef]
- Khan, J.A.; He, X.; Khan, H.M.; Shah, N.S.; Dionysiou, D.D. Oxidative Degradation of Atrazine in Aqueous Solution by UV/H2O2/Fe2+, UV/S2/Fe2+ and UV/HS/Fe2+ Processes: A Comparative Study. Chem. Eng. J. 2013, 218, 376–383. [Google Scholar] [CrossRef]
- Stadlmair, L.F.; Letzel, T.; Drewes, J.E.; Grassmann, J. Enzymes in Removal of Pharmaceuticals from Wastewater: A Critical Review of Challenges, Applications and Screening Methods for Their Selection. Chemosphere 2018, 205, 649–661. [Google Scholar] [CrossRef] [PubMed]
- Burke, V.; Duennbier, U.; Massmann, G. The Effect of Aeration on the Removal of Wastewater-Derived Pharmaceutical Residues from Groundwater ⋯ A Laboratory Study. Water Sci. Technol. 2013, 67, 658–666. [Google Scholar] [CrossRef] [PubMed]
- Adams, C.; Asce, M.; Wang, Y.; Loftin, K.; Meyer, M. Removal of Antibiotics from Surface and Distilled Water in Conventional Water Treatment Processes. J. Environ. Eng. 2002, 128, 253–260. [Google Scholar] [CrossRef]
- Ternes, T.A.; Meisenheimer, M.; McDowell, D.; Sacher, F.; Brauch, H.J.; Haist-Gulde, B.; Preuss, G.; Wilme, U.; Zulei-Seibert, N. Removal of Pharmaceuticals during Drinking Water Treatment. Environ. Sci. Technol. 2002, 36, 3855–3863. [Google Scholar] [CrossRef]
- Westerhoff, P.; Yoon, Y.; Snyder, S.; Wert, E. Fate of Endocrine-Disruptor, Pharmaceutical, and Personal Care Product Chemicals during Simulated Drinking Water Treatment Processes. Environ. Sci. Technol. 2005, 39, 6649–6663. [Google Scholar] [CrossRef]
- Ferreira, R.C.; De Lima, H.H.C.; Cândido, A.A.; Junior, O.C.; Arroyo, P.A.; De Carvalho, K.Q.; Gauze, G.F.; Barros, M.A.S.D. Adsorption of Paracetamol Using Activated Carbon of Dende and Babassu Coconut Mesocarp. Int. J. Biotechnol. Bioeng. 2015, 9, 717–722. [Google Scholar] [CrossRef]
- Mukoko, T.; Mupa, M.; Guyo, U.; Dziike, F. Preparation of Rice Hull Activated Carbon for the Removal of Selected Pharmaceutical Waste Compounds in Hospital Effluent. J. Environ. Anal. Toxicol. 2015, s7, 1–9. [Google Scholar] [CrossRef]
- Nazari, G.; Abolghasemi, H.; Esmaieli, M. Batch Adsorption of Cephalexin Antibiotic from Aqueous Solution by Walnut Shell-Based Activated Carbon. J. Taiwan. Inst. Chem. Eng. 2016, 58, 357–365. [Google Scholar] [CrossRef]
- Ahmadi, S.; Banach, A.; Kord Mostafapour, F.; Balarak, D. Study Survey of Cupric Oxide Nanoparticles in Removal Efficiency of Ciprofloxacin Antibiotic from Aqueous Solution: Adsorption Isotherm Study. Desalination Water Treat. 2017, 89, 297–303. [Google Scholar] [CrossRef]
- Prasannamedha, G.; Kumar, P.S.; Mehala, R.; Sharumitha, T.J.; Surendhar, D. Enhanced Adsorptive Removal of Sulfamethoxazole from Water Using Biochar Derived from Hydrothermal Carbonization of Sugarcane Bagasse. J. Hazard. Mater. 2021, 407, 124825. [Google Scholar] [CrossRef]
- Richter, D.; Massmann, G.; Dünnbier, U. Behaviour and Biodegradation of Sulfonamides (p-TSA, o-TSA, BSA) during Drinking Water Treatment. Chemosphere 2008, 71, 1574–1581. [Google Scholar] [CrossRef] [PubMed]
- Meffe, R.; Kohfahl, C.; Holzbecher, E.; Massmann, G.; Richter, D.; Dünnbier, U.; Pekdeger, A. Modelling the Removal of P-TSA (Para-Toluenesulfonamide) during Rapid Sand Filtration Used for Drinking Water Treatment. Water Res. 2010, 44, 205–213. [Google Scholar] [CrossRef] [PubMed]
- Prasse, C.; Wagner, M.; Schulz, R.; Ternes, T.A. Biotransformation of the Antiviral Drugs Acyclovir and Penciclovir in Activated Sludge Treatment. Environ. Sci. Technol. 2011, 45, 2761–2769. [Google Scholar] [CrossRef]
- Zearley, T.L.; Summers, R.S. Removal of Trace Organic Micropollutants by Drinking Water Biological Filters. Environ. Sci. Technol. 2012, 46, 9412–9419. [Google Scholar] [CrossRef]
- Helbling, D.E.; Hollender, J.; Kohler, H.P.E.; Singer, H.; Fenner, K. High-Throughput Identification of Microbial Transformation Products of Organic Micropollutants. Environ. Sci. Technol. 2010, 44, 6621–6627. [Google Scholar] [CrossRef]
- Zhao, S.; Ba, C.; Yao, Y.; Zheng, W.; Economy, J.; Wang, P. Removal of Antibiotics Using Polyethylenimine Cross-Linked Nanofiltration Membranes: Relating Membrane Performance to Surface Charge Characteristics. Chem. Eng. J. 2018, 335, 101–109. [Google Scholar] [CrossRef]
- Mallakpour, S.; Azadi, E. Nanofiltration Membranes for Food and Pharmaceutical Industries. Emergent Mater. 2022, 5, 1329–1343. [Google Scholar] [CrossRef]
- Nayak, V.; Cuhorka, J.; Mikulášek, P. Separation of Drugs by Commercial Nanofiltration Membranes and Their Modelling. Membranes 2022, 12, 528. [Google Scholar] [CrossRef] [PubMed]
- Radjenović, J.; Petrović, M.; Ventura, F.; Barceló, D. Rejection of Pharmaceuticals in Nanofiltration and Reverse Osmosis Membrane Drinking Water Treatment. Water Res. 2008, 42, 3601–3610. [Google Scholar] [CrossRef] [PubMed]
- Urtiaga, A.M.; Pérez, G.; Ibáñez, R.; Ortiz, I. Removal of Pharmaceuticals from a WWTP Secondary Effluent by Ultrafiltration/Reverse Osmosis Followed by Electrochemical Oxidation of the RO Concentrate. Desalination 2013, 331, 26–34. [Google Scholar] [CrossRef]
- Hollman, J.; Khan, M.F.; Dominic, J.A.; Achari, G. Pilot-Scale Treatment of Neutral Pharmaceuticals in Municipal Wastewater Using Reverse Osmosis and Ozonation. J. Environ. Eng. 2020, 146, 04020121. [Google Scholar] [CrossRef]
- Urase, T.; Kikuta, T. Separate Estimation of Adsorption and Degradation of Pharmaceutical Substances and Estrogens in the Activated Sludge Process. Water Res. 2005, 39, 1289–1300. [Google Scholar] [CrossRef]
- Krah, D.; Ghattas, A.K.; Wick, A.; Bröder, K.; Ternes, T.A. Micropollutant Degradation via Extracted Native Enzymes from Activated Sludge. Water Res. 2016, 95, 348–360. [Google Scholar] [CrossRef]
- Hena, S.; Znad, H. Membrane Bioreactor for Pharmaceuticals and Personal Care Products Removal from Wastewater. In Comprehensive Analytical Chemistry; Elsevier B.V.: Amsterdam, The Netherlands, 2018; Volume 81, pp. 201–256. ISBN 9780444640642. [Google Scholar]
- de Wilt, A.; Butkovskyi, A.; Tuantet, K.; Leal, L.H.; Fernandes, T.V.; Langenhoff, A.; Zeeman, G. Micropollutant Removal in an Algal Treatment System Fed with Source Separated Wastewater Streams. J. Hazard. Mater. 2016, 304, 84–92. [Google Scholar] [CrossRef]
- Peng, F.Q.; Ying, G.G.; Yang, B.; Liu, S.; Lai, H.J.; Liu, Y.S.; Chen, Z.F.; Zhou, G.J. Biotransformation of Progesterone and Norgestrel by Two Freshwater Microalgae (Scenedesmus obliquus and Chlorella pyrenoidosa): Transformation Kinetics and Products Identification. Chemosphere 2014, 95, 581–588. [Google Scholar] [CrossRef]
- Sathishkumar, P.; Chae, J.C.; Unnithan, A.R.; Palvannan, T.; Kim, H.Y.; Lee, K.J.; Cho, M.; Kamala-Kannan, S.; Oh, B.T. Laccase-Poly(Lactic-Co-Glycolic Acid) (PLGA) Nanofiber: Highly Stable, Reusable, and Efficacious for the Transformation of Diclofenac. Enzym. Microb. Technol. 2012, 51, 113–118. [Google Scholar] [CrossRef] [PubMed]
- Sathishkumar, P.; Mythili, A.; Hadibarata, T.; Jayakumar, R.; Kanthimathi, M.S.; Palvannan, T.; Ponraj, M.; Salim, M.R.; Mohd Yusoff, A.R. Laccase Mediated Diclofenac Transformation and Cytotoxicity Assessment on Mouse Fibroblast 3T3-L1 Preadipocytes. RSC Adv. 2014, 4, 11689–11697. [Google Scholar] [CrossRef]
- Touahar, I.E.; Haroune, L.; Ba, S.; Bellenger, J.P.; Cabana, H. Characterization of Combined Cross-Linked Enzyme Aggregates from Laccase, Versatile Peroxidase and Glucose Oxidase, and Their Utilization for the Elimination of Pharmaceuticals. Sci. Total Environ. 2014, 481, 90–99. [Google Scholar] [CrossRef] [PubMed]
- Acero, J.L.; Benitez, F.J.; Real, F.J.; Roldan, G. Kinetics of Aqueous Chlorination of Some Pharmaceuticals and Their Elimination from Water Matrices. Water Res. 2010, 44, 4158–4170. [Google Scholar] [CrossRef]
- Quintana, J.B.; Rodil, R.; López-Mahía, P.; Muniategui-Lorenzo, S.; Prada-Rodríguez, D. Investigating the Chlorination of Acidic Pharmaceuticals and By-Product Formation Aided by an Experimental Design Methodology. Water Res. 2010, 44, 243–255. [Google Scholar] [CrossRef]
- Gagnon, C.; Lajeunesse, A.; Cejka, P.; Gagné, F.; Hausler, R. Degradation of Selected Acidic and Neutral Pharmaceutical Products in a Primary-Treated Wastewater by Disinfection Processes. Ozone Sci. Eng. 2008, 30, 387–392. [Google Scholar] [CrossRef]
- Broséus, R.; Vincent, S.; Aboulfadl, K.; Daneshvar, A.; Sauvé, S.; Barbeau, B.; Prévost, M. Ozone Oxidation of Pharmaceuticals, Endocrine Disruptors and Pesticides during Drinking Water Treatment. Water Res. 2009, 43, 4707–4717. [Google Scholar] [CrossRef]
- Guo, Y.; Zhu, S.; Wang, B.; Huang, J.; Deng, S.; Yu, G.; Wang, Y. Modelling of Emerging Contaminant Removal during Heterogeneous Catalytic Ozonation Using Chemical Kinetic Approaches. J. Hazard. Mater. 2019, 380, 120888. [Google Scholar] [CrossRef]
- Elmolla, E.S.; Chaudhuri, M. The Feasibility of Using Combined TiO2 Photocatalysis-SBR Process for Antibiotic Wastewater Treatment. Desalination 2011, 272, 218–224. [Google Scholar] [CrossRef]
- Malato, S.; Giménez, J.; Oller, I.; Agüera, A.; Sánchez Pérez, J.A. Removal and Degradation of Pharmaceutically Active Compounds (PhACs) in Wastewaters by Solar Advanced Oxidation Processes. In The Handbook of Environmental Chemistry; Rodriguez-Mozaz, S., Blánquez Cano, P., Adroguer, M.S., Eds.; Springer: Berlin/Heidelberg, Germany, 2020; Volume 108, pp. 299–326. [Google Scholar]
- Singh, S.; Kumar, V.; Anil, A.G.; Kapoor, D.; Khasnabis, S.; Shekar, S.; Pavithra, N.; Samuel, J.; Subramanian, S.; Singh, J.; et al. Adsorption and Detoxification of Pharmaceutical Compounds from Wastewater Using Nanomaterials: A Review on Mechanism, Kinetics, Valorization and Circular Economy. J. Environ. Manag. 2021, 300, 113569. [Google Scholar] [CrossRef]
- Torres-Pinto, A.; Díez, A.M.; Silva, C.G.; Faria, J.L.; Sanromán, M.Á.; Silva, A.M.T.; Pazos, M. Photoelectrocatalytic Degradation of Pharmaceuticals Promoted by a Metal-Free G-C3N4 Catalyst. Chem. Eng. J. 2023, 476, 146761. [Google Scholar] [CrossRef]
- Liu, R.; Gao, C.; Zhao, Y.G.; Wang, A.; Lu, S.; Wang, M.; Maqbool, F.; Huang, Q. Biological Treatment of Steroidal Drug Industrial Effluent and Electricity Generation in the Microbial Fuel Cells. Bioresour. Technol. 2012, 123, 86–91. [Google Scholar] [CrossRef]
- Li, H.; Zhang, S.; Yang, X.L.; Yang, Y.L.; Xu, H.; Li, X.N.; Song, H.L. Enhanced Degradation of Bisphenol A and Ibuprofen by an Up-Flow Microbial Fuel Cell-Coupled Constructed Wetland and Analysis of Bacterial Community Structure. Chemosphere 2019, 217, 599–608. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Song, H.L.; Cao, X.; Li, H.; Guo, J.; Yang, X.L.; Singh, R.P.; Liu, S. Inhibition of Methanogens Decreased Sulfadiazine Removal and Increased Antibiotic Resistance Gene Development in Microbial Fuel Cells. Bioresour. Technol. 2019, 281, 188–194. [Google Scholar] [CrossRef] [PubMed]
- Ondon, B.S.; Li, S.; Zhou, Q.; Li, F. Simultaneous Removal and High Tolerance of Norfloxacin with Electricity Generation in Microbial Fuel Cell and Its Antibiotic Resistance Genes Quantification. Bioresour. Technol. 2020, 304, 122984. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.; Ge, J.; Gu, F.; Bai, L. Electrochemical Oxidation Technique to Pharmaceutical Pollutants Removal. Chemosphere 2023, 337, 139373. [Google Scholar] [CrossRef]
- Branchet, P.; Arpin-Pont, L.; Piram, A.; Boissery, P.; Wong-Wah-Chung, P.; Doumenq, P. Pharmaceuticals in the Marine Environment: What Are the Present Challenges in Their Monitoring? Sci. Total Environ. 2021, 766, 142644. [Google Scholar] [CrossRef] [PubMed]
- Rathi, B.S.; Kumar, P.S.; Vo, D.V.N. Critical Review on Hazardous Pollutants in Water Environment: Occurrence, Monitoring, Fate, Removal Technologies and Risk Assessment. Sci. Total Environ. 2021, 797, 149134. [Google Scholar] [CrossRef]
- Aminot, Y.; Litrico, X.; Chambolle, M.; Arnaud, C.; Pardon, P.; Budzinski, H. Development and Application of a Multi-Residue Method for the Determination of 53 Pharmaceuticals in Water, Sediment, and Suspended Solids Using Liquid Chromatography-Tandem Mass Spectrometry. Anal. Bioanal. Chem. 2015, 407, 8585–8604. [Google Scholar] [CrossRef]
- Justo-Vega, A.; Jinadasa, K.K.; Jayasinghe, G.D.T.M.; Álvarez-Freire, I.; Bermejo, A.M.; Bermejo-Barrera, P.; Moreda-Piñeiro, A. Ultrasound Assisted Membrane-Assisted Solvent Extraction for the Simultaneous Assessment of Some Drugs Involved in Drug-Facilitated Sexual Assaults by Liquid Chromatography-Tandem Mass Spectrometry. J. Chromatogr. A 2023, 1706, 464284. [Google Scholar] [CrossRef]
- Díaz, A.; Peña-Alvarez, A. A Simple Method for the Simultaneous Determination of Pharmaceuticals and Personal Care Products in River Sediment by Ultrasound-Assisted Extraction Followed by Solid-Phase Microextraction Coupled with Gas Chromatography-Mass Spectrometry. J. Chromatogr. Sci. 2017, 55, 946–953. [Google Scholar] [CrossRef] [PubMed]
- Qin, H.; Liu, H.; Liu, Y.; Di, S.; Bao, Y.; Zhai, Y.; Zhu, S. Recent Advances in Sample Preparation and Chromatographic Analysis of Pharmaceuticals and Personal Care Products in Environment. TrAC—Trends Anal. Chem. 2023, 164, 117112. [Google Scholar] [CrossRef]
- Ramos, S.; Homem, V.; Santos, L. Simultaneous Determination of Synthetic Musks and UV-Filters in Water Matrices by Dispersive Liquid-Liquid Microextraction Followed by Gas Chromatography Tandem Mass-Spectrometry. J. Chromatogr. A 2019, 1590, 47–57. [Google Scholar] [CrossRef]
- Li, Y.; Niu, X.; Yao, C.; Yang, W.; Lu, G. Distribution, Removal, and Risk Assessment of Pharmaceuticals and Their Metabolites in Five Sewage Plants. Int. J. Environ. Res. Public Health 2019, 16, 4729. [Google Scholar] [CrossRef] [PubMed]
- Rice, S.L.; Mitra, S. Microwave-Assisted Solvent Extraction of Solid Matrices and Subsequent Detection of Pharmaceuticals and Personal Care Products (PPCPs) Using Gas Chromatography-Mass Spectrometry. Anal. Chim. Acta 2007, 589, 125–132. [Google Scholar] [CrossRef]
- Trujillo-Rodríguez, M.J.; Nan, H.; Anderson, J.L. Expanding the Use of Polymeric Ionic Liquids in Headspace Solid-Phase Microextraction: Determination of Ultraviolet Filters in Water Samples. J. Chromatogr. A 2018, 1540, 11–20. [Google Scholar] [CrossRef]
- Fawzy, M.G.; Said, M.A. Valuation of Environmental Influence of Recently Invented High-Performance Liquid Chromatographic Method for Hypoglycemic Mixtures of Gliflozins and Metformin in the Presence of Melamine Impurities: Application of Molecular Modeling Simulation Approach. J. Sep. Sci. 2023, 46, e2300267. [Google Scholar] [CrossRef] [PubMed]
- Goeury, K.; Vo Duy, S.; Munoz, G.; Prévost, M.; Sauvé, S. Assessment of Automated Off-Line Solid-Phase Extraction LC-MS/MS to Monitor EPA Priority Endocrine Disruptors in Tap Water, Surface Water, and Wastewater. Talanta 2022, 241, 123216. [Google Scholar] [CrossRef]
- Chen, L.; Yan, X.; Zhou, X.; Peng, P.; Sun, Q.; Zhao, F. Advances in the On-Line Solid-Phase Extraction-Liquid Chromatography-Mass Spectrometry Analysis of Emerging Organic Contaminants. TrAC—Trends Anal. Chem. 2023, 160, 116976. [Google Scholar] [CrossRef]
- Mthiyane, Z.L.; Makhubela, N.; Nyoni, H.; Madikizela, L.M.; Maseko, B.R.; Ncube, S. Determination of Antibiotics during Treatment of Hospital Wastewater Using Automated Solid-Phase Extraction Followed by UHPLC-MS: Occurrence, Removal and Environmental Risks. Environ. Technol. 2023, 1–11. [Google Scholar] [CrossRef]
- Restrepo-Vieira, L.H.; Busetti, F.; Linge, K.L.; Joll, C.A. Development and Validation of a Direct Injection Liquid Chromatography-Tandem Mass Spectrometry Method for the Analysis of Illicit Drugs and Psychopharmaceuticals in Wastewater. J. Chromatogr. A 2022, 1685, 463562. [Google Scholar] [CrossRef]
- Zhi, S.; Zhou, J.; Zhang, Z.; Zhang, K. Determination of 38 Antibiotics in Raw and Treated Wastewater from Swine Farms Using Liquid Chromatography-Mass Spectrometry. J. Sep. Sci. 2022, 45, 1525–1537. [Google Scholar] [CrossRef]
- Hernández, F.; Ibáñez, M.; Bade, R.; Bijlsma, L.; Sancho, J.V. Investigation of Pharmaceuticals and Illicit Drugs in Waters by Liquid Chromatography-High-Resolution Mass Spectrometry. TrAC—Trends Anal. Chem. 2014, 63, 140–157. [Google Scholar] [CrossRef]
- Martín-Pozo, L.; de Alarcón-Gómez, B.; Rodríguez-Gómez, R.; García-Córcoles, M.T.; Çipa, M.; Zafra-Gómez, A. Analytical Methods for the Determination of Emerging Contaminants in Sewage Sludge Samples. A Review. Talanta 2019, 192, 508–533. [Google Scholar] [CrossRef] [PubMed]
- Poole, C.F. Alkylsilyl Derivatives for Gas Chromatography. J. Chromatogr. A 2013, 1296, 2–14. [Google Scholar] [CrossRef] [PubMed]
- Jones, S.J.; Lassiter, M.G. Environmental Toxicology: Aquatic. In Information Resources in Toxicology; Wexler, P., Ed.; Elsevier: Amsterdam, The Netherlands, 2020; pp. 263–278. [Google Scholar]
- Sumudumali, R.G.I.; Jayawardana, J.M.C.K. A Review of Biological Monitoring of Aquatic Ecosystems Approaches: With Special Reference to Macroinvertebrates and Pesticide Pollution. Environ. Manag. 2021, 67, 263–276. [Google Scholar] [CrossRef] [PubMed]
- Adeyemo, O.K.; Sogbanmu, T.O.; Alarape, S.A.; Denslow, N.D. Biomonitoring of Aquatic Pollution: Status and Trends from Genomics to Populations. Proc. Niger. Acad. Sci. 2021, 13, 2s. [Google Scholar] [CrossRef]
- de Mello, K.; Taniwaki, R.H.; Macedo, D.R.; Leal, C.G.; Randhir, T.O. Biomonitoring for Watershed Protection from a Multiscale Land-Use Perspective. Diversity 2023, 15, 636. [Google Scholar] [CrossRef]
- Fabbri, E.; Valbonesi, P.; Moon, T.W. Pharmaceuticals in the Marine Environment: Occurrence, Fate, and Biological Effects. In Contaminants of Emerging Concern in the Marine Environment; León, V.M., Bellas, J., Eds.; Elsevier: Amsterdam, The Netherlands, 2023; pp. 11–71. [Google Scholar]
- Kuehne, L.M.; Dickens, C.; Tickner, D.; Messager, M.L.; Olden, J.D.; O’Brien, G.; Lehner, B.; Eriyagama, N. The Future of Global River Health Monitoring. PLOS Water 2023, 2, e0000101. [Google Scholar] [CrossRef]
- Jeppe, K.J.; Yang, J.; Long, S.M.; Carew, M.E.; Zhang, X.; Pettigrove, V.; Hoffmann, A.A. Detecting Copper Toxicity in Sediments: From the Subindividual Level to the Population Level. J. Appl. Ecol. 2017, 54, 1331–1342. [Google Scholar] [CrossRef]
- King, R.S.; Brain, R.A.; Back, J.A.; Becker, C.; Wright, M.V.; Toteu Djomte, V.; Scott, W.C.; Virgil, S.R.; Brooks, B.W.; Hosmer, A.J.; et al. Effects of Pulsed Atrazine Exposures on Autotrophic Community Structure, Biomass, and Production in Field-Based Stream Mesocosms. Environ. Toxicol. Chem. 2016, 35, 660–675. [Google Scholar] [CrossRef] [PubMed]
- Van Straalen, N.M. Ecotoxicology Becomes Stress Ecology. Environ. Sci. Technol. 2003, 37, 324A–330A. [Google Scholar] [CrossRef] [PubMed]
- Bundschuh, M.; Mesquita-Joanes, F.; Rico, A.; Camacho, A. Understanding Ecological Complexity in a Chemical Stress Context: A Reflection on Recolonization, Recovery, and Adaptation of Aquatic Populations and Communities. Environ. Toxicol. Chem. 2023, 42, 1857–1866. [Google Scholar] [CrossRef] [PubMed]
- Morin, S.; Artigas, J. Twenty Years of Research in Ecosystem Functions in Aquatic Microbial Ecotoxicology. Environ. Toxicol. Chem. 2023, 42, 1867–1888. [Google Scholar] [CrossRef] [PubMed]
- Enns, D.; Cunze, S.; Baker, N.J.; Oehlmann, J.; Jourdan, J. Flushing Away the Future: The Effects of Wastewater Treatment Plants on Aquatic Invertebrates. Water Res. 2023, 243, 120388. [Google Scholar] [CrossRef]
- Mcmahon, T.A.; Halstead, N.T.; Johnson, S.; Raffel, T.R.; Romansic, J.M.; Crumrine, P.W.; Rohr, J.R. Fungicide-Induced Declines of Freshwater Biodiversity Modify Ecosystem Functions and Services. Ecol. Lett. 2012, 15, 714–722. [Google Scholar] [CrossRef]
- Halstead, N.T.; Mcmahon, T.A.; Johnson, S.A.; Raffel, T.R.; Romansic, J.M.; Crumrine, P.W.; Rohr, J.R. Community Ecology Theory Predicts the Effects of Agrochemical Mixtures on Aquatic Biodiversity and Ecosystem Properties. Ecol. Lett. 2014, 17, 932–941. [Google Scholar] [CrossRef]
- Rodrigues, A.C.M.; Machado, A.L.; Bordalo, M.D.; Saro, L.; Simão, F.C.P.; Rocha, R.J.M.; Golovko, O.; Žlábek, V.; Barata, C.; Soares, A.M.V.M.; et al. Invasive Species Mediate Insecticide Effects on Community and Ecosystem Functioning. Environ. Sci. Technol. 2018, 52, 4889–4900. [Google Scholar] [CrossRef]
- De Laender, F.; Rohr, J.R.; Ashauer, R.; Baird, D.J.; Berger, U.; Eisenhauer, N.; Grimm, V.; Hommen, U.; Maltby, L.; Meliàn, C.J.; et al. Reintroducing Environmental Change Drivers in Biodiversity–Ecosystem Functioning Research. Trends Ecol. Evol. 2016, 31, 905–915. [Google Scholar] [CrossRef]
- Hermens, J.L.M.; Ankley, G.T.; Sumpter, J.P. Ecotoxicology—A Multidisciplinary, Problem-Driven Science. Environ. Sci. Technol. 2004, 38, 446A–447A. [Google Scholar] [CrossRef][Green Version]
- Zaghloul, A.; Saber, M.; Gadow, S.; Awad, F. Biological Indicators for Pollution Detection in Terrestrial and Aquatic Ecosystems. Bull. Natl. Res. Cent. 2020, 44, 127. [Google Scholar] [CrossRef]
- Huang, C.W.; Lin, C.; Nguyen, M.K.; Hussain, A.; Bui, X.T.; Ngo, H.H. A Review of Biosensor for Environmental Monitoring: Principle, Application, and Corresponding Achievement of Sustainable Development Goals. Bioengineered 2023, 14, 58–80. [Google Scholar] [CrossRef]
- Kulik, K.; Lenart-Boroń, A.; Wyrzykowska, K. Impact of Antibiotic Pollution on the Bacterial Population within Surface Water with Special Focus on Mountain Rivers. Water 2023, 15, 975. [Google Scholar] [CrossRef]
- Yusuf, A.; O’Flynn, D.; White, B.; Holland, L.; Parle-Mcdermott, A.; Lawler, J.; McCloughlin, T.; Harold, D.; Huerta, B.; Regan, F. Monitoring of Emerging Contaminants of Concern in the Aquatic Environment: A Review of Studies Showing the Application of Effect-Based Measures. Anal. Methods 2021, 13, 5120–5143. [Google Scholar] [CrossRef] [PubMed]
- Arroyo, C.L.; Dionela, M.; Ann Bautista, M.G.; Concepcion, R.; Vicerra, R.R.; Duarte, B. Biomonitoring Technologies Used in Aquatic Ecosystems: A Systematic and Trend Analysis. In Proceedings of the 2022 IEEE 14th International Conference on Humanoid, Nanotechnology, Information Technology, Communication and Control, Environment, and Management, HNICEM 2022, Boracay Island, Philippines, 1–4 December 2022. [Google Scholar]
- Justino, C.I.L.; Duarte, A.C.; Rocha-Santos, T.A.P. Recent Progress in Biosensors for Environmental Monitoring: A Review. Sensors 2017, 17, 2918. [Google Scholar] [CrossRef]
- Daunert, S.; Barrett, G.; Feliciano, J.S.; Shetty, R.S.; Shrestha, S.; Smith-Spencer, W. Genetically Engineered Whole-Cell Sensing Systems: Coupling Biological Recognition with Reporter Genes. Chem. Rev. 2000, 100, 2705–2738. [Google Scholar] [CrossRef] [PubMed]
- Rogers, K.R. Recent Advances in Biosensor Techniques for Environmental Monitoring. Anal. Chim. Acta 2006, 568, 222–231. [Google Scholar] [CrossRef] [PubMed]
- Kaur, H.; Kumar, R.; Babu, J.N.; Mittal, S. Advances in Arsenic Biosensor Development—A Comprehensive Review. Biosens. Bioelectron. 2015, 63, 533–545. [Google Scholar] [CrossRef]
- Rather, R.A.; Ara, S.; Padder, S.A.; Sharma, S.; Pathak, S.P.; Baba, T.R. Seasonal Fluctuation of Water Quality and Ecogenomic Phylogeny of Novel Potential Microbial Pollution Indicators of Veshaw River Kashmir-Western Himalaya. Environ. Pollut. 2023, 320, 121104. [Google Scholar] [CrossRef]
- Machuca-Sepúlveda, J.; Miranda, J.; Lefin, N.; Pedroso, A.; Beltrán, J.F.; Farias, J.G. Current Status of Omics in Biological Quality Elements for Freshwater Biomonitoring. Biology 2023, 12, 923. [Google Scholar] [CrossRef]
- Mielecki, D.; Grzesiuk, E.; Bednarska, A.; Garbicz, D.; Świderska, B.; Grzesiuk, M. Contamination of Aquatic Environment with Anticancer Reagents Influences Daphnia Magna—Ecotoxicogenomics Approach. Ecotoxicol. Environ. Saf. 2023, 249, 114372. [Google Scholar] [CrossRef] [PubMed]
- Pham, K.; Ho, L.; D’Incal, C.P.; De Cock, A.; Berghe, W.V.; Goethals, P. Epigenetic Analytical Approaches in Ecotoxicological Aquatic Research. Environ. Pollut. 2023, 330, 121737. [Google Scholar] [CrossRef] [PubMed]
- Brack, W.; Escher, B.I.; Müller, E.; Schmitt-Jansen, M.; Schulze, T.; Slobodnik, J.; Hollert, H. Towards a Holistic and Solution-Oriented Monitoring of Chemical Status of European Water Bodies: How to Support the EU Strategy for a Non-Toxic Environment? Environ. Sci. Eur. 2018, 30, 33. [Google Scholar] [CrossRef] [PubMed]
- Amorim, C.L.; Alves, M.; Castro, P.M.L.; Henriques, I. Bacterial Community Dynamics within an Aerobic Granular Sludge Reactor Treating Wastewater Loaded with Pharmaceuticals. Ecotoxicol. Environ. Saf. 2018, 147, 905–912. [Google Scholar] [CrossRef] [PubMed]
- Marchlewicz, A.; Domaradzka, D.; Guzik, U.; Wojcieszyńska, D. Bacillus Thuringiensis B1(2015b) Is a Gram-Positive Bacteria Able to Degrade Naproxen and Ibuprofen. Water Air Soil. Pollut. 2016, 227, 197. [Google Scholar] [CrossRef]
- Zhou, N.A.; Lutovsky, A.C.; Andaker, G.L.; Gough, H.L.; Ferguson, J.F. Cultivation and Characterization of Bacterial Isolates Capable of Degrading Pharmaceutical and Personal Care Products for Improved Removal in Activated Sludge Wastewater Treatment. Biodegradation 2013, 24, 813–827. [Google Scholar] [CrossRef]
- Lin, B.; Lyu, J.; Lyu, X.J.; Yu, H.Q.; Hu, Z.; Lam, J.C.W.; Lam, P.K.S. Characterization of Cefalexin Degradation Capabilities of Two Pseudomonas Strains Isolated from Activated Sludge. J. Hazard. Mater. 2015, 282, 158–164. [Google Scholar] [CrossRef]
- Esplugas, M.; González, O.; Sans, C. Bacterial Community Characterization of a Sequencing Batch Reactor Treating Pre-Ozonized Sulfamethoxazole in Water. Environ. Technol. 2013, 34, 1583–1591. [Google Scholar] [CrossRef]
- Sauvêtre, A.; Schröder, P. Uptake of Carbamazepine by Rhizomes and Endophytic Bacteria of Phragmites Australis. Front. Plant Sci. 2015, 6, 83. [Google Scholar] [CrossRef]
- Hirth, N.; Topp, E.; Dörfler, U.; Stupperich, E.; Munch, J.C.; Schroll, R. An Effective Bioremediation Approach for Enhanced Microbial Degradation of the Veterinary Antibiotic Sulfamethazine in an Agricultural Soil. Chem. Biol. Technol. Agric. 2016, 3, 29. [Google Scholar] [CrossRef]
- D’Alessio, M.; Yoneyama, B.; Kirs, M.; Kisand, V.; Ray, C. Pharmaceutically Active Compounds: Their Removal during Slow Sand Filtration and Their Impact on Slow Sand Filtration Bacterial Removal. Sci. Total Environ. 2015, 524–525, 124–135. [Google Scholar] [CrossRef] [PubMed]
- Forrez, I.; Carballa, M.; Boon, N.; Verstraete, W. Biological Removal of 17α-Ethinylestradiol (EE2) in an Aerated Nitrifying Fixed Bed Reactor during Ammonium Starvation. J. Chem. Technol. Biotechnol. 2009, 84, 119–125. [Google Scholar] [CrossRef]
- Pauwels, B.; Wille, K.; Noppe, H.; De Brabander, H.; Van De Wiele, T.; Verstraete, W.; Boon, N. 17α-Ethinylestradiol Cometabolism by Bacteria Degrading Estrone, 17β-Estradiol and Estriol. Biodegradation 2008, 19, 683–693. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Hu, C.; Shen, Y.; Shi, B.; Zhao, D.; Xing, X. Response of Microorganisms in Biofilm to Sulfadiazine and Ciprofloxacin in Drinking Water Distribution Systems. Chemosphere 2019, 218, 197–204. [Google Scholar] [CrossRef] [PubMed]
- Proia, L.; Lupini, G.; Osorio, V.; Pérez, S.; Barceló, D.; Schwartz, T.; Amalfitano, S.; Fazi, S.; Romaní, A.M.; Sabater, S. Response of Biofilm Bacterial Communities to Antibiotic Pollutants in a Mediterranean River. Chemosphere 2013, 92, 1126–1135. [Google Scholar] [CrossRef] [PubMed]
- Rosi-Marshall, E.J.; Kincaid, D.W.; Bechtold, H.A.; Royer, T.V.; Rojas, M.; Kelly, J.J. Pharmaceuticals Suppress Algal Growth and Microbial Respiration and Alter Bacterial Communities in Stream Biofilms. Ecol. Appl. 2013, 23, 583–593. [Google Scholar] [CrossRef]
- Suleiman, M.; Demaria, F.; Zimmardi, C.; Kolvenbach, B.A.; Corvini, P.F.X. Analyzing Microbial Communities and Their Biodegradation of Multiple Pharmaceuticals in Membrane Bioreactors. Appl. Microbiol. Biotechnol. 2023, 107, 5545–5554. [Google Scholar] [CrossRef] [PubMed]
- Leiviskä, T.; Risteelä, S. Analysis of Pharmaceuticals, Hormones and Bacterial Communities in a Municipal Wastewater Treatment Plant—Comparison of Parallel Full-Scale Membrane Bioreactor and Activated Sludge Systems. Environ. Pollut. 2022, 292, 118433. [Google Scholar] [CrossRef]
- Yang, Y.; Li, B.; Zou, S.; Fang, H.H.; Zhang, T. Fate of Antibiotic Resistance Genes in Sewage Treatment Plant Revealed by Metagenomic Approach. Water Res. 2014, 62, 97–106. [Google Scholar] [CrossRef] [PubMed]
- Berendonk, T.U.; Manaia, C.M.; Merlin, C.; Fatta-Kassinos, D.; Cytryn, E.; Walsh, F.; Bürgmann, H.; Sørum, H.; Norström, M.; Pons, M.N.; et al. Tackling Antibiotic Resistance: The Environmental Framework. Nat. Rev. Microbiol. 2015, 13, 310–317. [Google Scholar] [CrossRef]
- Araújo, S.; Silva, I.A.T.; Tacão, M.; Patinha, C.; Alves, A.; Henriques, I. Characterization of Antibiotic Resistant and Pathogenic Escherichia Coli in Irrigation Water and Vegetables in Household Farms. Int. J. Food Microbiol. 2017, 257, 192–200. [Google Scholar] [CrossRef] [PubMed]
- Heitzer, A.; Malachowsky, K.; Thonnard, J.E.; Bienkowski, P.R.; White, D.C.; Sayleri, G.S. Optical Biosensor for Environmental On-Line Monitoring of Naphthalene and Salicylate Bioavailability with an Immobilized Bioluminescent Catabolic Reporter Bacterium. Appl. Environ. Microbiol. 1994, 60, 1487–1494. [Google Scholar] [CrossRef]
- Valtonen, S.J.; Kurittu, J.S.; Karp, M.T. A Luminescent Escherichia Coli Biosensor for the High Throughput Detection of β-Lactams. J. Biomol. Screen. 2002, 7, 127–134. [Google Scholar] [CrossRef]
- Shemer, B.; Belkin, S. Microbial Biosensors for the Detection of Organic Pollutants. In Handbook of Cell Biosensors; Thouand, G., Ed.; Springer International Publishing: Cham, Switzerland, 2019; pp. 1–24. [Google Scholar]
- Lautenschläger, N.; Popp, P.F.; Mascher, T. Development of a Novel Heterologous β-Lactam-Specific Whole-Cell Biosensor in Bacillus Subtilis. J. Biol. Eng. 2020, 14, 21. [Google Scholar] [CrossRef]
- Yin, J.; Zhu, Y.; Liang, Y.; Luo, Y.; Lou, J.; Hu, X.; Meng, Q.; Zhu, T.; Yu, Z. Development of Whole-Cell Biosensors for Screening of Peptidoglycan-Targeting Antibiotics in a Gram-Negative Bacterium. Appl. Environ. Microbiol. 2022, 88, e0084622. [Google Scholar] [CrossRef]
- Yin, J.; Cheng, D.; Zhu, Y.; Liang, Y.; Yu, Z. Development of a Whole-Cell Biosensor for Detection of Antibiotics Targeting Bacterial Cell Envelope in Bacillus Subtilis. Appl. Microbiol. Biotechnol. 2022, 106, 789–798. [Google Scholar] [CrossRef]
- Minamoto, T.; Yamanaka, H.; Takahara, T.; Honjo, M.N.; Kawabata, Z. Surveillance of Fish Species Composition Using Environmental DNA. Limnology 2012, 13, 193–197. [Google Scholar] [CrossRef]
- Wilcox, T.M.; McKelvey, K.S.; Young, M.K.; Jane, S.F.; Lowe, W.H.; Whiteley, A.R.; Schwartz, M.K. Robust Detection of Rare Species Using Environmental DNA: The Importance of Primer Specificity. PLoS ONE 2013, 8, e59520. [Google Scholar] [CrossRef]
- Ardura, A.; Zaiko, A.; Martinez, J.L.; Samulioviene, A.; Semenova, A.; Garcia-Vazquez, E. eDNA and Specific Primers for Early Detection of Invasive Species—A Case Study on the Bivalve Rangia Cuneata, Currently Spreading in Europe. Mar. Environ. Res. 2015, 112, 48–55. [Google Scholar] [CrossRef] [PubMed]
- Roy, M.; Belliveau, V.; Mandrak, N.E.; Gagné, N. Development of Environmental DNA (eDNA) Methods for Detecting High-Risk Freshwater Fishes in Live Trade in Canada. Biol. Invasions 2018, 20, 299–314. [Google Scholar] [CrossRef]
- Stat, M.; Huggett, M.J.; Bernasconi, R.; Dibattista, J.D.; Berry, T.E.; Newman, S.J.; Harvey, E.S.; Bunce, M. Ecosystem Biomonitoring with eDNA: Metabarcoding across the Tree of Life in a Tropical Marine Environment. Sci. Rep. 2017, 7, 12240. [Google Scholar] [CrossRef]
- Djurhuus, A.; Closek, C.J.; Kelly, R.P.; Pitz, K.J.; Michisaki, R.P.; Starks, H.A.; Walz, K.R.; Andruszkiewicz, E.A.; Olesin, E.; Hubbard, K.; et al. Environmental DNA Reveals Seasonal Shifts and Potential Interactions in a Marine Community. Nat. Commun. 2020, 11, 254. [Google Scholar] [CrossRef] [PubMed]
- Lyu, Y.; Xu, X.; Yuan, Y.; Wang, Z.; Hu, J.; Chen, Q.; Sun, W. Antibiotic Profiles and Their Relationships with Multitrophic Aquatic Communities in an Urban River. Sci. Total Environ. 2023, 868, 161678. [Google Scholar] [CrossRef] [PubMed]
- Taberlet, P.; Coissac, E.; Hajibabaei, M.; Rieseberg, L.H. Environmental DNA. Mol. Ecol. 2012, 21, 1789–1793. [Google Scholar] [CrossRef] [PubMed]
- Ruppert, K.M.; Kline, R.J.; Rahman, M.S. Past, Present, and Future Perspectives of Environmental DNA (eDNA) Metabarcoding: A Systematic Review in Methods, Monitoring, and Applications of Global eDNA. Glob. Ecol. Conserv. 2019, 17, e00547. [Google Scholar] [CrossRef]
- Tsuji, S.; Takahara, T.; Doi, H.; Shibata, N.; Yamanaka, H. The Detection of Aquatic Macroorganisms Using Environmental DNA Analysis—A Review of Methods for Collection, Extraction, and Detection. Environ. DNA 2019, 1, 99–108. [Google Scholar] [CrossRef]
- Beng, K.C.; Corlett, R.T. Applications of Environmental DNA (eDNA) in Ecology and Conservation: Opportunities, Challenges and Prospects. Biodivers. Conserv. 2020, 29, 2089–2121. [Google Scholar] [CrossRef]
- Pont, D.; Meulenbroek, P.; Bammer, V.; Dejean, T.; Erős, T.; Jean, P.; Lenhardt, M.; Nagel, C.; Pekarik, L.; Schabuss, M.; et al. Quantitative Monitoring of Diverse Fish Communities on a Large Scale Combining eDNA Metabarcoding and QPCR. Mol. Ecol. Resour. 2023, 23, 396–409. [Google Scholar] [CrossRef]
- Yang, J.; Zhang, L.; Mu, Y.; Wang, J.; Yu, H.; Zhang, X. Unsupervised Biological Integrity Assessment by eDNA Biomonitoring of Multi-Trophic Aquatic Taxa. Environ. Int. 2023, 175, 107950. [Google Scholar] [CrossRef]
- Deiner, K.; Yamanaka, H.; Bernatchez, L. The Future of Biodiversity Monitoring and Conservation Utilizing Environmental DNA. Environ. DNA 2021, 3, 3–7. [Google Scholar] [CrossRef]
- Deiner, K.; Bik, H.M.; Mächler, E.; Seymour, M.; Lacoursière-Roussel, A.; Altermatt, F.; Creer, S.; Bista, I.; Lodge, D.M.; de Vere, N.; et al. Environmental DNA Metabarcoding: Transforming How We Survey Animal and Plant Communities. Mol. Ecol. 2017, 26, 5872–5895. [Google Scholar] [CrossRef] [PubMed]
- Gold, Z.; Sprague, J.; Kushner, D.J.; Marin, E.Z.; Barber, P.H. eDNA Metabarcoding as a Biomonitoring Tool for Marine Protected Areas. PLoS ONE 2021, 16, e0238557. [Google Scholar] [CrossRef]
- McElroy, M.E.; Dressler, T.L.; Titcomb, G.C.; Wilson, E.A.; Deiner, K.; Dudley, T.L.; Eliason, E.J.; Evans, N.T.; Gaines, S.D.; Lafferty, K.D.; et al. Calibrating Environmental DNA Metabarcoding to Conventional Surveys for Measuring Fish Species Richness. Front. Ecol. Evol. 2020, 8, 276. [Google Scholar] [CrossRef]
- Prié, V.; Valentini, A.; Lopes-Lima, M.; Froufe, E.; Rocle, M.; Poulet, N.; Taberlet, P.; Dejean, T. Environmental DNA Metabarcoding for Freshwater Bivalves Biodiversity Assessment: Methods and Results for the Western Palearctic (European Sub-Region). Hydrobiologia 2021, 848, 2931–2950. [Google Scholar] [CrossRef]
- Svenningsen, A.K.N.; Pertoldi, C.; Bruhn, D. eDNA Metabarcoding Benchmarked towards Conventional Survey Methods in Amphibian Monitoring. Animals 2022, 12, 763. [Google Scholar] [CrossRef]
- Rees, H.C.; Maddison, B.C.; Middleditch, D.J.; Patmore, J.R.M.; Gough, K.C. The Detection of Aquatic Animal Species Using Environmental DNA—A Review of eDNA as a Survey Tool in Ecology. J. Appl. Ecol. 2014, 51, 1450–1459. [Google Scholar] [CrossRef]
- Senapati, D.; Bhattacharya, M.; Kar, A.; Chini, D.S.; Das, B.K.; Patra, B.C. Environmental DNA (eDNA): A Promising Biological Survey Tool for Aquatic Species Detection. Proc. Zool. Soc. 2019, 72, 211–228. [Google Scholar] [CrossRef]
- Rey, A.; Carney, K.J.; Quinones, L.E.; Pagenkopp Lohan, K.M.; Ruiz, G.M.; Basurko, O.C.; Rodríguez-Ezpeleta, N. Environmental DNA Metabarcoding: A Promising Tool for Ballast Water Monitoring. Environ. Sci. Technol. 2019, 53, 11849–11859. [Google Scholar] [CrossRef]
- Majaneva, M.; Diserud, O.H.; Eagle, S.H.C.; Boström, E.; Hajibabaei, M.; Ekrem, T. Environmental DNA Filtration Techniques Affect Recovered Biodiversity. Sci. Rep. 2018, 8, 4682. [Google Scholar] [CrossRef]
- Takasaki, K.; Aihara, H.; Imanaka, T.; Matsudaira, T.; Tsukahara, K.; Usui, A.; Osaki, S.; Doi, H. Water Pre-Filtration Methods to Improve Environmental DNA Detection by Real-Time PCR and Metabarcoding. PLoS ONE 2021, 16, e0250162. [Google Scholar] [CrossRef]
- Alexander, J.B.; Bunce, M.; White, N.; Wilkinson, S.P.; Adam, A.A.S.; Berry, T.; Stat, M.; Thomas, L.; Newman, S.J.; Dugal, L.; et al. Development of a Multi-Assay Approach for Monitoring Coral Diversity Using eDNA Metabarcoding. Coral Reefs 2020, 39, 159–171. [Google Scholar] [CrossRef]
- Sepulveda, A.J.; Birch, J.M.; Barnhart, E.P.; Merkes, C.M.; Yamahara, K.M.; Marin, R.; Kinsey, S.M.; Wright, P.R.; Schmidt, C. Robotic Environmental DNA Bio-Surveillance of Freshwater Health. Sci. Rep. 2020, 10, 14389. [Google Scholar] [CrossRef]
- Hendricks, A.; Mackie, C.M.; Luy, E.; Sonnichsen, C.; Smith, J.; Grundke, I.; Tavasoli, M.; Furlong, A.; Beiko, R.G.; LaRoche, J.; et al. Compact and Automated eDNA Sampler for in Situ Monitoring of Marine Environments. Sci. Rep. 2023, 13, 5210. [Google Scholar] [CrossRef]
- Xiong, W.; Huang, X.; Chen, Y.; Fu, R.; Du, X.; Chen, X.; Zhan, A. Zooplankton Biodiversity Monitoring in Polluted Freshwater Ecosystems: A Technical Review. Environ. Sci. Ecotechnology 2020, 1, 100008. [Google Scholar] [CrossRef]
- Lawson Handley, L. How Will the “molecular Revolution” Contribute to Biological Recording? Biol. J. Linn. Soc. 2015, 115, 750–766. [Google Scholar] [CrossRef]
- Gold, Z.; Wall, A.R.; Schweizer, T.M.; Pentcheff, N.D.; Curd, E.E.; Barber, P.H.; Meyer, R.S.; Wayne, R.; Stolzenbach, K.; Prickett, K.; et al. A Manager’s Guide to Using eDNA Metabarcoding in Marine Ecosystems. PeerJ 2022, 10, e14071. [Google Scholar] [CrossRef]
- García-Machado, E.; Normandeau, E.; Côté, G.; Bernatchez, L. How eDNA Data Filtration, Sequence Coverage, and Primer Selection Influence Assessment of Fish Communities in Northern Temperate Lakes. Environ. DNA 2023, 00, 1–18. [Google Scholar] [CrossRef]
- Wang, Z.; Liu, X.; Liang, D.; Wang, Q.; Zhang, L.; Zhang, P. VertU: Universal Multilocus Primer Sets for eDNA Metabarcoding of Vertebrate Diversity, Evaluated by Both Artificial and Natural Cases. Front. Ecol. Evol. 2023, 11, 1164206. [Google Scholar] [CrossRef]
- Weigand, H.; Beermann, A.J.; Čiampor, F.; Costa, F.O.; Csabai, Z.; Duarte, S.; Geiger, M.F.; Grabowski, M.; Rimet, F.; Rulik, B.; et al. DNA Barcode Reference Libraries for the Monitoring of Aquatic Biota in Europe: Gap-Analysis and Recommendations for Future Work. Sci. Total Environ. 2019, 678, 499–524. [Google Scholar] [CrossRef]
- Abbott, C.; Coulson, M.; Gagné, N.; Lacoursière-Roussel, A.; Parent, G.J.; Bajno, R.; Dietrich, C.; May-Mcnally, S. Guidance on the Use of Targeted Environmental DNA (eDNA) Analysis for the Management of Aquatic Invasive Species and Species at Risk; Canadian Science Advisory Secretariat (CSAS): Ottawa, ON, Canada, 2021.
- Huszarik, M.; Röder, N.; Eberhardt, L.; Kennedy, S.; Krehenwinkel, H.; Schwenk, K.; Entling, M.H. External DNA Contamination and Efficiency of Bleach Decontamination for Arthropod Diet Analysis. Environ. DNA 2023, 5, 540–550. [Google Scholar] [CrossRef]
- Beentjes, K.K.; Speksnijder, A.G.C.L.; Schilthuizen, M.; Hoogeveen, M.; Van Der Hoorn, B.B. The Effects of Spatial and Temporal Replicate Sampling on eDNA Metabarcoding. PeerJ 2019, 2019, e7335. [Google Scholar] [CrossRef] [PubMed]
- Troth, C.R.; Sweet, M.J.; Nightingale, J.; Burian, A. Seasonality, DNA Degradation and Spatial Heterogeneity as Drivers of eDNA Detection Dynamics. Sci. Total Environ. 2021, 768, 144466. [Google Scholar] [CrossRef] [PubMed]
- Wee, A.K.S.; Salmo, S.G.; Sivakumar, K.; Then, A.Y.H.; Basyuni, M.; Fall, J.; Habib, K.A.; Isowa, Y.; Leopardas, V.; Peer, N.; et al. Prospects and Challenges of Environmental DNA (eDNA) Metabarcoding in Mangrove Restoration in Southeast Asia. Front. Mar. Sci. 2023, 10, 1033258. [Google Scholar] [CrossRef]
- Spens, J.; Evans, A.R.; Halfmaerten, D.; Knudsen, S.W.; Sengupta, M.E.; Mak, S.S.T.; Sigsgaard, E.E.; Hellström, M. Comparison of Capture and Storage Methods for Aqueous Macrobial eDNA Using an Optimized Extraction Protocol: Advantage of Enclosed Filter. Methods Ecol. Evol. 2017, 8, 635–645. [Google Scholar] [CrossRef]
- Jarman, S.N.; Berry, O.; Bunce, M. The Value of Environmental DNA Biobanking for Long-Term Biomonitoring. Nat. Ecol. Evol. 2018, 2, 1192–1193. [Google Scholar] [CrossRef]
- Xing, Y.; Gao, W.; Shen, Z.; Zhang, Y.; Bai, J.; Cai, X.; Ouyang, J.; Zhao, Y. A Review of Environmental DNA Field and Laboratory Protocols Applied in Fish Ecology and Environmental Health. Front. Environ. Sci. 2022, 10, 725360. [Google Scholar] [CrossRef]
- Evans, N.T.; Lamberti, G.A. Freshwater Fisheries Assessment Using Environmental DNA: A Primer on the Method, Its Potential, and Shortcomings as a Conservation Tool. Fish. Res. 2018, 197, 60–66. [Google Scholar] [CrossRef]
- Pascher, K.; Švara, V.; Jungmeier, M. Environmental DNA-Based Methods in Biodiversity Monitoring of Protected Areas: Application Range, Limitations, and Needs. Diversity 2022, 14, 463. [Google Scholar] [CrossRef]
- Fonseca, V.G.; Davison, P.I.; Creach, V.; Stone, D.; Bass, D.; Tidbury, H.J. The Application of eDNA for Monitoring Aquatic Non-Indigenous Species: Practical and Policy Considerations. Diversity 2023, 15, 631. [Google Scholar] [CrossRef]
- Valentini, A.; Taberlet, P.; Miaud, C.; Civade, R.; Herder, J.; Thomsen, P.F.; Bellemain, E.; Besnard, A.; Coissac, E.; Boyer, F.; et al. Next-Generation Monitoring of Aquatic Biodiversity Using Environmental DNA Metabarcoding. Mol. Ecol. 2016, 25, 929–942. [Google Scholar] [CrossRef]
- Rishan, S.T.; Kline, R.J.; Rahman, M.S. Applications of Environmental DNA (eDNA) to Detect Subterranean and Aquatic Invasive Species: A Critical Review on the Challenges and Limitations of eDNA Metabarcoding. Environ. Adv. 2023, 12, 100370. [Google Scholar] [CrossRef]
- Macher, T.-H.; Schütz, R.; Yildiz, A.; Beermann, A.J.; Leese, F. Evaluating Five Primer Pairs for Environmental DNA Metabarcoding of Central European Fish Species Based on Mock Communities. Metabarcoding Metagenom 2023, 7, e103856. [Google Scholar] [CrossRef]
- Bálint, M.; Nowak, C.; Márton, O.; Pauls, S.U.; Wittwer, C.; Aramayo, J.L.; Schulze, A.; Chambert, T.; Cocchiararo, B.; Jansen, M. Accuracy, Limitations and Cost Efficiency of eDNA-Based Community Survey in Tropical Frogs. Mol. Ecol. Resour. 2018, 18, 1415–1426. [Google Scholar] [CrossRef] [PubMed]
- Bani, A.; Randall, K.C.; Clark, D.R.; Gregson, B.H.; Henderson, D.K.; Losty, E.C.; Ferguson, R.M.W. Mind the Gaps: What Do We Know about How Multiple Chemical Stressors Impact Freshwater Aquatic Microbiomes? In Advances in Ecological Research; Academic Press Inc.: Cambridge, MA, USA, 2022; Volume 67, pp. 331–377. ISBN 9780323985932. [Google Scholar]
- Hansen, B.K.; Bekkevold, D.; Clausen, L.W.; Nielsen, E.E. The Sceptical Optimist: Challenges and Perspectives for the Application of Environmental DNA in Marine Fisheries. Fish. Fish. 2018, 19, 751–768. [Google Scholar] [CrossRef]
- Harper, L.R.; Buxton, A.S.; Rees, H.C.; Bruce, K.; Brys, R.; Halfmaerten, D.; Read, D.S.; Watson, H.V.; Sayer, C.D.; Jones, E.P.; et al. Prospects and Challenges of Environmental DNA (eDNA) Monitoring in Freshwater Ponds. Hydrobiologia 2019, 826, 25–41. [Google Scholar] [CrossRef]
- Hunter, M.E.; Ferrante, J.A.; Meigs-Friend, G.; Ulmer, A. Improving eDNA Yield and Inhibitor Reduction through Increased Water Volumes and Multi-Filter Isolation Techniques. Sci. Rep. 2019, 9, 5259. [Google Scholar] [CrossRef]
- Bernos, T.A.; Yates, M.C.; Docker, M.F.; Fitzgerald, A.; Hanner, R.; Heath, D.; Imrit, A.; Livernois, J.; Myler, E.; Patel, K.; et al. Environmental DNA (eDNA) Applications in Freshwater Fisheries Management and Conservation in Canada: Overview of Current Challenges and Opportunities. Can. J. Fish. Aquat. Sci. 2023, 80, 1170–1186. [Google Scholar] [CrossRef]
- Capurso, G.; Carroll, B.; Stewart, K.A. Transforming Marine Monitoring: Using eDNA Metabarcoding to Improve the Monitoring of the Mediterranean Marine Protected Areas Network. Mar. Policy 2023, 156, 105807. [Google Scholar] [CrossRef]
- Frontalini, F.; Greco, M.; Di Bella, L.; Lejzerowicz, F.; Reo, E.; Caruso, A.; Cosentino, C.; Maccotta, A.; Scopelliti, G.; Nardelli, M.P.; et al. Assessing the Effect of Mercury Pollution on Cultured Benthic Foraminifera Community Using Morphological and eDNA Metabarcoding Approaches. Mar. Pollut. Bull. 2018, 129, 512–524. [Google Scholar] [CrossRef] [PubMed]
- Chang, J. Monitoring Biodiversity and Water Pollution via High-Throughput eDNA Metabarcoding. Berkeley Sci. J. 2020, 24, 50–58. [Google Scholar] [CrossRef]
- Lee, V.M. Validation of eDNA Metabarcoding: A Comparison to Traditional Survey Methods in Ozark Streams. Master’s Thesis, Missouri University of Science and Technology, Rolla, MI, USA, 2022. [Google Scholar]
- Miya, M. Environmental DNA Metabarcoding: A Novel Method for Biodiversity Monitoring of Marine Fish Communities. Annu. Rev. Mar. Sci. 2021, 14, 161–185. [Google Scholar] [CrossRef] [PubMed]
- Kelly, R.P.; Lodge, D.M.; Lee, K.N.; Theroux, S.; Sepulveda, A.J.; Scholin, C.A.; Craine, J.M.; Andruszkiewicz Allan, E.; Nichols, K.M.; Parsons, K.M.; et al. Toward a National eDNA Strategy for the United States. Environ. DNA 2023, 1–10. [Google Scholar] [CrossRef]
- Ebenstein, A. The Consequences of Industrialization: Evidence from Water Pollution and Digestive Cancers in China. Rev. Econ. Stat. 2012, 94, 186–201. [Google Scholar] [CrossRef]
- Chaudhry, F.N.; Malik, M.F. Factors Affecting Water Pollution: A Review. J. Ecosyst. Ecography 2017, 7, 225. [Google Scholar] [CrossRef]
- Schmidt, J.R.; Shaskus, M.; Estenik, J.F.; Oesch, C.; Khidekel, R.; Boyer, G.L. Variations in the Microcystin Content of Different Fish Species Collected from a Eutrophic Lake. Toxins 2013, 5, 992–1009. [Google Scholar] [CrossRef] [PubMed]
- Ebele, A.J.; Abou-Elwafa Abdallah, M.; Harrad, S. Pharmaceuticals and Personal Care Products (PPCPs) in the Freshwater Aquatic Environment. Emerg. Contam. 2017, 3, 1–16. [Google Scholar] [CrossRef]
- Brix, K.V.; Blust, R.; Mertens, J.; Baken, S.; Middleton, E.T.; Cooper, C. Evaluation of Effects-Based Methods as Monitoring Tools for Assessing Ecological Impacts of Metals in Aquatic Ecosystems. Integr. Environ. Assess. Manag. 2023, 19, 24–31. [Google Scholar] [CrossRef]
- Chonova, T.; Kurmayer, R.; Rimet, F.; Labanowski, J.; Vasselon, V.; Keck, F.; Illmer, P.; Bouchez, A. Benthic Diatom Communities in an Alpine River Impacted by Waste Water Treatment Effluents as Revealed Using DNA Metabarcoding. Front. Microbiol. 2019, 10, 653. [Google Scholar] [CrossRef]
- Kajtar, A. Environmental DNA Plumes: Linking Fish Farm eDNA to Microbial Communities and Novel Detection of Transgenic eDNA. Master’s Thesis, Great Lakes Institute for Environmental Research, Windsor, ON, Canada, 2021. [Google Scholar]
- Shi, P.; Jia, S.; Zhang, X.X.; Zhang, T.; Cheng, S.; Li, A. Metagenomic Insights into Chlorination Effects on Microbial Antibiotic Resistance in Drinking Water. Water Res. 2013, 47, 111–120. [Google Scholar] [CrossRef]
- Pinto, I.; Simões, M.; Gomes, I.B. An Overview of the Impact of Pharmaceuticals on Aquatic Microbial Communities. Antibiotics 2022, 11, 1700. [Google Scholar] [CrossRef]
- Ebert, I.; Bachmann, J.; Kühnen, U.; Küster, A.; Kussatz, C.; Maletzki, D.; Schlüter, C. Toxicity of the Fluoroquinolone Antibiotics Enrofloxacin and Ciprofloxacin to Photoautotrophic Aquatic Organisms. Environ. Toxicol. Chem. 2011, 30, 2786–2792. [Google Scholar] [CrossRef] [PubMed]
- Sehnal, L.; Brammer-Robbins, E.; Wormington, A.M.; Blaha, L.; Bisesi, J.; Larkin, I.; Martyniuk, C.J.; Simonin, M.; Adamovsky, O. Microbiome Composition and Function in Aquatic Vertebrates: Small Organisms Making Big Impacts on Aquatic Animal Health. Front. Microbiol. 2021, 12, 567408. [Google Scholar] [CrossRef] [PubMed]
- Binh, V.N.; Dang, N.; Anh, N.T.K.; Ky, L.X.; Thai, P.K. Antibiotics in the Aquatic Environment of Vietnam: Sources, Concentrations, Risk and Control Strategy. Chemosphere 2018, 197, 438–450. [Google Scholar] [CrossRef]
- Välitalo, P.; Kruglova, A.; Mikola, A.; Vahala, R. Toxicological Impacts of Antibiotics on Aquatic Micro-Organisms: A Mini-Review. Int. J. Hyg. Environ. Health 2017, 220, 558–569. [Google Scholar] [CrossRef]
- Xie, Y.; Wang, J.; Yang, J.; Giesy, J.P.; Yu, H.; Zhang, X. Environmental DNA Metabarcoding Reveals Primary Chemical Contaminants in Freshwater Sediments from Different Land-Use Types. Chemosphere 2017, 172, 201–209. [Google Scholar] [CrossRef] [PubMed]
- Stoeck, T.; Pan, H.; Dully, V.; Forster, D.; Jung, T. Towards an eDNA Metabarcode-Based Performance Indicator for Full-Scale Municipal Wastewater Treatment Plants. Water Res. 2018, 144, 322–331. [Google Scholar] [CrossRef]
- Cruz, A.F.; Wijesekara, R.G.S.; Jinadasa, K.B.S.N.; Gonzales, B.J.; Ohura, T.; Guruge, K.S. Preliminary Investigation of Microbial Community in Wastewater and Surface Waters in Sri Lanka and the Philippines. Front. Water 2021, 3, 730124. [Google Scholar] [CrossRef]
- Romero, P.E.; Calla-Quispe, E.; Castillo-Vilcahuaman, C.; Yokoo, M.; Fuentes-Rivera, H.L.; Ramirez, J.L.; Ampuero, A.; Ibáñez, A.J.; Wong, P. From the Andes to the Desert: 16S RRNA Metabarcoding Characterization of Aquatic Bacterial Communities in the Rimac River, the Main Source of Water for Lima, Peru. PLoS ONE 2021, 16, e0250401. [Google Scholar] [CrossRef]
- Bouchali, R.; Marjolet, L.; Mondamert, L.; Chonova, T.; Ribun, S.; Laurent, E.; Bouchez, A.; Labanowski, J.; Cournoyer, B. Evidence of Bacterial Community Coalescence between Freshwater and Discharged Tpm-Harboring Bacterial Taxa from Hospital and Domestic Wastewater Treatment Plants among Epilithic Biofilms. Microorganisms 2023, 11, 922. [Google Scholar] [CrossRef] [PubMed]
- Pathak, V.M.; Verma, V.K.; Rawat, B.S.; Kaur, B.; Babu, N.; Sharma, A.; Dewali, S.; Yadav, M.; Kumari, R.; Singh, S.; et al. Current Status of Pesticide Effects on Environment, Human Health and It’s Eco-Friendly Management as Bioremediation: A Comprehensive Review. Front. Microbiol. 2022, 13, 962619. [Google Scholar] [CrossRef]
- Brain, R.A.; Hanson, M.L.; Solomon, K.R.; Brooks, B.W. Aquatic Plants Exposed to Pharmaceuticals: Effects and Risks. In Reviews of Environmental Contamination and Toxicology; Springer: New York, NY, USA, 2008; Volume 192, pp. 67–115. [Google Scholar]
- Pawlowski, J.; Kelly-Quinn, M.; Altermatt, F.; Apothéloz-Perret-Gentil, L.; Beja, P.; Boggero, A.; Borja, A.; Bouchez, A.; Cordier, T.; Domaizon, I.; et al. The Future of Biotic Indices in the Ecogenomic Era: Integrating (e)DNA Metabarcoding in Biological Assessment of Aquatic Ecosystems. Sci. Total Environ. 2018, 637–638, 1295–1310. [Google Scholar] [CrossRef] [PubMed]
- Ji, F.; Yan, L.; Yan, S.; Qin, T.; Shen, J.; Zha, J. Estimating Aquatic Plant Diversity and Distribution in Rivers from Jingjinji Region, China, Using Environmental DNA Metabarcoding and a Traditional Survey Method. Environ. Res. 2021, 199, 111348. [Google Scholar] [CrossRef]
- Liu, Q.; Zhang, Y.; Wu, H.; Liu, F.; Peng, W.; Zhang, X.; Chang, F.; Xie, P.; Zhang, H. A Review and Perspective of Edna Application to Eutrophication and Hab Control in Freshwater and Marine Ecosystems. Microorganisms 2020, 8, 417. [Google Scholar] [CrossRef] [PubMed]
- Park, J.-W.; Park, K.; Kwak, I.-S. Surveillance Spilled Chironomidae (Diptera) Larvae from Drinking Water Treatment Plants in South Korea Using Morphogenetic Species Analysis and eDNA Metabarcoding. Sci. Total Environ. 2023, 896, 165241. [Google Scholar] [CrossRef] [PubMed]
- Pilliod, D.S.; Goldberg, C.S.; Arkle, R.S.; Waits, L.P. Factors Influencing Detection of eDNA from a Stream-Dwelling Amphibian. Mol. Ecol. Resour. 2014, 14, 109–116. [Google Scholar] [CrossRef] [PubMed]
- Uchida, N.; Kubota, K.; Aita, S.; Kazama, S. Aquatic Insect Community Structure Revealed by eDNA Metabarcoding Derives Indices for Environmental Assessment. PeerJ 2020, 2020, e9176. [Google Scholar] [CrossRef]
- Xie, R.; Zhao, G.; Yang, J.; Wang, Z.; Xu, Y.; Zhang, X.; Wang, Z. eDNA Metabarcoding Revealed Differential Structures of Aquatic Communities in a Dynamic Freshwater Ecosystem Shaped by Habitat Heterogeneity. Environ. Res. 2021, 201, 111602. [Google Scholar] [CrossRef]
- Pinna, M.; Zangaro, F.; Saccomanno, B.; Scalone, C.; Bozzeda, F.; Fanini, L.; Specchia, V. An Overview of Ecological Indicators of Fish to Evaluate the Anthropogenic Pressures in Aquatic Ecosystems: From Traditional to Innovative DNA-Based Approaches. Water 2023, 15, 949. [Google Scholar] [CrossRef]
- Yao, M.; Zhang, S.; Lu, Q.; Chen, X.; Zhang, S.Y.; Kong, Y.; Zhao, J. Fishing for Fish Environmental DNA: Ecological Applications, Methodological Considerations, Surveying Designs, and Ways Forward. Mol. Ecol. 2022, 31, 5132–5164. [Google Scholar] [CrossRef]
- Davison, H.; Macadam, C.R.; Smith, D. Pharmaceuticals in Freshwater Environments and Their Potential. Effects on Freshwater Invertebrates Saving the Small Things That Run the Planet; Buglife: Peterborough, UK, 2021. [Google Scholar]
- Biswas, C.; Maity, S.; Adhikari, M.; Chatterjee, A.; Guchhait, R.; Pramanick, K. Pharmaceuticals in the Aquatic Environment and Their Endocrine Disruptive Effects in Fish. Proc. Zool. Soc. 2021, 74, 507–522. [Google Scholar] [CrossRef]
- Khan, A.H.A.; Barros, R. Pharmaceuticals in Water: Risks to Aquatic Life and Remediation Strategies. Hydrobiology 2023, 2, 395–409. [Google Scholar] [CrossRef]
- Kitamura, R.S.A.; Vicentini, M.; Perussolo, M.C.; Lirola, J.R.; Cirilo dos Santos, C.F.; Moreira Brito, J.C.; Cestari, M.M.; Prodocimo, M.M.; Gomes, M.P.; Silva de Assis, H.C. Sublethal Biochemical, Histopathological and Genotoxicological Effects of Short-Term Exposure to Ciprofloxacin in Catfish Rhamdia Quelen. Environ. Pollut. 2022, 300, 118935. [Google Scholar] [CrossRef]
- Srivastava, B.; Reddy, P.B. Impacts of Human Pharmaceuticals on Fish Health. Int. J. Pharm. Sci. Res. 2021, 12, 5185. [Google Scholar] [CrossRef]
- Brodin, T.; Fick, J.; Jonsson, M.; Klaminder, J. Dilute Concentrations of a Psychiatric Drug Alter Behavior of Fish from Natural Populations. Science (1979) 2013, 339, 814–815. [Google Scholar] [CrossRef]
- Hubená, P.; Horký, P.; Grabic, R.; Grabicová, K.; Douda, K.; Slavík, O.; Randák, T. Prescribed Aggression of Fishes: Pharmaceuticals Modify Aggression in Environmentally Relevant Concentrations. Ecotoxicol. Environ. Saf. 2021, 227, 112944. [Google Scholar] [CrossRef]
- Mennigen, J.A.; Stroud, P.; Zamora, J.M.; Moon, T.W.; Trudeau, V.L. Pharmaceuticals as Neuroendocrine Disruptors: Lessons Learned from Fish on Prozac. J. Toxicol. Environ. Health B Crit. Rev. 2011, 14, 387–412. [Google Scholar] [CrossRef]
- Jenila, J.S.; Issac, P.K.; Lam, S.S.; Oviya, J.C.; Jones, S.; Munusamy-Ramanujam, G.; Chang, S.W.; Ravindran, B.; Mannacharaju, M.; Ghotekar, S.; et al. Deleterious Effect of Gestagens from Wastewater Effluent on Fish Reproduction in Aquatic Environment: A Review. Environ. Res. 2023, 236, 116810. [Google Scholar] [CrossRef]
- Du, B.; Haddad, S.P.; Luek, A.; Scott, W.C.; Saari, G.N.; Kristofco, L.A.; Connors, K.A.; Rash, C.; Rasmussen, J.B.; Chambliss, C.K.; et al. Bioaccumulation and Trophic Dilution of Human Pharmaceuticals across Trophic Positions of an Effluent-Dependent Wadeable Stream. Philos. Trans. R. Soc. B Biol. Sci. 2014, 369, 20140058. [Google Scholar] [CrossRef]
- Xie, Z.; Lu, G.; Yan, Z.; Liu, J.; Wang, P.; Wang, Y. Bioaccumulation and Trophic Transfer of Pharmaceuticals in Food Webs from a Large Freshwater Lake. Environ. Pollut. 2017, 222, 356–366. [Google Scholar] [CrossRef]
- Yang, H.; Lu, G.; Yan, Z.; Liu, J.; Dong, H.; Bao, X.; Zhang, X.; Sun, Y. Residues, Bioaccumulation, and Trophic Transfer of Pharmaceuticals and Personal Care Products in Highly Urbanized Rivers Affected by Water Diversion. J. Hazard. Mater. 2020, 391, 122245. [Google Scholar] [CrossRef]
- Seymour, M.; Edwards, F.K.; Cosby, B.J.; Kelly, M.G.; de Bruyn, M.; Carvalho, G.R.; Creer, S. Executing Multi-Taxa eDNA Ecological Assessment via Traditional Metrics and Interactive Networks. Sci. Total Environ. 2020, 729, 138801. [Google Scholar] [CrossRef]
- Zhang, X.; Xia, P.; Wang, P.; Yang, J.; Baird, D.J. Omics Advances in Ecotoxicology. Environ. Sci. Technol. 2018, 52, 3842–3851. [Google Scholar] [CrossRef]
- Anagnostopoulos, D.A.; Parlapani, F.F.; Natoudi, S.; Syropoulou, F.; Kyritsi, M.; Vergos, I.; Hadjichristodoulou, C.; Kagalou, I.; Boziaris, I.S. Bacterial Communities and Antibiotic Resistance of Potential Pathogens Involved in Food Safety and Public Health in Fish and Water of Lake Karla, Thessaly, Greece. Pathogens 2022, 11, 1473. [Google Scholar] [CrossRef]
- Li, Y.; Chen, X.; Wang, X.; Shang, J.; Niu, L.; Wang, L.; Zhang, H.; Zhang, W. The Effects of Paroxetine on Benthic Microbial Food Web and Nitrogen Transformation in River Sediments. Int. J. Environ. Res. Public Health 2022, 19, 14602. [Google Scholar] [CrossRef]
- Bessey, C.; Neil Jarman, S.; Simpson, T.; Miller, H.; Stewart, T.; Kenneth Keesing, J.; Berry, O. Passive eDNA Collection Enhances Aquatic Biodiversity Analysis. Commun. Biol. 2021, 4, 236. [Google Scholar] [CrossRef]
- Traugott, M.; Thalinger, B.; Wallinger, C.; Sint, D. Fish as Predators and Prey: DNA-Based Assessment of Their Role in Food Webs. J. Fish. Biol. 2021, 98, 367–382. [Google Scholar] [CrossRef]
- Berry, T.E.; Osterrieder, S.K.; Murray, D.C.; Coghlan, M.L.; Richardson, A.J.; Grealy, A.K.; Stat, M.; Bejder, L.; Bunce, M. DNA Metabarcoding for Diet Analysis and Biodiversity: A Case Study Using the Endangered Australian Sea Lion (Neophoca cinerea). Ecol. Evol. 2017, 7, 5435–5453. [Google Scholar] [CrossRef]
- D’Alessandro, S.; Mariani, S. Sifting Environmental DNA Metabarcoding Data Sets for Rapid Reconstruction of Marine Food Webs. Fish. Fish. 2021, 22, 822–833. [Google Scholar] [CrossRef]
- Naselli-Flores, L.; Padisák, J. Ecosystem Services Provided by Marine and Freshwater Phytoplankton. Hydrobiologia 2023, 850, 2691–2706. [Google Scholar] [CrossRef]
- Bergeron, S.; Boopathy, R.; Nathaniel, R.; Corbin, A.; LaFleur, G. Presence of Antibiotic Resistant Bacteria and Antibiotic Resistance Genes in Raw Source Water and Treated Drinking Water. Int. Biodeterior. Biodegrad. 2015, 102, 370–374. [Google Scholar] [CrossRef]
- Li, J.; Cheng, W.; Xu, L.; Strong, P.J.; Chen, H. Antibiotic-Resistant Genes and Antibiotic-Resistant Bacteria in the Effluent of Urban Residential Areas, Hospitals, and a Municipal Wastewater Treatment Plant System. Environ. Sci. Pollut. Res. 2015, 22, 4587–4596. [Google Scholar] [CrossRef] [PubMed]
- Pallares-Vega, R.; Blaak, H.; van der Plaats, R.; de Roda Husman, A.M.; Hernandez Leal, L.; van Loosdrecht, M.C.M.; Weissbrodt, D.G.; Schmitt, H. Determinants of Presence and Removal of Antibiotic Resistance Genes during WWTP Treatment: A Cross-Sectional Study. Water Res. 2019, 161, 319–328. [Google Scholar] [CrossRef] [PubMed]
- Xu, L.; Ouyang, W.; Qian, Y.; Su, C.; Su, J.; Chen, H. High-Throughput Profiling of Antibiotic Resistance Genes in Drinking Water Treatment Plants and Distribution Systems. Environ. Pollut. 2016, 213, 119–126. [Google Scholar] [CrossRef]
- Szekeres, E.; Chiriac, C.M.; Baricz, A.; Szőke-Nagy, T.; Lung, I.; Soran, M.L.; Rudi, K.; Dragos, N.; Coman, C. Investigating Antibiotics, Antibiotic Resistance Genes, and Microbial Contaminants in Groundwater in Relation to the Proximity of Urban Areas. Environ. Pollut. 2018, 236, 734–744. [Google Scholar] [CrossRef]
- Wang, Z.; Chen, Q.; Zhang, J.; Guan, T.; Chen, Y.; Shi, W. Critical Roles of Cyanobacteria as Reservoir and Source for Antibiotic Resistance Genes. Environ. Int. 2020, 144, 106034. [Google Scholar] [CrossRef]
- Jiang, L.; Hu, X.; Xu, T.; Zhang, H.; Sheng, D.; Yin, D. Prevalence of Antibiotic Resistance Genes and Their Relationship with Antibiotics in the Huangpu River and the Drinking Water Sources, Shanghai, China. Sci. Total Environ. 2013, 458–460, 267–272. [Google Scholar] [CrossRef]
- Zheng, J.; Gao, R.; Wei, Y.; Chen, T.; Fan, J.; Zhou, Z.; Makimilua, T.B.; Jiao, Y.; Chen, H. High-Throughput Profiling and Analysis of Antibiotic Resistance Genes in East Tiaoxi River, China. Environ. Pollut. 2017, 230, 648–654. [Google Scholar] [CrossRef] [PubMed]
- Zhou, M.; Li, Q.; Yu, S.; Han, H.; Osborne, N.J. Co-Proliferation of Antimicrobial Resistance Genes in Tilapia Farming Ponds Associated with Use of Antimicrobials. Sci. Total Environ. 2023, 887, 164046. [Google Scholar] [CrossRef] [PubMed]
- Saxena, G.; Marzinelli, E.M.; Naing, N.N.; He, Z.; Liang, Y.; Tom, L.; Mitra, S.; Ping, H.; Joshi, U.M.; Reuben, S.; et al. Ecogenomics Reveals Metals and Land-Use Pressures on Microbial Communities in the Waterways of a Megacity. Environ. Sci. Technol. 2015, 49, 1462–1471. [Google Scholar] [CrossRef]
- Cavicchioli, R.; Ripple, W.J.; Timmis, K.N.; Azam, F.; Bakken, L.R.; Baylis, M.; Behrenfeld, M.J.; Boetius, A.; Boyd, P.W.; Classen, A.T.; et al. Scientists’ Warning to Humanity: Microorganisms and Climate Change. Nat. Rev. Microbiol. 2019, 17, 569–586. [Google Scholar] [CrossRef]
- Wan, Y.; Ruan, X.; Zhang, Y.; Li, R. Illumina Sequencing-Based Analysis of Sediment Bacteria Community in Different Trophic Status Freshwater Lakes. Microbiologyopen 2017, 6, e00450. [Google Scholar] [CrossRef] [PubMed]
- Oetken, M.; Nentwig, G.; Löffler, D.; Ternes, T.; Oehlmann, J. Effects of Pharmaceuticals on Aquatic Invertebrates. Part I. The Antiepileptic Drug Carbamazepine. Arch. Environ. Contam. Toxicol. 2005, 49, 353–361. [Google Scholar] [CrossRef]
- Barznji, D.A.M. Role of Aquatic Plants in Improving Water Quality. Unique J. Pharm. Biol. Sci. 2014, 2, 12–16. [Google Scholar]
- Couto, E.; Assemany, P.P.; Assis Carneiro, G.C.; Ferreira Soares, D.C. The Potential of Algae and Aquatic Macrophytes in the Pharmaceutical and Personal Care Products (PPCPs) Environmental Removal: A Review. Chemosphere 2022, 302, 134808. [Google Scholar] [CrossRef]
- Kafle, A.; Timilsina, A.; Gautam, A.; Adhikari, K.; Bhattarai, A.; Aryal, N. Phytoremediation: Mechanisms, Plant Selection and Enhancement by Natural and Synthetic Agents. Environ. Adv. 2022, 8, 100203. [Google Scholar] [CrossRef]
- Bala, S.; Garg, D.; Thirumalesh, B.V.; Sharma, M.; Sridhar, K.; Inbaraj, B.S.; Tripathi, M. Recent Strategies for Bioremediation of Emerging Pollutants: A Review for a Green and Sustainable Environment. Toxics 2022, 10, 484. [Google Scholar] [CrossRef]
- Wei, H.; Tang, M.; Xu, X. Mechanism of Uptake, Accumulation, Transport, Metabolism and Phytotoxic Effects of Pharmaceuticals and Personal Care Products within Plants: A Review. Sci. Total Environ. 2023, 892, 164413. [Google Scholar] [CrossRef]
- Nieto, C.; Ovando, X.M.C.; Loyola, R.; Izquierdo, A.; Romero, F.; Molineri, C.; Rodríguez, J.; Rueda Martín, P.; Fernández, H.; Manzo, V.; et al. The Role of Macroinvertebrates for Conservation of Freshwater Systems. Ecol. Evol. 2017, 7, 5502–5513. [Google Scholar] [CrossRef]
- Saad El-Din, M.I. A Review of the Effects of Pharmaceuticals on Marine Invertebrates. Biomed. J. Sci. Tech. Res. 2023, 48, 40196–40198. [Google Scholar] [CrossRef]
- Grabicová, K.; Grabic, R.; Blaha, M.; Kumar, V.; Cerveny, D.; Fedorova, G.; Randak, T. Presence of Pharmaceuticals in Benthic Fauna Living in a Small Stream Affected by Effluent from a Municipal Sewage Treatment Plant. Water Res. 2015, 72, 145–153. [Google Scholar] [CrossRef]
- Grabicová, K.; Vojs Staňová, A.; Švecová, H.; Nováková, P.; Kodeš, V.; Leontovyčová, D.; Brooks, B.W.; Grabic, R. Invertebrates Differentially Bioaccumulate Pharmaceuticals: Implications for Routine Biomonitoring. Environ. Pollut. 2022, 309, 119715. [Google Scholar] [CrossRef]
- Rodrigues, S.; Antunes, S.C.; Nunes, B.; Correia, A.T. Histological Alterations in Gills and Liver of Rainbow Trout (Oncorhynchus mykiss) after Exposure to the Antibiotic Oxytetracycline. Environ. Toxicol. Pharmacol. 2017, 53, 164–176. [Google Scholar] [CrossRef]
- Rodrigues, S.; Antunes, S.C.; Nunes, B.; Correia, A.T. Histopathological Effects in Gills and Liver of Sparus Aurata Following Acute and Chronic Exposures to Erythromycin and Oxytetracycline. Environ. Sci. Pollut. Res. 2019, 26, 15481–15495. [Google Scholar] [CrossRef]
- Bereketoglu, C.; Pradhan, A.; Olsson, P.E. Nonsteroidal Anti-Inflammatory Drugs (NSAIDs) Cause Male-Biased Sex Differentiation in Zebrafish. Aquat. Toxicol. 2020, 223, 105476. [Google Scholar] [CrossRef]
- Kayode-Afolayan, S.D.; Ahuekwe, E.F.; Nwinyi, O.C. Impacts of Pharmaceutical Effluents on Aquatic Ecosystems. Sci. Afr. 2022, 17, e01288. [Google Scholar] [CrossRef]
- Hassan, S.; Sabreena; Poczai, P.; Ganai, B.A.; Almalki, W.H.; Gafur, A.; Sayyed, R.Z. Environmental DNA Metabarcoding: A Novel Contrivance for Documenting Terrestrial Biodiversity. Biology 2022, 11, 1297. [Google Scholar] [CrossRef]
- Su, G.; Logez, M.; Xu, J.; Tao, S.; Villéger, S.; Brosse, S. Human Impacts on Global Freshwater Fish Biodiversity. Science (1979) 2021, 371, 835–838. [Google Scholar] [CrossRef]
- Takahashi, M.; Saccò, M.; Kestel, J.H.; Nester, G.; Campbell, M.A.; van der Heyde, M.; Heydenrych, M.J.; Juszkiewicz, D.J.; Nevill, P.; Dawkins, K.L.; et al. Aquatic Environmental DNA: A Review of the Macro-Organismal Biomonitoring Revolution. Sci. Total Environ. 2023, 873, 162322. [Google Scholar] [CrossRef]
- Blackman, R.C.; Mächler, E.; Altermatt, F.; Arnold, A.; Beja, P.; Boets, P.; Egeter, B.; Elbrecht, V.; Filipe, A.F.; Iwan Jones, J.; et al. Advancing the Use of Molecular Methods for Routine Freshwater Macroinvertebrate Biomonitoring—The Need for Calibration Experiments. Metabarcoding Metagenomics 2019, 3, 49–57. [Google Scholar] [CrossRef]
- Espinosa Prieto, A.; Beisel, J.N.; Verschuren, P.; Hardion, L. Toward Freshwater Plant Diversity Surveys with eDNA Barcoding and Metabarcoding. Environ. DNA 2023, 5, 648–670. [Google Scholar] [CrossRef]
- Fernández, S.; Rodríguez, S.; Martínez, J.L.; Borrell, Y.J.; Ardura, A.; García-Vázquez, E. Evaluating Freshwater Macroinvertebrates from eDNA Metabarcoding: A River Nalón Case Study. PLoS ONE 2018, 13, e0201741. [Google Scholar] [CrossRef]
- Liu, J.; Zhang, H. Combining Multiple Markers in Environmental DNA Metabarcoding to Assess Deep-Sea Benthic Biodiversity. Front. Mar. Sci. 2021, 8, 684955. [Google Scholar] [CrossRef]
- Wu, F.; Zou, Y.; Qin, S.; Li, F.; Zhang, Y. eDNA Biomonitoring of Macroinvertebrate Communities for the Bioassessment of a River’s Ecological Status. Water 2023, 15, 308. [Google Scholar] [CrossRef]
- Pawlowski, J.; Esling, P.; Lejzerowicz, F.; Cedhagen, T.; Wilding, T.A. Environmental Monitoring through Protist Next-Generation Sequencing Metabarcoding: Assessing the Impact of Fish Farming on Benthic Foraminifera Communities. Mol. Ecol. Resour. 2014, 14, 1129–1140. [Google Scholar] [CrossRef] [PubMed]
- Pochon, X.; Wood, S.A.; Keeley, N.B.; Lejzerowicz, F.; Esling, P.; Drew, J.; Pawlowski, J. Accurate Assessment of the Impact of Salmon Farming on Benthic Sediment Enrichment Using Foraminiferal Metabarcoding. Mar. Pollut. Bull. 2015, 100, 370–382. [Google Scholar] [CrossRef] [PubMed]
- Grzesiuk, M.; Gryglewicz, E.; Bentkowski, P.; Pijanowska, J. Impact of Fluoxetine on Herbivorous Zooplankton and Planktivorous Fish. Environ. Toxicol. Chem. 2023, 42, 385–392. [Google Scholar] [CrossRef] [PubMed]
- Lagesson, A.; Fahlman, J.; Brodin, T.; Fick, J.; Jonsson, M.; Byström, P.; Klaminder, J. Bioaccumulation of Five Pharmaceuticals at Multiple Trophic Levels in an Aquatic Food Web—Insights from a Field Experiment. Sci. Total Environ. 2016, 568, 208–215. [Google Scholar] [CrossRef]
- Yan, Z.; Zhou, Y.; Zhang, Y.; Zhang, X. Distribution, Bioaccumulation, and Risks of Pharmaceutical Metabolites and Their Parents: A Case Study in an Yunliang River, Nanjing City. Int. J. Environ. Res. Public Health 2023, 20, 2967. [Google Scholar] [CrossRef]
- Nilsen, E.; Smalling, K.L.; Ahrens, L.; Gros, M.; Miglioranza, K.S.B.; Picó, Y.; Schoenfuss, H.L. Critical Review: Grand Challenges in Assessing the Adverse Effects of Contaminants of Emerging Concern on Aquatic Food Webs. Environ. Toxicol. Chem. 2019, 38, 46–60. [Google Scholar] [CrossRef]
- Sparrow, B.D.; Edwards, W.; Munroe, S.E.M.; Wardle, G.M.; Guerin, G.R.; Bastin, J.F.; Morris, B.; Christensen, R.; Phinn, S.; Lowe, A.J. Effective Ecosystem Monitoring Requires a Multi-Scaled Approach. Biol. Rev. 2020, 95, 1706–1719. [Google Scholar] [CrossRef]
- Cordier, T.; Esling, P.; Lejzerowicz, F.; Visco, J.; Ouadahi, A.; Martins, C.; Cedhagen, T.; Pawlowski, J. Predicting the Ecological Quality Status of Marine Environments from eDNA Metabarcoding Data Using Supervised Machine Learning. Environ. Sci. Technol. 2017, 51, 9118–9126. [Google Scholar] [CrossRef]
- Prakash, S.; Verma, A.K. Anthropogenic Activities and Biodiversity Threats. Int. J. Biol. Innov. 2022, 4, 94–103. [Google Scholar] [CrossRef]
- Rangel-Buitrago, N. Human Epoch—Human Responsibility: Rethinking Coastal Zone Management in the Anthropocene. Ocean. Coast. Manag. 2023, 244, 106801. [Google Scholar] [CrossRef]
- Miettinen, M.; Khan, S.A. Pharmaceutical Pollution: A Weakly Regulated Global Environmental Risk. Rev. Eur. Comp. Int. Environ. Law. 2022, 31, 75–88. [Google Scholar] [CrossRef]
- Küster, A.; Adler, N. Pharmaceuticals in the Environment: Scientific Evidence of Risks and Its Regulation. Philos. Trans. R. Soc. B Biol. Sci. 2014, 369, 20130587. [Google Scholar] [CrossRef]
- Paut Kusturica, M.; Jevtic, M.; Ristovski, J.T. Minimizing the Environmental Impact of Unused Pharmaceuticals: Review Focused on Prevention. Front. Environ. Sci. 2022, 10, 1077974. [Google Scholar] [CrossRef]
- Guo, M.; Chen, H.; Dong, S.; Zhang, Z.; Luo, H. CRISPR-Cas Gene Editing Technology and Its Application Prospect in Medicinal Plants. Chin. Med. 2022, 17, 33. [Google Scholar] [CrossRef]
- Rafeeq, H.; Afsheen, N.; Rafique, S.; Arshad, A.; Intisar, M.; Hussain, A.; Bilal, M.; Iqbal, H.M.N. Genetically Engineered Microorganisms for Environmental Remediation. Chemosphere 2023, 310, 136751. [Google Scholar] [CrossRef]
- Andújar, C.; Arribas, P.; Gray, C.; Bruce, C.; Woodward, G.; Yu, D.W.; Vogler, A.P. Metabarcoding of Freshwater Invertebrates to Detect the Effects of a Pesticide Spill. Mol. Ecol. 2018, 27, 146–166. [Google Scholar] [CrossRef]
- Zhang, X. Environmental DNA Shaping a New Era of Ecotoxicological Research. Environ. Sci. Technol. 2019, 53, 5605–5612. [Google Scholar] [CrossRef] [PubMed]
- Blaxter, M.; Mann, J.; Chapman, T.; Thomas, F.; Whitton, C.; Floyd, R.; Abebe, E. Defining Operational Taxonomic Units Using DNA Barcode Data. Philos. Trans. R. Soc. B Biol. Sci. 2005, 360, 1935–1943. [Google Scholar] [CrossRef]
- Wang, S.; Yan, Z.; Hänfling, B.; Zheng, X.; Wang, P.; Fan, J.; Li, J. Methodology of Fish eDNA and Its Applications in Ecology and Environment. Sci. Total Environ. 2021, 755, 142622. [Google Scholar] [CrossRef] [PubMed]
- Nagarajan, R.P.; Bedwell, M.; Holmes, A.E.; Sanches, T.; Acuña, S.; Baerwald, M.; Barnes, M.A.; Blankenship, S.; Connon, R.E.; Deiner, K.; et al. Environmental DNA Methods for Ecological Monitoring and Biodiversity Assessment in Estuaries. Estuaries Coasts 2022, 45, 2254–2273. [Google Scholar] [CrossRef]
- Harper, L.R.; Lawson Handley, L.; Carpenter, A.I.; Ghazali, M.; Di Muri, C.; Macgregor, C.J.; Logan, T.W.; Law, A.; Breithaupt, T.; Read, D.S.; et al. Environmental DNA (eDNA) Metabarcoding of Pond Water as a Tool to Survey Conservation and Management Priority Mammals. Biol. Conserv. 2019, 238, 108225. [Google Scholar] [CrossRef]
- Deiner, K.; Altermatt, F. Transport Distance of Invertebrate Environmental DNA in a Natural River. PLoS ONE 2014, 9, e88786. [Google Scholar] [CrossRef] [PubMed]
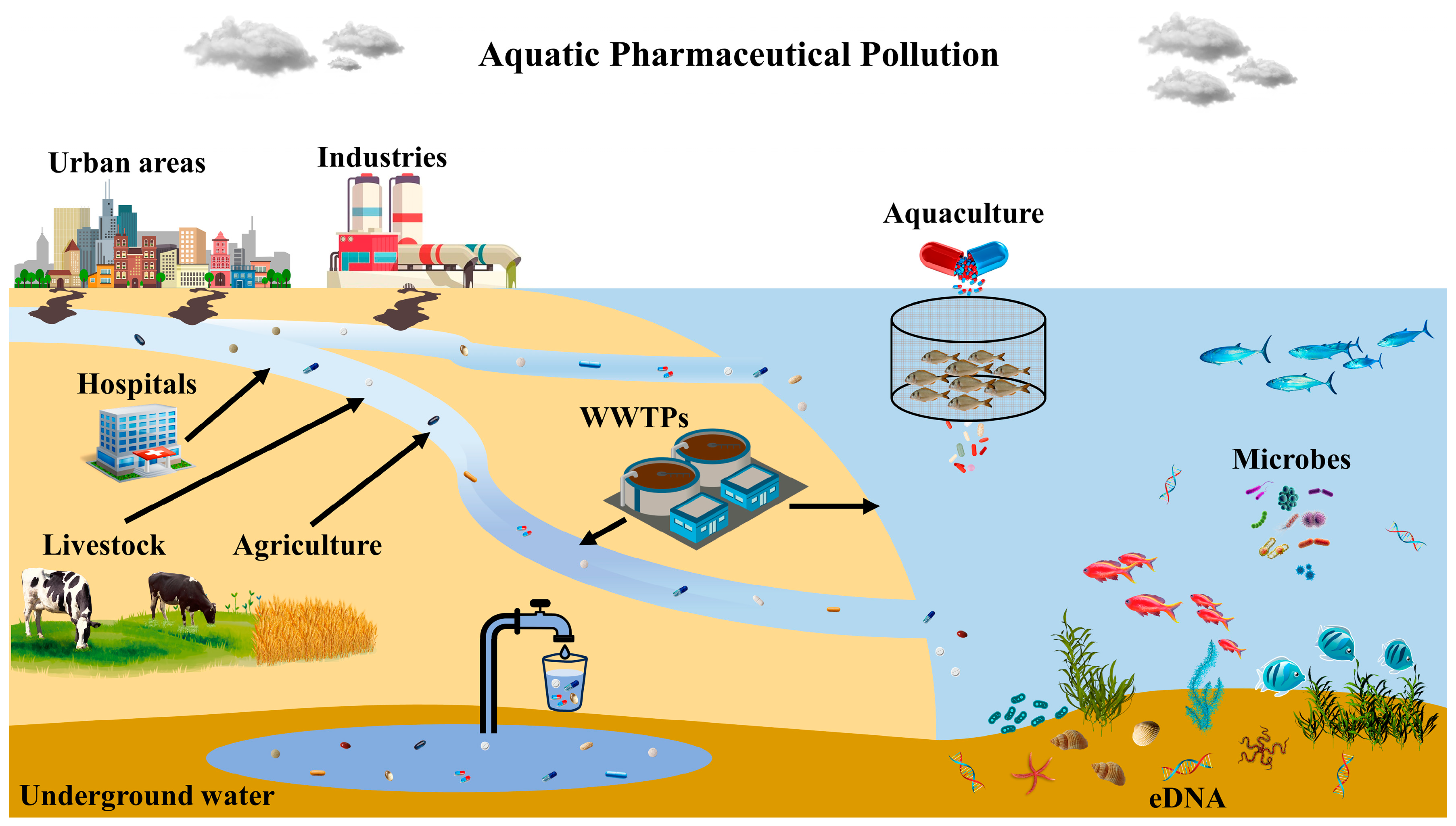
| Pharmaceuticals | Sources | Effects | References |
|---|---|---|---|
| Antibiotics | Contamination of water bodies from human and veterinary medicine wastes. |
| [81,82,83,84,85,86,87,88,89,90,91] |
| Hormones and endocrine-disrupting chemicals (EDCs) | Enter the aquatic environment through agricultural and livestock manure, excretion (e.g., urine and feces), improper disposal. |
| [92,93,94,95,96,97,98,99,100] |
| Analgesics and nonsteroidal anti-inflammatory drugs (NSAIDs) | Main pathways to the aquatic environment: human excretion, the inappropriate disposal of unused medications, wastewater discharges from pharmaceutical and healthcare facilities. |
| [27,101,102,103,104,105] |
| Psychotropic and antiepileptic drugs | Enter water bodies through human excretion, and wastewater systems via sewage or septic tanks |
| [43,44,106,107,108] |
| β-blockers | Infiltrate the aquatic environment through human excretion, and wastewater systems via sewage or septic tanks |
| [109,110,111,112] |
| Chemotherapy and anticancer drugs | Introduction to the aquatic environment via human excretion and the incorrect disposal of unused medicines |
| [113,114,115] |
| Type of Treatment Method | Treatment Method | Efficiency | Pharmaceutical Compounds | References |
|---|---|---|---|---|
| Physico-chemical Treatment | Aeration | Low | Analgesics and antibiotics | [153] |
| Coagulation, flocculation, and sedimentation | Very low | Antibiotics, antidepressants, AEDs, analgesics, NSAIDs | [154,155,156] | |
| Adsorption | High | Antibiotics and NSAIDs | [157,158,159,160,161] | |
| Filtration | Contaminant dependent | AEDs, NSAIDs, antibiotics, EDCs | [162,163,164,165] | |
| Nanofiltration | Moderate to high | EDCs, β-blockers, psychotropics and AEDs, antibiotics | [166,167,168,169] | |
| Reverse osmosis | High | Analgesics, NSAIDs, β-blockers, AEDs, psychotropic drugs | [170,171,172] | |
| Biological Treatment | Conventional activated sludge | Low to moderate | Analgesics, EDCs, antibiotics, β-blockers, AEDs | [173,174] |
| Membrane bioreactors | Moderate to high | EDCs, psychotropic drugs, NSAIDs, anti-diabetic drugs, β-blockers, | [108,140,175] | |
| Microalgal bioremediation process | Low to moderate | NSAIDs, β-blockers, AEDs, antibiotics, EDCs | [176,177] | |
| Enzyme-based treatment | Moderate to high | NSAIDs | [152,178,179,180] | |
| Oxidation Treatment | Chlorination | Contaminant-dependent | Antibiotics, EDCs, β-blockers, analgesics, NSAIDs | [181,182] |
| Ozonation | High | Antibiotics, EDCs, AEDs, NSAIDs, psychotropic drugs | [172,183,184,185] | |
| Advanced oxidation processes (AOPs) | High | EDCs, antibiotics, NSAIDs, psychotropic drugs | [89,186,187,188,189] | |
| Electrochemical treatment | Electrochemical technologies | High | Antibiotics, EDCs, NSAIDs | [190,191,192,193,194] |
| Methods Associated with the Use of Bacteria for Pharmaceutical Pollution | Indicative Taxa (or Genus or Phylum) | Pharmaceutical Compounds | Method of Detection | Refs. | |
|---|---|---|---|---|---|
| Indirect Methods | Bioremediation | Actinobacteria, Chryseobacterium, Flavobacterium, Pseudoxanthomonas, | β-blockers | PCR-DGGE and pyrosequencing | [245] |
| Bacillus thuringiensis B1 Novosphingobium sp. Sphingomonas sp. Sphingopyxis sp. Sphingobium sp. Isoptericola sp. Nubsella sp. Rhodococcus sp. Bacillus sp. | NSAIDs | Gram staining, API CORYNE system analysis, FAMEs analysis, HPLC, cell cultures, PCR | [246,247] | ||
| Pseudomonas sp. CE21 Pseudomonas sp. CE22 Paucibacter Filomicrobium | Antibiotics | Cell cultures, PCR, LC-MS, degradation analysis with MS, BOD 5/COD Ratio, HPLC, TOC/TN analysis, ammonia and nitrate analysis, SEM | [248,249] | ||
| Chryseobacterium taeanense Rhizobium daejeonense Diaphorobacter nitroreducens Achromobacter mucicolens Pseudomonas veronii Pseudomonas lini | AEDs | PCR and HPLC | [250] | ||
| Microbacterium sp. C448 | Anti-cancer | Liquid scintillation counting, HPLC-MS/MS, LC-MS, solvent extraction (ASE 200), NGS | [251] | ||
| Flavobacterium Novosphingobium sp. Sphingomonas sp. Sphingopyxis sp. Sphingobium sp. Isoptericola sp. Nubsella sp. Rhodococcus sp. Bacillus sp. Nitrosomonas europaea Acinetobacter sp. Phyllobacterium myrsinacearum Ralstonia pickettii Pseudomonas | EDCs | HPLC, IC, TOC analysis, oxygen probe analysis, rep-PCR, NGS, fluorescence detection, colorimetric analysis, UV/fluorescence detection, GC-MS/MS and LC-MS/MS, ATP/OD measurement | [247,252,253,254] | ||
| Communities’ function and structure | Actinobacteria, Bacteroidetes, Cyanobacteria, Flavobacteria, Firmicutes, Proteobacteria, Fusobacteria | Hormones, antibiotics, antipsychotic drugs, AEDs, NSAIDs, β-blockers, antihistamines, antidiabetics, analgesics, H2 blockers, ACE inhibitors | HPLC, UPLC-MS/MS, FTIR, LC-MS/MS, enzyme assays, MBR and batch cultures, qPCR, PCR-DGGE, NGS, metagenomics | [255,256,257,258,259] | |
| Detection of ARBs (and ARGs) | Escherichia coli, Klebsiella pneumoniae, Aeromonas spp., Pseudomonas aeruginosa, Enterococcus faecalis, Enterococcus faecium, Acinetobacter baumannii, Flavobacterium, Poriferibacter, Bacteroides, Acinetobacter, Actinobaculum, Streptococcus | Antibiotics | Metagenomics-metatranscriptomics, qPCR, rep-PCR | [260,261,262] | |
| Direct Methods | Whole-cell Biosensors | Escherichia coli, Pseudomonas fluorescen, Bacillus subtilis | Antibiotics, NSAIDs, EDCs | Biosensor (optical, fluorescence, electrochemical, etc.) | [263,264,265,266,267,268] |
| Affected Aquatic Organisms | Summary of PhACs Implications | Evaluation of eDNA Metabarcoding in Pharmaceutical Pollution Assessment |
|---|---|---|
| Microbial communities |
| |
| Plants | ||
| Invertebrates |
| |
| Vertebrates |
| |
| Aquatic food webs |
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Papaioannou, C.; Geladakis, G.; Kommata, V.; Batargias, C.; Lagoumintzis, G. Insights in Pharmaceutical Pollution: The Prospective Role of eDNA Metabarcoding. Toxics 2023, 11, 903. https://doi.org/10.3390/toxics11110903
Papaioannou C, Geladakis G, Kommata V, Batargias C, Lagoumintzis G. Insights in Pharmaceutical Pollution: The Prospective Role of eDNA Metabarcoding. Toxics. 2023; 11(11):903. https://doi.org/10.3390/toxics11110903
Chicago/Turabian StylePapaioannou, Charikleia, George Geladakis, Vasiliki Kommata, Costas Batargias, and George Lagoumintzis. 2023. "Insights in Pharmaceutical Pollution: The Prospective Role of eDNA Metabarcoding" Toxics 11, no. 11: 903. https://doi.org/10.3390/toxics11110903
APA StylePapaioannou, C., Geladakis, G., Kommata, V., Batargias, C., & Lagoumintzis, G. (2023). Insights in Pharmaceutical Pollution: The Prospective Role of eDNA Metabarcoding. Toxics, 11(11), 903. https://doi.org/10.3390/toxics11110903









