Application of a High-Throughput Amplicon Sequencing Method to Chart the Bacterial Communities that Are Associated with European Fermented Meats from Different Origins
Abstract
1. Introduction
2. Materials and Methods
2.1. Sampling and Experimental Set-Up
2.2. Extraction of Bacterial Genomic DNA
2.3. PCR Assay and Sequencing
2.4. Bioinformatic Analysis
2.5. Volatile Organic Compound Profiling
2.6. Statistics
3. Results
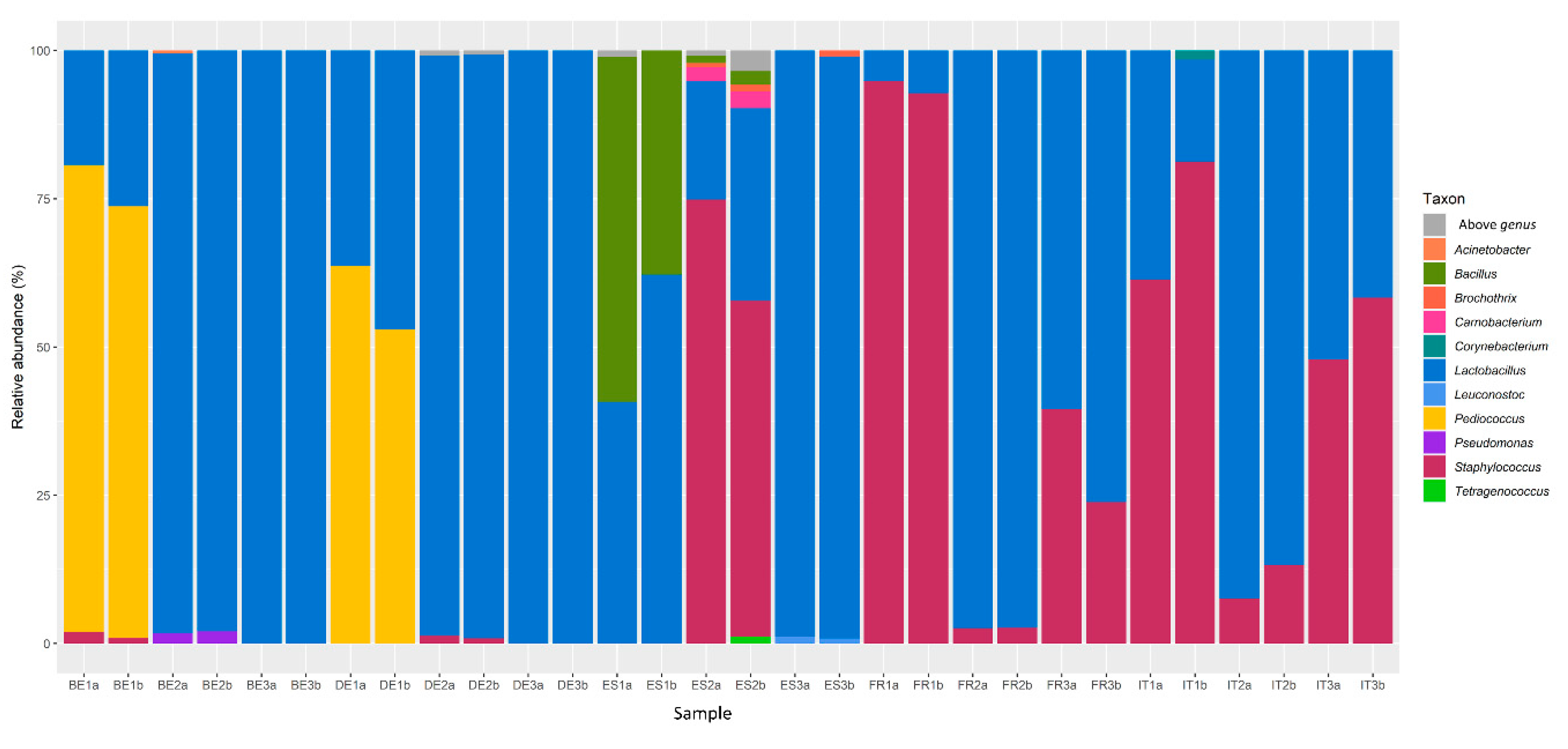
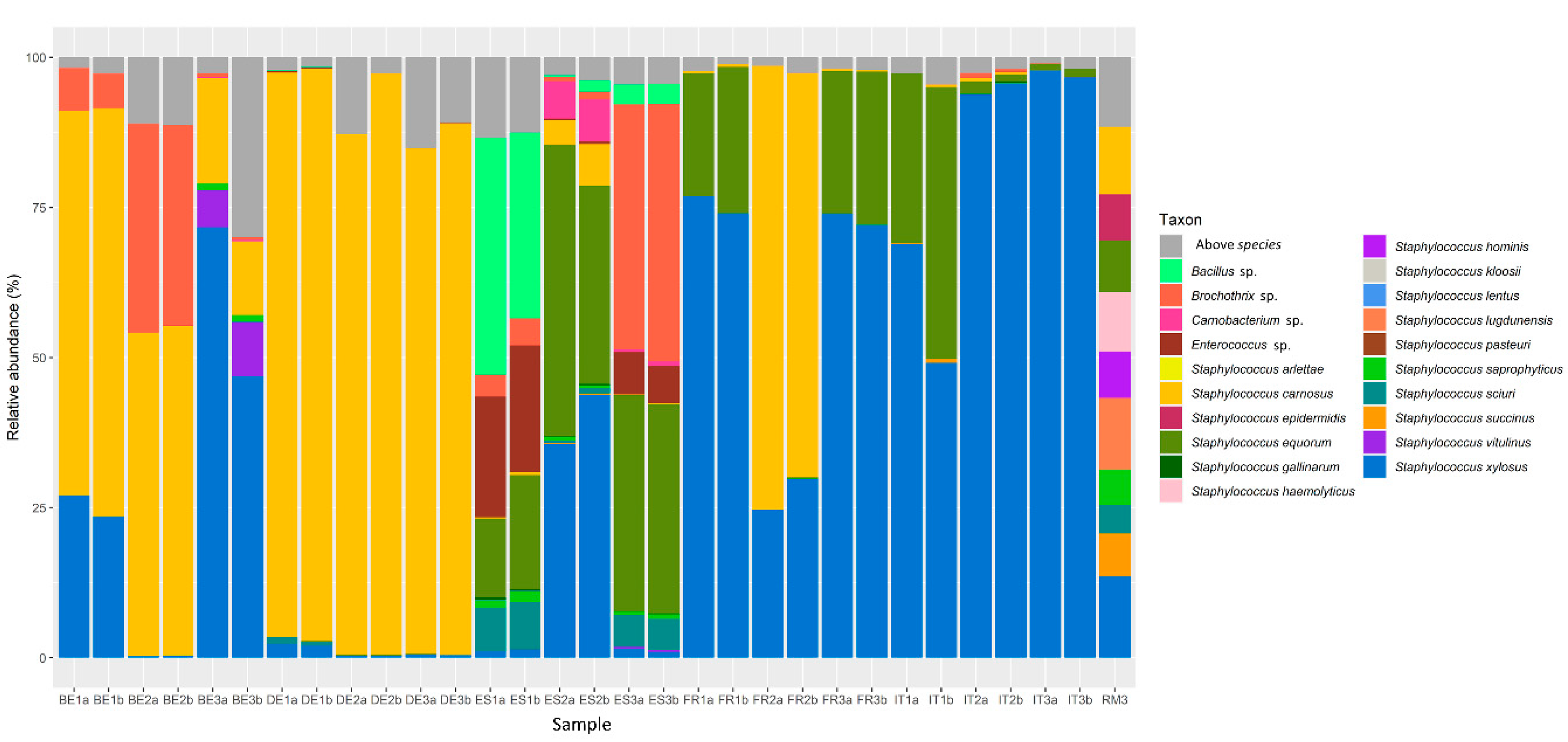
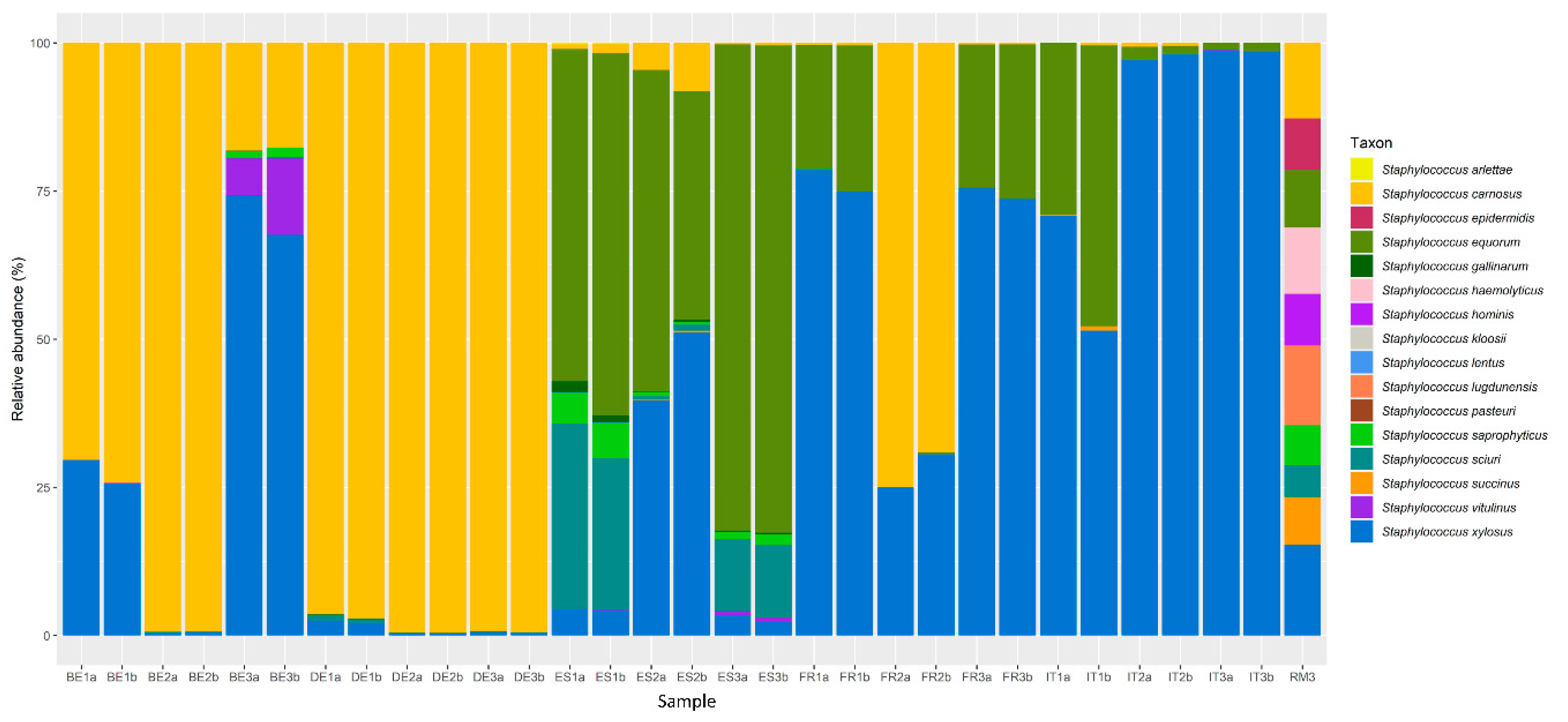
3.1. Bacterial Communities in Fermented Meat Products from Different Origins
3.2. Volatile Organic Compound Profiling
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
Data Availability
References
- Guerrero, L.; Guàrdia, M.D.; Xicola, J.; Verbeke, W.; Vanhonacker, F.; Zakowska-Biemans, S.; Sajdakowska, M.; Sulmont-Rossé, C.; Issanchou, S.; Contel, M.; et al. Consumer-driven definition of traditional food products and innovation in traditional foods. A qualitative cross-cultural study. Appetite 2009, 52, 345–354. [Google Scholar]
- Leroy, F.; Geyzen, A.; Janssens, M.; De Vuyst, L.; Scholliers, P. Meat fermentation at the crossroads of innovation and tradition: A historical outlook. Trends Food Sci. Technol. 2013, 31, 130–137. [Google Scholar] [CrossRef]
- Fenger, M.H.; Aschemann-Witzel, J.; Hansen, F.; Grunert, K.G. Delicious words—Assessing the impact of short storytelling messages on consumer preferences for variations of a new processed meat product. Food Qual. Pref. 2015, 41, 237–244. [Google Scholar] [CrossRef]
- Zhong, Z.; Hou, Q.; Kwok, L.; Yu, Z.; Zheng, Y.; Sun, Z.; Menghe, B.; Zhang, H. Bacterial microbiota compositions of naturally fermented milk are shaped by both geographic origin and sample type. J. Dairy Sci. 2016, 99, 7832–7841. [Google Scholar] [CrossRef]
- Leroy, F.; Scholliers, P.; Amilien, V. Elements of innovation and tradition in meat fermentation: Conflicts and synergies. Int. J. Food Microbiol. 2015, 212, 2–8. [Google Scholar] [CrossRef]
- Van Reckem, E.; Geeraerts, W.; Charmpi, C.; Van der Veken, D.; De Vuyst, L.; Leroy, F. Exploring the link between the geographical origin of European fermented foods and the diversity of their bacterial communities: The case of fermented meats. Front. Microbiol. 2019, 10, 2302. [Google Scholar] [CrossRef]
- Leroy, F.; Verluyten, J.; De Vuyst, L. Functional meat starter cultures for improved sausage fermentation. Int. J. Food Microbiol. 2006, 106, 270–285. [Google Scholar] [CrossRef]
- Aquilanti, L.; Garofalo, C.; Osimani, A.; Clementi, F. Ecology of lactic acid bacteria and coagulase-negative cocci in fermented dry sausages manufactured in Italy and other Mediterranean countries: An overview. Int. Food Res. J. 2016, 23, 429–445. [Google Scholar]
- Holck, A.; Heir, E.; Johannessen, T.C.; Axelsson, L. Northern European products. In Handbook of Fermented Meat and Poultry, 2nd ed.; Toldrà, F., Hui, Y.H., Astiasarán, I., Sebranek, J.G., Talon, R., Eds.; John Wiley and Sons: Chichester, UK, 2015; pp. 313–320. [Google Scholar]
- Janssens, M.; Myter, N.; De Vuyst, L.; Leroy, F. Community dynamics of coagulase-negative staphylococci during spontaneous artisan-type meat fermentations differ between smoking and moulding treatments. Int. J. Food Microbiol. 2013, 166, 168–175. [Google Scholar] [CrossRef]
- Hierro, E.; Fernández, M.; de la Hoz, L.; Ordóñez, J.A. Mediterranean products. In Handbook of Fermented Meat and Poultry, 2nd ed.; Toldrà, F., Hui, Y.H., Astiasarán, I., Sebranek, J.G., Talon, R., Eds.; John Wiley and Sons: Chichester, UK, 2015; pp. 301–308. [Google Scholar]
- Janssens, M.; Myter, N.; De Vuyst, L.; Leroy, F. Species diversity and metabolic impact of the microbiota are low in spontaneously acidified Belgian sausages with an added starter culture of Staphylococcus carnosus. Food Microbiol. 2012, 29, 167–177. [Google Scholar] [CrossRef]
- Fonseca, S.; Cachaldora, A.; Gómez, M.; Franco, I.; Carballo, J. Monitoring the bacterial population dynamics during the ripening of Galician chorizo, a traditional dry fermented Spanish sausage. Food Microbiol. 2013, 33, 77–84. [Google Scholar] [CrossRef] [PubMed]
- Greppi, A.; Ferrocino, I.; La Storia, A.; Rantsiou, K.; Ercolini, D.; Cocolin, L. Monitoring of the microbiota of fermented sausages by culture independent rRNA based approaches. Int. J. Food Microbiol. 2015, 212, 67–75. [Google Scholar] [CrossRef] [PubMed]
- Stavropoulou, D.A.; De Maere, H.; Berardo, A.; Janssens, B.; Filippou, P.; De Vuyst, L.; De Smet, S.; Leroy, F. Species pervasiveness within the group of coagulase-negative staphylococci associated with meat fermentation is modulated by pH. Front. Microbiol. 2018, 9, 2232. [Google Scholar] [CrossRef] [PubMed]
- Ojha, K.S.; Kerry, J.P.; Duffy, G.; Beresford, T.; Tiwari, B.K. Technological advances for enhancing quality and safety of fermented meat products. Trends Food Sci. Tech. 2015, 44, 105–116. [Google Scholar] [CrossRef]
- Stavropoulou, D.A.; De Vuyst, L.; Leroy, F. Nonconventional starter cultures of coagulase-negative staphylococci to produce animal-derived fermented foods, a SWOT analysis. J. Appl. Microbiol. 2018, 125, 1570–1586. [Google Scholar] [CrossRef]
- Laranjo, M.; Potes, M.E.; Elias, M. Role of starter cultures on the safety of fermented meat products. Front. Microbiol. 2019, 10, 853. [Google Scholar] [CrossRef]
- Belleggia, L.; Ferrocino, I.; Reale, A.; Boscaino, F.; Di Renzo, T.; Corvaglia, M.R.; Cocolin, L.; Milanović, V.; Cardinali, F.; Garofalo, C.; et al. Portuguese cacholeira blood sausage: A first taste of its microbiota and volatile organic compounds. Food Res. Int. 2020, 136, 109567. [Google Scholar] [CrossRef]
- Settanni, L.; Barbaccia, P.; Bonanno, A.; Ponte, M.; Di Gerlando, R.; Franciosi, E.; Di Grigoli, A.; Gaglio, R. Evolution of indigenous starter microorganisms and physicochemical parameters in spontaneously fermented beef, horse, wild boar and pork salamis produced under controlled conditions. Food Microbiol. 2020, 87, 103385. [Google Scholar] [CrossRef]
- Charmpi, C.; Van der Veken, D.; Van Reckem, E.; De Vuyst, L.; Leroy, F. Raw meat quality and salt levels affect the bacterial species diversity and community dynamics during the fermentation of pork mince. Food Microbiol. 2020, 89, 103434. [Google Scholar] [CrossRef]
- Rantsiou, K.; Urso, R.; Iacumin, L.; Cantoni, C.; Cattaneo, P.; Comi, G.; Cocolin, L. Culture-dependent and -independent methods to investigate the microbial ecology of Italian fermented sausages. Appl. Environ. Microbiol. 2005, 71, 1977–1986. [Google Scholar] [CrossRef]
- Cocolin, L.; Alessandria, V.; Dolci, P.; Gorra, R.; Rantsiou, K. Culture independent methods to assess the diversity and dynamics of microbiota during food fermentation. Int. J. Food Microbiol. 2013, 167, 29–43. [Google Scholar] [CrossRef] [PubMed]
- Giraffa, G.; Carminati, D. Molecular techniques in food fermentation: Principles and applications. In Molecular Techniques in the Microbial Ecology of Fermented Foods; Cocolin, L., Ercolini, D., Eds.; Springer: New York, NY, USA, 2008; pp. 1–30. [Google Scholar]
- Połka, J.; Rebecchi, A.; Pisacane, V.; Morelli, L.; Puglisi, E. Bacterial diversity in typical Italian salami at different ripening stages as revealed by high-throughput sequencing of 16S rRNA amplicons. Food Microbiol. 2015, 46, 342–356. [Google Scholar] [CrossRef] [PubMed]
- Ercolini, D.; De Filippis, F.; La Storia, A.; Iacono, M. “Remake” by high-throughput sequencing of the microbiota involved in the production of water buffalo mozzarella cheese. Appl. Environ. Microbiol. 2012, 78, 8142–8145. [Google Scholar] [CrossRef] [PubMed]
- De Filippis, F.; Parente, E.; Ercolini, D. Recent past, present, and future of the food microbiome. Annu. Rev. Food Sci. Technol. 2018, 9, 589–608. [Google Scholar] [CrossRef]
- Maidana, S.D.; Ficoseco, C.A.; Bassi, D.; Cocconcelli, P.S.; Puglisi, E.; Savoy, G.; Vignolo, G.; Fontana, C. Biodiversity and technological-functional potential of lactic acid bacteria isolated from spontaneously fermented chia sourdough. Int. J. Food Microbiol. 2020, 316, 108425. [Google Scholar] [CrossRef]
- dos Santos Cruxen, C.E.; Funck, G.D.; Haubert, L.; da Silva Dannenberg, G.; de Lima Marques, J.; Chaves, F.C.; Padilha da Silva, W.; Fiorentini, Â.M. Selection of native bacterial starter culture in the production of fermented meat sausages: Application potential, safety aspects, and emerging technologies. Food Res. Int. 2019, 122, 371–382. [Google Scholar] [CrossRef]
- Heikens, E.; Fleer, A.; Paauw, A.; Florijn, A.; Fluit, A.C. Comparison of genotypic and phenotypic methods for species-level identification of clinical isolates of coagulase-negative staphylococci. J. Clin. Microbiol. 2005, 43, 2286–2290. [Google Scholar] [CrossRef]
- Ghebremedhin, B.; Layer, F.; König, W.; König, B. Genetic classification and distinguishing of Staphylococcus species based on different partial gap, 16S rRNA, hsp60, rpoB, sodA, and tuf gene sequences. J. Clin. Microbiol. 2008, 46, 1019–1025. [Google Scholar] [CrossRef]
- McMurray, C.L.; Hardy, K.J.; Calus, S.T.; Loman, N.J.; Hawkey, P.M. Staphylococcal species heterogeneity in the nasal microbiome following antibiotic prophylaxis revealed by tuf gene deep sequencing. Microbiome 2016, 4, 63. [Google Scholar] [CrossRef]
- Van Reckem, E.; De Vuyst, L.; Leroy, F.; Weckx, S. Amplicon-based high-throughput sequencing method capable of species-level identification of coagulase-negative staphylococci in diverse communities. Microorganisms 2020, 8, 897. [Google Scholar] [CrossRef]
- Vermote, L.; Verce, M.; De Vuyst, L.; Weckx, S. Amplicon and shotgun metagenomic sequencing indicates that microbial ecosystems present in cheese brines reflect environmental inoculation during the cheese production process. Int. Dairy J. 2018, 87, 44–53. [Google Scholar] [CrossRef]
- Caporaso, J.G.; Lauber, C.L.; Walters, W.A.; Berg-Lyons, D.; Lozupone, C.A.; Turnbaugh, P.J.; Fierer, N.; Knight, R. Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc. Natl. Acad. Sci. USA 2011, 108, 4516–4522. [Google Scholar] [CrossRef] [PubMed]
- De Bruyn, F.; Zhang, S.J.; Pothakos, V.; Torres, J.; Lambot, C.; Moroni, A.V.; Callanan, M.; Sybesma, W.; Weckx, S.; De Vuyst, L. Exploring the impacts of postharvest processing on the microbiota and metabolite profiles during green coffee bean production. Appl. Environ. Microbiol. 2017, 83, e02398-16. [Google Scholar] [CrossRef]
- Zhang, S.J.; De Bruyn, F.; Pothakos, V.; Torres, J.; Falconi, C.; Moccand, C.; Weckx, S.; De Vuyst, L. Following coffee production from cherries to cup: Microbiological and metabolomic analysis of wet processing of Coffea arabica. Appl. Environ. Microbiol. 2019, 85, e02635-18. [Google Scholar] [CrossRef]
- Callahan, B.J.; McMurdie, P.J.; Holmes, S.P. Exact sequence variants should replace operational taxonomic units in marker-gene data analysis. ISME J. 2017, 11, 2639–2643. [Google Scholar] [CrossRef] [PubMed]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2012, 41, D590–D596. [Google Scholar] [CrossRef] [PubMed]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Geeraerts, W.; De Vuyst, L.; Leroy, F.; Van Kerrebroeck, S. Monitoring of volatile production in cooked poultry products using selected ion flow tube-mass spectrometry. Food Res. Int. 2019, 119, 196–206. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2018. [Google Scholar]
- Oksanen, J.; Blanchet, F.G.; Friendly, M.; Kindt, R.; Legendre, P.; McGlinn, D.; Minchin, P.R.; O’Hara, R.B.; Simpson, G.L.; Solymos, P.; et al. Vegan: Community Ecology Package. R package version 2.5-4. 2019. Available online: https://CRAN.R-project.org/package=vegan (accessed on 10 June 2020).
- Hervé, M. RVAideMemoire: Testing and Plotting Procedures for Biostatistics. R package version 0.9-73. 2019. Available online: https://CRAN.R-project.org/package=RVAideMemoire (accessed on 10 June 2020).
- Gu, Z.; Eils, R.; Schlesner, M. Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics 2016, 32, 2847–2849. [Google Scholar] [CrossRef]
- Gu, Z.; Gu, L.; Eils, R.; Schlesner, M.; Brors, B. Circlize implements and enhances circular visualization in R. Bioinformatics 2014, 30, 2811–2812. [Google Scholar] [CrossRef]
- Delgado-Pando, G.; Fischer, E.; Allen, P.; Kerry, J.P.; O’Sullivan, M.; Hamill, R.M. Salt content and minimum acceptable levels in whole-muscle cured meat products. Meat Sci. 2018, 139, 179–186. [Google Scholar] [CrossRef] [PubMed]
- Zheng, J.; Wittouck, S.; Salvetti, E.; Franz, C.M.; Harris, H.M.; Mattarelli, P.; O’Toole, W.; Pot, B.; Vandamme, P.; Walter, J.; et al. A taxonomic note on the genus Lactobacillus: Description of 23 novel genera, emended description of the genus Lactobacillus Beijerinck 1901, and union of Lactobacillaceae and Leuconostocaceae. Int. J. Syst. Evol. Microbiol. 2020, 70, 2782–2858. [Google Scholar] [CrossRef] [PubMed]
- Chaillou, S.; Champomier-Vergès, M.C.; Cornet, M.; Crutz-Le Coq, A.M.; Dudez, A.M.; Martin, V.; Beaufils, S.; Darbon-Rongère, E.; Bossy, R.; Loux, V.; et al. The complete genome sequence of the meat-borne lactic acid bacterium Lactobacillus sakei 23K. Nat. Biotechnol. 2005, 23, 1527–1533. [Google Scholar] [CrossRef] [PubMed]
- Kumar, P.; Chatli, M.K.; Verma, A.K.; Mehta, N.; Malav, O.P.; Kumar, D.; Sharma, N. Quality, functionality, and shelf life of fermented meat and meat products: A review. Crit. Rev. Food Sci. Nutr. 2017, 57, 2844–2856. [Google Scholar] [CrossRef]
- Masson, F.; Johansson, G.; Montel, M.C. Tyramine production by a strain of Carnobacterium divergens inoculated in meat–fat mixture. Meat Sci. 1999, 52, 65–69. [Google Scholar] [CrossRef]
- Amadoro, C.; Rossi, F.; Piccirilli, M.; Colavita, G. Tetragenococcus koreensis is part of the microbiota in a traditional Italian raw fermented sausage. Food Microbiol. 2015, 50, 78–82. [Google Scholar] [CrossRef]
- Pisacane, V.; Callegari, M.L.; Puglisi, E.; Dallolio, G.; Rebecchi, A. Microbial analyses of traditional Italian salami reveal microorganisms transfer from the natural casing to the meat matrix. Int. J. Food Microbiol. 2015, 207, 57–65. [Google Scholar] [CrossRef]
- Stavropoulou, D.A.; Filippou, P.; De Smet, S.; De Vuyst, L.; Leroy, F. Effect of temperature and pH on the community dynamics of coagulase-negative staphylococci during spontaneous meat fermentation in a model system. Food Microbiol. 2018, 76, 180–188. [Google Scholar] [CrossRef]
- Geeraerts, W.; Stavropoulou, D.A.; De Vuyst, L.; Leroy, F. Meat and Meat Products. In How Fermented Foods Feed a Healthy Gut Microbiota; Azcarate-Peril, M.A., Arnold, R.R., Bruno-Bárcena, J.M., Eds.; Springer Nature: Cham, Switzerland, 2019; pp. 57–90. [Google Scholar]
- Amend, A.S.; Seifert, K.A.; Bruns, T.D. Quantifying microbial communities with 454 pyrosequencing: Does read abundance count? Mol. Ecol. 2010, 19, 5555–5565. [Google Scholar] [CrossRef]
- Elbrecht, V.; Leese, F. Can DNA-based ecosystem assessments quantify species abundance? Testing primer bias and biomass—Sequence relationships with an innovative metabarcoding protocol. PLoS ONE 2015, 10, e0130324. [Google Scholar] [CrossRef]
- Baruzzi, F.; Matarante, A.; Caputo, L.; Morea, M. Molecular and physiological characterization of natural microbial communities isolated from a traditional Southern Italian processed sausage. Meat Sci. 2006, 72, 261–269. [Google Scholar] [CrossRef] [PubMed]
- Iacumin, L.; Comi, G.; Cantoni, C.; Cocolin, L. Ecology and dynamics of coagulase-negative cocci isolated from naturally fermented Italian sausages. Syst. Appl. Microbiol. 2006, 29, 480–486. [Google Scholar] [CrossRef] [PubMed]
- Aymerich, T.; Martin, B.; Garriga, M.; Hugas, M. Microbial quality and direct PCR identification of lactic acid bacteria and nonpathogenic staphylococci from artisanal low-acid sausages. Appl. Environ. Microbiol. 2003, 69, 4583–4594. [Google Scholar] [CrossRef] [PubMed]
- Drosinos, E.H.; Mataragas, M.; Xiraphi, N.; Moschonas, G.; Gaitis, F.; Metaxopoulos, J. Characterization of the microbial flora from a traditional Greek fermented sausage. Meat Sci. 2005, 69, 307–317. [Google Scholar] [CrossRef]
- Drosinos, E.H.; Paramithiotis, S.; Kolovos, G.; Tsikouras, I.; Metaxopoulos, I. Phenotypic and technological diversity of lactic acid bacteria and staphylococci isolated from traditionally fermented sausages in Southern Greece. Food Microbiol. 2007, 24, 260–270. [Google Scholar] [CrossRef]
- Janssens, M.; Van der Mijnsbrugge, A.; Mainar, M.S.; Balzarini, T.; De Vuyst, L.; Leroy, F. The use of nucleosides and arginine as alternative energy sources by coagulase-negative staphylococci in view of meat fermentation. Food Microbiol. 2014, 39, 53–60. [Google Scholar] [CrossRef]
- Sawitzki, M.C.; Fiorentini, Â.M.; Bertol, T.M.; Sant’Anna, E.S. Lactobacillus plantarum strains isolated from naturally fermented sausages and their technological properties for application as starter cultures. Food Sci. Technol. 2009, 29, 340–345. [Google Scholar] [CrossRef]
- Talon, R.; Leroy, S.; Lebert, I. Microbial ecosystems of traditional fermented meat products: The importance of indigenous starters. Meat Sci. 2007, 77, 55–62. [Google Scholar] [CrossRef]
- Ravyts, F.; De Vuyst, L.; Leroy, F. Bacterial diversity and functionalities in food fermentations. Eng. Life Sci. 2012, 12, 356–367. [Google Scholar] [CrossRef]
- Leroy, S.; Giammarinaro, P.; Chacornac, J.P.; Lebert, I.; Talon, R. Biodiversity of indigenous staphylococci of naturally fermented dry sausages and manufacturing environments of small-scale processing units. Food Microbiol. 2010, 27, 294–301. [Google Scholar] [CrossRef]
- Marty, E.; Buchs, J.; Eugster-Meier, E.; Lacroix, C.; Meile, L. Identification of staphylococci and dominant lactic acid bacteria in spontaneously fermented Swiss meat products using PCR–RFLP. Food Microbiol. 2012, 29, 157–166. [Google Scholar] [CrossRef] [PubMed]
- Djinović, J.; Popović, A.; Jira, W. Polycyclic aromatic hydrocarbons (PAHs) in different types of smoked meat products from Serbia. Meat Sci. 2008, 80, 449–456. [Google Scholar] [CrossRef] [PubMed]
- Casaburi, A.; Piombino, P.; Nychas, G.-J.; Villani, F.; Ercolini, D. Bacterial populations and the volatilome associated to meat spoilage. Food Microbiol. 2015, 45, 83–102. [Google Scholar] [CrossRef] [PubMed]
- Flores, M.; Olivares, A. Flavor. In Handbook of Fermented Meat and Poultry, 2nd ed.; Toldrà, F., Hui, Y.H., Astiasarán, I., Sebranek, J.G., Talon, R., Eds.; John Wiley and Sons: Chichester, UK, 2015; pp. 217–225. [Google Scholar]
- Montanari, C.; Bargossi, E.; Gardini, A.; Lanciotti, R.; Magnani, R.; Gardini, F.; Tabanelli, G. Correlation between volatile profiles of Italian fermented sausages and their size and starter culture. Food Chem. 2016, 192, 736–744. [Google Scholar] [CrossRef]
- Belleggia, L.; Milanović, V.; Ferrocino, I.; Cocolin, L.; Haouet, M.N.; Scuota, S.; Maolonia, A.; Garofalo, C.; Cardinalia, F.; Aquilantia, L.; et al. Is there any still undisclosed biodiversity in Ciauscolo salami? A new glance into the microbiota of an artisan production as revealed by high-throughput sequencing. Meat Sci. 2020. [Google Scholar] [CrossRef]
| Primer | Primer Sequence | Amplicon Size (bp) |
|---|---|---|
| Tuf387 | 5’-YCCAATGCCWCAAACKCGTGA-3’ | 379 |
| Tuf765 | 5’-RAYTTGHCCACGTTCAACAC-3’ |
| Sample | MSA Counts [log(CFU/g)] | MRS Agar Counts [log(CFU/g)] | pH | Salt Content [g/100g] | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Individual | Average | SD | Individual | Average | SD | Individual | Average | SD | Individual | Average | SD | |
| BE1 | 5.40 | 6.75 | 5.12 | 4.10 | ||||||||
| BE2 | 5.13 | 5.18 | 0.20 | 8.27 | 7.76 | 0.87 | 5.00 | 5.27 | 0.37 | 4.60 | 4.30 | 0.26 |
| BE3 | 5.02 | 8.25 | 5.70 | 4.20 | ||||||||
| DE1 | 5.57 | 7.14 | 4.78 | 3.80 | ||||||||
| DE2 | 5.00 | 5.17 | 0.35 | 7.91 | 7.64 | 0.43 | 4.57 | 4.66 | 0.11 | 3.25 | 3.43 | 0.32 |
| DE3 | 4.94 | 7.88 | 4.63 | 3.25 | ||||||||
| ES1 | 5.72 | 6.53 | 5.82 | 3.00 | ||||||||
| ES2 | 5.80 | 5.83 | 0.12 | 5.60 | 6.83 | 1.41 | 6.04 | 5.85 | 0.18 | 3.00 | 3.27 | 0.46 |
| ES3 | 5.95 | 8.37 | 5.69 | 3.80 | ||||||||
| FR1 | 8.16 | 7.70 | 5.70 | 5.70 | ||||||||
| FR2 | 6.52 | 7.31 | 0.82 | 8.26 | 8.16 | 0.42 | 5.51 | 5.59 | 0.10 | 4.60 | 4.83 | 0.78 |
| FR3 | 7.26 | 8.52 | 5.56 | 4.20 | ||||||||
| IT1 | 8.63 | 8.44 | 5.66 | 3.40 | ||||||||
| IT2 | 6.68 | 7.34 | 1.12 | 8.56 | 8.34 | 0.30 | 5.26 | 5.57 | 0.28 | 3.30 | 3.67 | 0.55 |
| IT3 | 6.70 | 8.00 | 5.80 | 4.30 | ||||||||
| Sample | Culture-Dependent Method (Total Number of Isolates) | Amplicon-Based HTS Method |
|---|---|---|
| BE1 | S. carnosus (18) | S. carnosus, S. vitulinus, S. xylosus |
| BE2 | S. carnosus (29) | S. carnosus, S. saprophyticus, S. xylosus |
| BE3 | n.a. 1 | S. carnosus, S. pasteuri, S. saprophyticus, S. sciuri, S. xylosus |
| DE1 | S. carnosus, S. xylosus (13) | S. carnosus, S. gallinarum, S. sciuri, S. xylosus |
| DE2 | S. carnosus (20) | S. carnosus, S. equorum, S. xylosus |
| DE3 | S. carnosus (15) | S. carnosus, S. xylosus |
| ES1 | S. equorum, S. saprophyticus (21) | S. arlettae, S. carnosus, S. equorum, S. gallinarum, S. lentus, S. saprophyticus, S. sciuri, S. xylosus |
| ES2 | S. carnosus, S. equorum, S. xylosus (15) | S. carnosus, S. equorum, S. gallinarum, S. saprophyticus, S. sciuri, S. succinus, S. xylosus |
| ES3 | S. equorum (14) | S. carnosus, S. equorum, S. gallinarum, S. saprophyticus, S. sciuri, S. vitulinus, S. xylosus |
| FR1 | S. equorum, S. xylosus (23) | S. carnosus, S. equorum, S. saprophyticus, S. xylosus |
| FR2 | S. equorum, S. xylosus (13) | S. carnosus, S. vitulinus, S. xylosus |
| FR3 | S. equorum, S. xylosus (10) | S. carnosus, S. equorum, S. saprophyticus, S. xylosus |
| IT1 | S. equorum, S. xylosus (12) | S. equorum, S. succinus, S. xylosus |
| IT2 | S. xylosus (13) | S. carnosus, S. equorum, S. gallinarum, S. kloosii, S. saprophyticus, S. sciuri, S. xylosus |
| IT3 | S. xylosus (20) | S. equorum, S. saprophyticus, S. vitulinus, S. xylosus |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Van Reckem, E.; Charmpi, C.; Van der Veken, D.; Borremans, W.; De Vuyst, L.; Weckx, S.; Leroy, F. Application of a High-Throughput Amplicon Sequencing Method to Chart the Bacterial Communities that Are Associated with European Fermented Meats from Different Origins. Foods 2020, 9, 1247. https://doi.org/10.3390/foods9091247
Van Reckem E, Charmpi C, Van der Veken D, Borremans W, De Vuyst L, Weckx S, Leroy F. Application of a High-Throughput Amplicon Sequencing Method to Chart the Bacterial Communities that Are Associated with European Fermented Meats from Different Origins. Foods. 2020; 9(9):1247. https://doi.org/10.3390/foods9091247
Chicago/Turabian StyleVan Reckem, Emiel, Christina Charmpi, David Van der Veken, Wim Borremans, Luc De Vuyst, Stefan Weckx, and Frédéric Leroy. 2020. "Application of a High-Throughput Amplicon Sequencing Method to Chart the Bacterial Communities that Are Associated with European Fermented Meats from Different Origins" Foods 9, no. 9: 1247. https://doi.org/10.3390/foods9091247
APA StyleVan Reckem, E., Charmpi, C., Van der Veken, D., Borremans, W., De Vuyst, L., Weckx, S., & Leroy, F. (2020). Application of a High-Throughput Amplicon Sequencing Method to Chart the Bacterial Communities that Are Associated with European Fermented Meats from Different Origins. Foods, 9(9), 1247. https://doi.org/10.3390/foods9091247






