The Role of Yeasts and Lactic Acid Bacteria on the Metabolism of Organic Acids during Winemaking
Abstract
1. Introduction
2. Organic Acids of Grape Juice
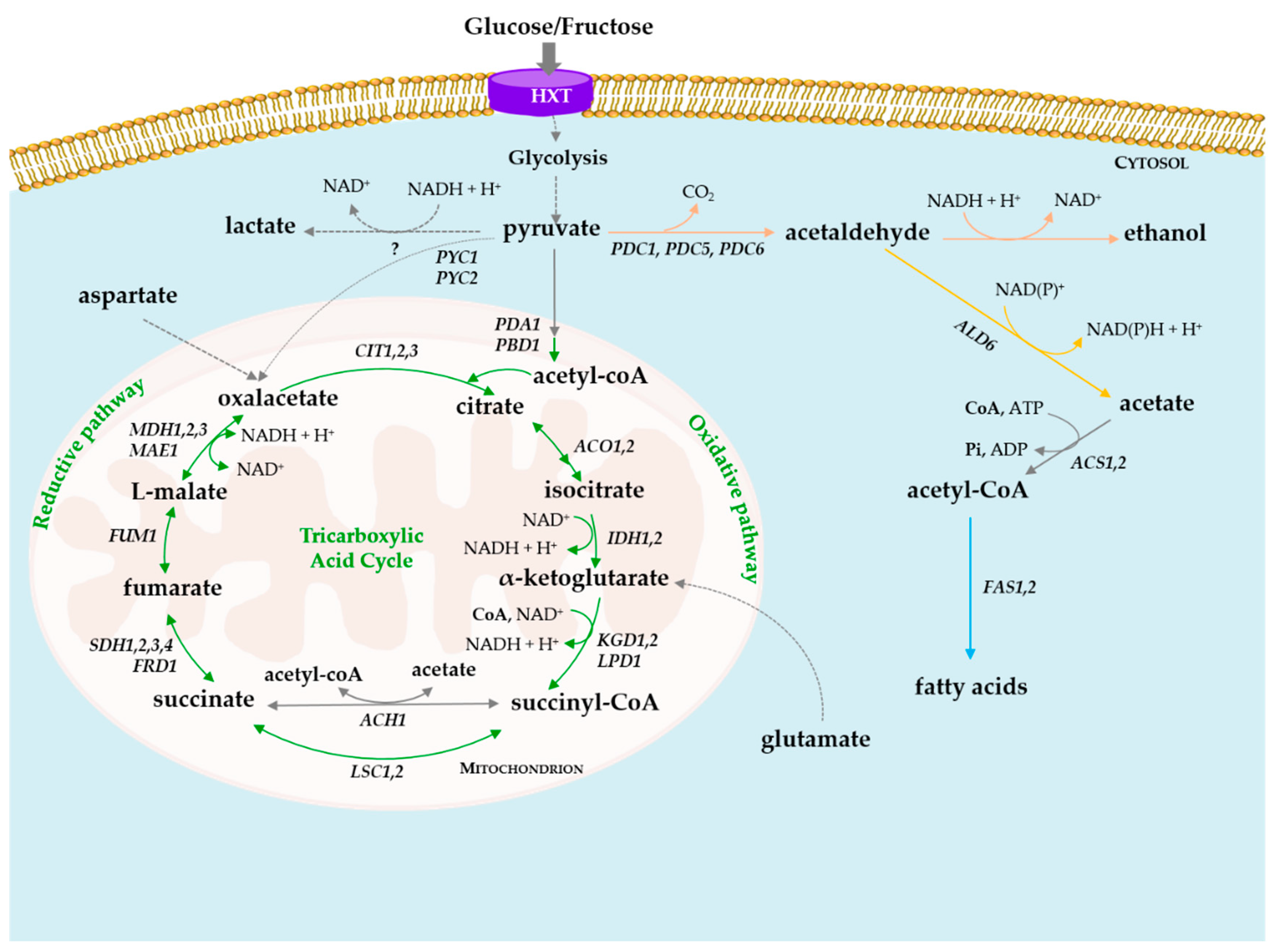
3. Degradation of Organic Acids by Yeasts
4. De Novo Synthesis of Organic Acids
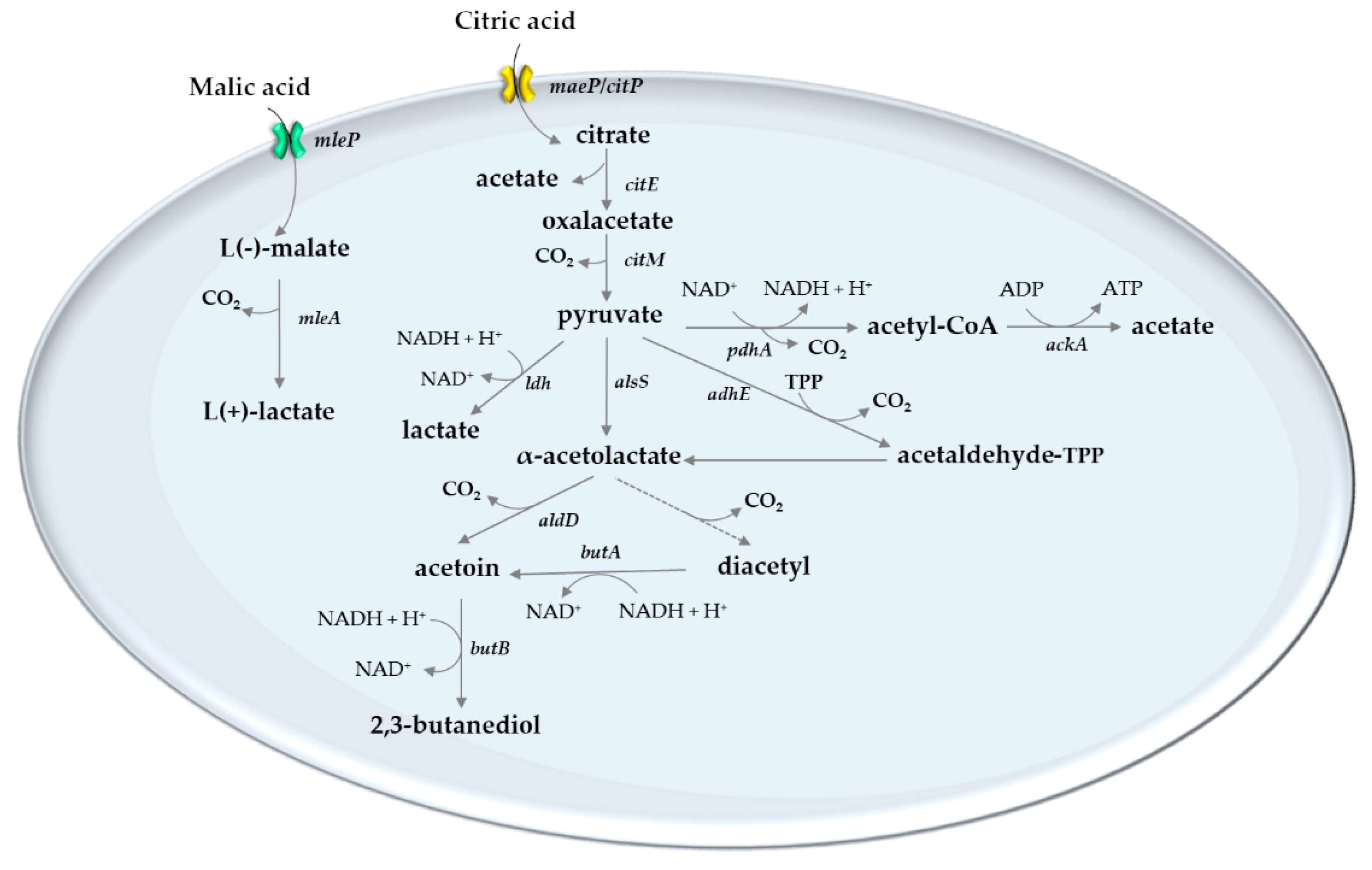
5. Lactic Acid Bacteria of the Wine
6. Role of Lactic Acid Bacteria on Acid Modulation
7. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Boulton, B.; Singleton, V.L.; Bisson, L.F.; Kunkee, R.E. Principles and Practices of Winemaking; Chapman and Hall: New York, NY, USA, 1996. [Google Scholar]
- Volschenk, H.; van Vuuren, H.J.J.; Viljoen-Bloom, M. Malic Acid in Wine: Origin, Function and Metabolism during Vinification. S. Afr. J. Enol. Vitic. 2006, 27, 123–136. [Google Scholar] [CrossRef]
- Ribéreau-Gayon, P.; Dubourdieu, D.; Donéche, B.; Lonvaud, A. Handbook of Enology, Volume 1: The Microbiology of Wine and Vinifications, 2nd ed.; John Wiley & Sons Ltd.: Chichester, UK, 2006. [Google Scholar]
- Sweetman, C.; Deluc, L.G.; Cramer, G.R.; Ford, C.M.; Soole, K.L. Regulation of malate metabolism in grape berry and other developing fruits. Phytochemistry 2009, 70, 1329–1344. [Google Scholar] [CrossRef] [PubMed]
- DeBolt, S.; Cook, D.R.; Ford, C.M. l-tartaric acid synthesis from vitamin C in higher plants. Proc. Natl. Acad. Sci. USA 2006, 103, 5608–5613. [Google Scholar] [CrossRef] [PubMed]
- Cholet, C.; Claverol, S.; Claisse, O.; Rabot, A.; Osowsky, A.; Dumot, V.; Ferrari, G.; Gény, L. Tartaric acid pathways in Vitis vinifera L. (cv. Ugni blanc): A comparative study of two vintages with contrasted climatic conditions. BMC Plant Biol. 2016, 16, 144. [Google Scholar] [CrossRef]
- Berbegal, C.; Fragasso, M.; Russo, P.; Bimbo, F.; Grieco, F.; Spano, G.; Capozzi, V. Climate Changes and Food Quality: The Potential of Microbial Activities as Mitigating Strategies in the Wine Sector. Fermentation 2019, 5, 85. [Google Scholar] [CrossRef]
- Fonseca, A. Utilization of tartaric acid and related compounds by yeasts: Taxonomic implications. Can. J. Microbiol. 1992, 38, 1242–1251. [Google Scholar] [CrossRef]
- Gao, C.; Fleet, G.H. Degradation of malic and tartaric acids by high density cell suspensions of wine yeasts. Food Microbiol. 1995, 12, 65–71. [Google Scholar] [CrossRef]
- Radler, F. Yeasts-metabolism of organic acids. In Wine Microbiology and Biotechnology; Fleet, G.H., Ed.; Harwood Academic Publishers: Chur, Switzerland, 1993; pp. 165–182. [Google Scholar]
- Peinado, R.A.; Mauricio, J.C.; Moreno, J. Aromatic series in sherry wines with gluconic acid subjected to different biological aging conditions by Saccharomyces cerevisiae var. capensis. Food Chem. 2006, 94, 232–239. [Google Scholar] [CrossRef]
- Zhong, W.; Chen, T.; Yang, H.; Li, E. Isolation and Selection of Non-Saccharomyces Yeasts Being Capable of Degrading Citric acid and Evaluation Its Effect on Kiwifruit Wine Fermentation. Fermentation 2020, 6, 25. [Google Scholar] [CrossRef]
- Barnett, J.A.; Kornberg, H.L. The utilisation by yeast of acids of the tricarboxylic acid cycle. J. Gen. Microbiol. 1960, 23, 65–82. [Google Scholar] [CrossRef]
- Côrte-Real, M.; Leão, C.; van Uden, N. Transport of L(−)malic acid and other dicarboxylic acids in the yeast Candida sphaerica. Appl. Microbiol. Biotechnol. 1989, 31, 551–555. [Google Scholar] [CrossRef]
- Cássio, F.; Leão, C. A comparative study on the transport of L(-)malic acid and other short-chain carboxylic acids in the yeast Candida utilis: Evidence for a general organic acid permease. Yeast 1993, 9, 743–752. [Google Scholar] [CrossRef] [PubMed]
- Côrte-Real, M.; Leão, C. Transport of malic acid and other dicarboxylic acids in the yeast Hansenula anomala. Appl. Environ. Microbiol. 1990, 56, 1109–1113. [Google Scholar] [CrossRef] [PubMed]
- Camarasa, C.; Bidard, F.; Bony, M.; Barre, P.; Dequin, S. Characterization of Schizosaccharomyces pombe malate permease by expression in Saccharomyces cerevisiae. Appl. Environ. Microbiol. 2001, 67, 4144–4151. [Google Scholar] [CrossRef] [PubMed]
- Osothsilp, C.; Subden, R.E. Malate transport in Schizosaccharomyces pombe. J. Bacteriol. 1986, 168, 1439–1443. [Google Scholar] [CrossRef] [PubMed]
- Baranowski, K.; Radler, F. The glucose-dependent transport of l-malate in Zygosaccharomyces bailii. Antonie van Leeuwenhoek 1984, 50, 329–340. [Google Scholar] [CrossRef]
- Ansanay, V.; Dequin, S.; Camarasa, C.; Schaeffer, V.; Grivet, J.; Blondin, B.; Salmon, J.; Barre, P. Malolactic fermentation by engineered Saccharomyces cerevisiae as compared with engineered Schizosaccharomyces pombe. Yeast 1996, 12, 215–225. [Google Scholar] [CrossRef]
- Volschenk, H.; Viljoen, M.; Grobler, J.; Petzold, B.; Bauer, F.; Subden, R.E.; Young, R.A.; Lonvaud, A.; Denayrolles, M.; van Vuuren, H.J.J. Engineering pathways for malate degradation in Saccharomyces cerevisiae. Nat. Biotechnol. 1997, 15, 253–257. [Google Scholar] [CrossRef]
- Sousa, M.J.; Mota, M.; Leão, C. Transport of malic acid in the yeast Schizosaccharomyces pombe: Evidence for a proton-dicarboxylate symport. Yeast 1992, 8, 1025–1031. [Google Scholar] [CrossRef]
- Grobler, J.; Bauer, F.; Subden, R.E.; Van Vuuren, H.J. The mael gene of Schizosaccharomyces pombe encodes a permease for malate and other C4 dicarboxylic acids. Yeast 1995, 11, 1485–1491. [Google Scholar] [CrossRef]
- Saayman, M.; Viljoen-Bloom, M. The Biochemistry of Malic Acid Metabolism by Wine Yeasts—A Review. S. Afr. J. Enol. Vitic. 2006, 27, 113–122. [Google Scholar] [CrossRef]
- Palmieri, L.; Vozza, A.; Hönlinger, A.; Dietmeier, K.; Palmisano, A.; Zara, V.; Palmieri, F. The mitochondrial dicarboxylate carrier is essential for the growth of Saccharomyces cerevisiae on ethanol or acetate as the sole carbon source. Mol. Microbiol. 1999, 31, 569–577. [Google Scholar] [CrossRef] [PubMed]
- Salmon, J.M. l-Malic acid permeation in resting cells of anaerobically grown Saccharomyces cerevisae. Biochim. Biophys. Acta 1987, 901, 30–34. [Google Scholar] [CrossRef]
- Chang, G.G.; Tong, L. Structure and function of malic enzymes, a new class of oxidative decarboxylases. Biochemistry 2003, 42, 12721–12733. [Google Scholar] [CrossRef] [PubMed]
- Volschenk, H.; van Vuuren, H.J.; Viljoen-Bloom, M. Malo-ethanolic fermentation in Saccharomyces and Schizosaccharomyces. Curr. Genet. 2003, 43, 379–391. [Google Scholar] [CrossRef]
- Tronconi, M.A.; Andreo, C.S.; Drincovich, M.F. Chimeric Structure of Plant Malic Enzyme Family: Different Evolutionary Scenarios for NAD- and NADP-Dependent Isoforms. Front. Plant Sci. 2018, 9, 565. [Google Scholar] [CrossRef]
- Zelle, R.M.; Harrison, J.C.; Pronk, J.T.; van Maris, A.J.A. Anaplerotic Role for Cytosolic Malic Enzyme in Engineered Saccharomyces cerevisiae Strains. Appl. Environ. Microbiol. 2011, 77, 732–738. [Google Scholar] [CrossRef]
- Boles, E.; de Jong-Gubbels, P.; Pronk, J.T. Identification and Characterization of MAE1, the Saccharomyces cerevisiae Structural Gene Encoding Mitochondrial Malic Enzyme. J. Bacteriol. 1998, 180, 2875–2882. [Google Scholar] [CrossRef]
- Xu, Y.; Bhargava, G.; Wu, H.; Loeber, G.; Tong, L. Crystal structure of human mitochondrial NAD(P)+-dependent malic enzyme: A new class of oxidative decarboxylases. Structure 1999, 7, 877–889. [Google Scholar] [CrossRef]
- Voegele, R.T.; Mitsch, M.J.; Finan, T.M. Characterization of two members of a novel malic enzyme class. Biochim. Biophys. Acta 1999, 1432, 275–285. [Google Scholar] [CrossRef]
- Gerrard-Wheeler, M.C.; Arias, C.L.; Tronconi, M.A.; Maurino, V.G.; Andreo, C.S.; Drincovich, M.F. Arabidopsis thaliana NADP-malic enzyme isoforms: High degree of identity but clearly distinct properties. Plant Mol. Biol. 2008, 67, 231–242. [Google Scholar] [CrossRef] [PubMed]
- Polakis, E.S.; Bartley, W. Changes in the enzyme activities of Saccharomyces cerevisiae during aerobic growth on different carbon sources. Biochem. J. 1965, 97, 284–297. [Google Scholar] [CrossRef] [PubMed]
- Stucka, R.; Dequin, S.; Salmon, J.; Gancedo, C. DNA sequences in chromosomes II and VII code for pyruvate carboxylase isoenzymes in Saccharomyces cerevisiae: Analysis of pyruvate carboxylase-deficient strains. Mol. Gen. Genet. 1991, 229, 307–315. [Google Scholar] [CrossRef] [PubMed]
- Maaheimo, H.; Fiaux, J.; Cakar, Z.; Bailey, J.; Sauer, U.; Szyperski, T. Central carbon metabolism of Saccharomyces cerevisiae explored by biosynthetic fractional 13C labeling of common amino acids. Eur. J. Biochem. 2001, 268, 2464–2479. [Google Scholar] [CrossRef] [PubMed]
- Minarik, P.; Tomaskova, N.; Kollarova, M.; Antalik, M. Malate dehydrogenases-structure and function. Gen. Physiol. Biophys. 2002, 21, 257–265. [Google Scholar]
- Minard, K.I.; McAlister-Henn, L. Isolation, nucleotide sequence analysis, and disruption of the MDH2 gene from Saccharomyces cerevisiae: Evidence for three isozymes of yeast malate dehydrogenase. Mol. Cell. Biol. 1991, 11, 370–380. [Google Scholar] [CrossRef]
- Steffan, J.S.; McAlister-Henn, L. Isolation and characterization of the yeast gene encoding the MDH3 isozyme of malate dehydrogenase. J. Biol. Chem. 1992, 267, 24708–24715. [Google Scholar]
- Moriyama, S.; Nishio, K.; Mizushima, T. Structure of Glyoxysomal Malate Dehydrogenase (MDH3) From Saccharomyces Cerevisiae. Acta Crystallogr. F Struct. Biol. Commun. 2018, 74, 617–624. [Google Scholar] [CrossRef]
- Sakihama, Y.; Hidese, R.; Hasunuma, T.; Kondo, A. Increased flux in acetyl-CoA synthetic pathway and TCA cycle of Kluyveromyces marxianus under respiratory conditions. Nat. Sci. Rep. 2019, 9, 5319. [Google Scholar] [CrossRef]
- Rankine, B.C. Decomposition of l-malic acid by wine yeasts. J. Sci. Food Agricult. 1966, 17, 312–316. [Google Scholar] [CrossRef]
- Redzepovic, S.; Orlic, S.; Majdak, A.; Kozina, B.; Volschenk, H.; Viljoen-Bloom, M. Differential malic acid degradation by selected strains of Saccharomyces during alcoholic fermentation. Int. J. Food Microbiol. 2003, 83, 49–61. [Google Scholar] [CrossRef]
- Belda, I.; Navascués, E.; Marquina, D.; Santos, A.; Calderon, F.; Benito, S. Dynamic analysis of physiological properties of Torulaspora delbrueckii in wine fermentations and its incidence on wine quality. Appl. Microbiol. Biotechnol. 2015, 99, 1911–1922. [Google Scholar] [CrossRef]
- Ruiz, J.; Belda, I.; Beisert, B.; Navascués, E.; Marquina, D.; Calderón, F.; Rauhut, D.; Santos, A.; Benito, S. Analytical impact of Metschnikowia pulcherrima in the volatile profile of Verdejo white wines. Appl. Microbiol. Biotechnol. 2018, 102, 8501–8509. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Hong, Y.; Park, H. Co-fermentation of grape must by Issatchenkia orientalis and Saccharomyces cerevisiae reduces the malic acid content in wine. Biotechnol. Lett. 2008, 30, 1633–1638. [Google Scholar] [CrossRef] [PubMed]
- Du Plessis, H.; Du Toit, M.; Hoff, J.; Hart, R.; Ndimba, B.; Jolly, N. Characterisation of Non-Saccharomyces Yeasts Using Different Methodologies and Evaluation of their Compatibility with Malolactic Fermentation. S. Afr. J. Enol. Vitic. 2017, 38, 46–63. [Google Scholar] [CrossRef]
- Benito, A.; Jeffares, D.C.; Palomero, F.; Calderón, F.; Bai, F.-Y.; Bähler, J.; Benito, S. Selected Schizosaccharomyces pombe Strains Have Characteristics That Are Beneficial for Winemaking. PLoS ONE 2016. [Google Scholar] [CrossRef]
- Kuczynski, J.T.; Radler, F. The anaerobic metabolism of malate of Saccharomyces bailii and the partial purification of malic enzyme. Arch. Microbiol. 1982, 131, 266–270. [Google Scholar] [CrossRef]
- Fuck, E.; Stärk, G.; Radler, F. Malic acid metabolism in Saccharomyces. II. Partial purification and characteristics of a “malic” enzyme. Arch. Mikrobiol. 1973, 89, 223–231. [Google Scholar] [CrossRef]
- Husnik, J.I.; Volschenk, H.; Bauer, J.; Colavizza, D.; Luo, Z.; van Vuuren, H.J. Metabolic engineering of malolactic wine yeast. Metab. Eng. 2006, 8, 315–323. [Google Scholar] [CrossRef]
- De Klerk, J.-L. Succinic Acid Production by Wine Yeasts. Master’s Thesis, University of Stellenbosch, Stellenbosch, South Africa, 2010. [Google Scholar]
- Whiting, G.S. Organic acid metabolism of yeasts during fermentation of alcoholic beverages—A review. J. Inst. Brew. 1976, 82, 84–92. [Google Scholar] [CrossRef]
- Chidi, B.S.; Rossouw, D.; Buica, A.S.; Bauer, F.F. Determining the Impact of Industrial Wine Yeast Strains on Organic Acid Production Under White and Red Wine-like Fermentation Conditions. S. Afr. J. Enol. Vitic. 2015, 36, 316–327. [Google Scholar] [CrossRef]
- Heerde, E.; Radler, F. Metabolism of the anaerobic formation of succinic acid by Saccharomyces cerevisiae. Arch. Microbiol. 1978, 117, 269–276. [Google Scholar] [CrossRef]
- Arikawa, Y.; Kobayashi, M.; Kodaira, R.; Shimosaka, M.; Muratsubaki, H.; Enomoto, K.; Okazaki, M. Isolation of sake yeast strains possessing various levels of succinate- and/or malate producing abilities by gene disruption or mutation. J. Biosci. Bioeng. 1999, 87, 333–339. [Google Scholar] [CrossRef]
- Morin, P.J.; Subramanian, G.S.; Gilmore, T.D. AAT1, a gene encoding a mitochondrial aspartate aminotransferase in Saccharomyces cerevisiae. Biochim. Biophys. Acta 1992, 1171, 211–214. [Google Scholar] [CrossRef]
- Cronin, V.B.; Maras, B.; Barra, D.; Doonan, S. The amino acid sequence of the aspartate aminotransferase from baker’s yeast (Saccharomyces cerevisiae). Biochem. J. 1991, 277, 335–340. [Google Scholar] [CrossRef] [PubMed]
- Peter, J.J.; Watson, T.L.; Walker, M.E.; Gardner, J.M.; Lang, T.A.; Borneman, A.; Forgan, A.; Tran, T.; Jiranek, V. Use of a wine yeast deletion collection reveals genes that influence fermentation performance under low-nitrogen conditions. FEMS Yeast Res. 2018, 18. [Google Scholar] [CrossRef]
- Miller, S.M.; Magasanik, B. Role of NAD-linked glutamate dehydrogenase in nitrogen metabolism in Saccharomyces cerevisiae. J. Bacteriol. 1990, 172, 4927–4935. [Google Scholar] [CrossRef]
- Cao, J.; Barbosa, J.M.; Singh, N.K.; Locy, R.D. GABA shunt mediates thermotolerance in Saccharomyces cerevisiae by reducing reactive oxygen production. Yeast 2013, 30, 129–144. [Google Scholar] [CrossRef]
- Wiebe, M.G.; Rintala, E.; Tamminen, A.; Simolin, H.; Salusjärvi, L.; Toivari, M.; Kokkonen, J.T.; Kiuru, J.; Ketola, R.A.; Jouhten, P.; et al. Central Carbon Metabolism of Saccharomyces cerevisiae in Anaerobic, Oxygen-Limited and Fully Aerobic Steady-State Conditions and Following a Shift to Anaerobic Conditions. FEMS Yeast Res. 2008, 8, 140–154. [Google Scholar] [CrossRef]
- Verduyn, C.; Postma, E.; Scheffers, W.A.; van Dijken, J.P. Physiology of Saccharomyces cerevisiae in anaerobic glucose-limited chemostat cultures. J. Gen. Microbiol. 1990, 136, 395–403. [Google Scholar] [CrossRef]
- Arikawa, Y.; Kuroyanagi, T.; Shimosaka, M.; Muratsubaki, H.; Enomoto, K.; Kodaira, R.; Okazaki, M. Effect of gene disruptions of the TCA Cycle on production of succinic acid in Saccharomyces cerevisiae. J. Biosci. Bioeng. 1999, 87, 28–36. [Google Scholar] [CrossRef]
- Camarasa, C.; Grivet, J.-P.; Dequin, S. Investigation by 13C-NMR and tricarboxylic acid (TCA) deletion mutant analysis of pathways for succinate formation in Saccharomyces cerevisiae during anaerobic fermentation. Microbiology 2003, 149, 2669–2678. [Google Scholar] [CrossRef] [PubMed]
- Bartowsky, E.J.; Pretorius, I.S. Microbial formation and modification of flavor and off-flavor compounds in wine. In Biology of Microorganisms on Grapes, in Must and in Wine; Konig, H., Unden, G., Frohlich, J., Eds.; Springer: Heidelberg, Germany, 2009; pp. 209–232. [Google Scholar]
- Erasmus, D.J.; Cliff, M.; van Vuuren, H.J.J. Impact of yeast strain on the production of acetic acid, glycerol, and the sensory attributes of Icewine. Am. J. Enol. Vitic. 2004, 55, 371–378. [Google Scholar]
- Torrens, J.; Urpí, P.; Riu-Aumatell, M.; Vichi, S.; López-Tamames, E.; Buxaderas, S. Different commercial yeast strains affecting the volatile and sensory profile of cava base wine. Int. J. Food Microbiol. 2008, 124, 48–57. [Google Scholar] [CrossRef]
- Beltran, G.; Novo, M.; Guillamón, J.M.; Mas, A.; Rozès, N. Effect of fermentation temperature and culture media on the yeast lipid composition and wine volatile compounds. Int. J. Food Microbiol. 2008, 121, 169–177. [Google Scholar] [CrossRef]
- Bely, M.; Rinaldi, A.; Dubourdieu, D. Influence of assimilable nitrogen on volatile acidity production by Saccharomyces cerevisiae during high sugar fermentation. J. Biosci. Bioeng. 2003, 96, 507–512. [Google Scholar] [CrossRef]
- Barbosa, C.; Falco, V.; Mendes-Faia, A.; Mendes-Ferreira, A. Nitrogen addition influences formation of aroma compounds, volatile acidity and ethanol in nitrogen deficient media fermented by Saccharomyces cerevisiae wine strains. J. Biosci. Bioeng. 2009, 108, 99–104. [Google Scholar] [CrossRef]
- Remize, F.; Andrieu, E.; Dequin, S. Engineering of the Pyruvate Dehydrogenase Bypass in Saccharomyces cerevisiae: Role of the Cytosolic Mg2+ and Mitochondrial K+ Acetaldehyde Dehydrogenases Ald6p and Ald4p in Acetate Formation during Alcoholic Fermentation. Appl. Environ. Microbiol. 2000, 66, 3151–3159. [Google Scholar] [CrossRef]
- Van Rossum, H.M.; Kozak, B.U.; Niemeijer, M.S.; Duine, H.J.; Luttik, M.A.; Boer, V.M.; Kötter, P.; Daran, J.-M.G.; van Maris, A.J.A.; Pronk, J.T. Alternative reactions at the interface of glycolysis and citric acid cycle in Saccharomyces cerevisiae. FEMS Yeast Res. 2016, 16. [Google Scholar] [CrossRef]
- Ding, J.; Holzwarth, G.; Penner, M.H.; Patton-Vogt, J.; Bakalinsky, A.T. Overexpression of acetyl-CoA synthetase in Saccharomyces cerevisiae increases acetic acid tolerance. FEMS Microbiol. Lett. 2015, 362, 1–7. [Google Scholar] [CrossRef]
- Benito, S. The impacts of Lachancea thermotolerans yeast strains on winemaking. Appl. Microbiol. Biotechnol. 2018, 102, 6775–6790. [Google Scholar] [CrossRef] [PubMed]
- Porter, T.J.; Divol, B.; Setati, M.E. Lachancea yeast species: Origin, biochemical characteristics and oenological significance. Food Res. Int. 2019, 119, 378–389. [Google Scholar] [CrossRef] [PubMed]
- Hranilovic, A.; Gambetta, J.M.; Schmidtke, L.; Boss, P.K.; Grbin, P.R.; Masneuf-Pomarede, I.; Bely, M.; Albertin, W.; Jiranek, V. Oenological traits of Lachancea thermotolerans show signs of domestication and allopatric differentiation. Sci. Rep. 2018, 8, 14812–14825. [Google Scholar] [CrossRef] [PubMed]
- Roullier-Gall, D.; Hemmler, D.; Schmitt-Kopplin, P.; Alexandre, H. Exploring yeast interactions through metabolic profiling. Sci. Rep. 2020, 10, 6073. [Google Scholar] [CrossRef] [PubMed]
- Englezos, V.; Cocolin, L.; Rantsiou, K.; Ortiz-Julien, A.; Bloem, A.; Dequin, S.; Camarasa, C. Specific Phenotypic Traits of Starmerella bacillaris Related to Nitrogen Source Consumption and Central Carbon Metabolite Production during Wine Fermentation. Appl. Environ. Microbiol. 2018, 84, e00797-18. [Google Scholar] [CrossRef] [PubMed]
- Mason, A.B.; Dufour, J.P. Alcohol acetyltransferases and the significance of ester synthesis in yeast. Yeast 2000, 16, 1287–1298. [Google Scholar] [CrossRef]
- Saerens, S.M.; Verstrepen, K.J.; Van Laere, S.D.; Voet, A.R.; Van Dijck, P.; Delvaux, F.R.; Thevelein, J.M. The Saccharomyces cerevisiae EHT1 and EEB1 genes encode novel enzymes with medium-chain fatty acid ethyl ester synthesis and hydrolysis capacity. J. Biol. Chem. 2006, 281, 4446–4456. [Google Scholar] [CrossRef]
- Knight, M.J.; Bull, I.D.; Curnow, P. The yeast enzyme Eht1 is an octanoyl-CoA:ethanol acyltransferase that also functions as a thioesterase. Yeast 2014, 31, 463–474. [Google Scholar] [CrossRef]
- Mendes-Ferreira, A.; Barbosa, C.; Falco, V.; Leão, C.; Mendes-Faia, A. The production hydrogen sulphide and other aroma compounds production by wine strains of Saccharomyces cerevisiae in synthetic media with different nitrogen concentrations. J. Ind. Microbiol. Biotechnol. 2009, 36, 571–583. [Google Scholar] [CrossRef]
- Peinado, R.A.; Moreno, J.; Bueno, J.E.; Moreno, J.A.; Mauricio, J.C. Comparative study of aromatic compounds in two young white wines subjected to pre-fermentative cryomaceration. Food Chem. 2004, 84, 585–590. [Google Scholar] [CrossRef]
- Beltran, G.; Novo, M.; Leberre, V.; Sokol, S.; Labourdette, D.; Guillamon, J.M.; Mas, A.; Francois, J.; Rozes, N. Integration of transcriptomic and metabolic analyses for understanding the global responses of low-temperature winemaking fermentations. FEMS Yeast Res. 2006, 6, 1167–1183. [Google Scholar] [CrossRef] [PubMed]
- Bisson, L.F.; Karpel, J.E. Genetics of Yeast Impacting Wine Quality. Annu. Rev. Food Sci. Technol. 2010, 1, 139–162. [Google Scholar] [CrossRef] [PubMed]
- Pretorius, I.S. Tasting the terroir of wine yeast innovation. FEMS Yeast Res. 2020, 20, foz084. [Google Scholar] [CrossRef] [PubMed]
- Mendes-Ferreira, A.; Barbosa, C.; Lage, P.; Mendes-Faia, A. The impact of nitrogen on yeast fermentation and wine quality. Cienc. Tec. Vitivinic. 2011, 26, 17–32. [Google Scholar]
- Tang, X.; Lee, J.; Chen, W.N. Engineering the fatty acid metabolic pathway in Saccharomyces cerevisiae for advanced biofuel production. Metab. Eng. Commun. 2015, 2, 58–66. [Google Scholar] [CrossRef]
- Schweizer, M.; Lebert, C.; Höltke, J.; Roberts, L.M.; Schweizer, E. Molecular cloning of the yeast fatty acid synthetase genes, FAS1 and FAS2: Illustrating the structure of the FAS1 cluster gene by transcript mapping and transformation studies. Mol. Gen. Genet. 1984, 194, 457. [Google Scholar] [CrossRef]
- Lilly, M.; Bauer, F.F.; Lambrechts, M.G.; Swiegers, J.H.; Cozzolino, D.; Pretorius, I.S. The effect of increased yeast alcohol acetyltransferase and esterase activity on the flavour profiles of wine and distillates. Yeast 2006, 23, 641–659. [Google Scholar] [CrossRef]
- Chen, Y.; Li, F.; Guo, J.; Liu, G.; Guo, X.; Xiao, D. Enhanced ethylcaproate production of Chinese liquor yeast by overexpressing EHT1 with deleted FAA1. J. Ind. Microbiol. Biotechnol. 2014, 41, 563–572. [Google Scholar] [CrossRef]
- Rossouw, D.; Næs, T.; Bauer, F.F. Linking gene regulation and the exo-metabolome: A comparative transcriptomics approach to identify genes that impact on the production of volatile aroma compounds in yeast. BMC Genom. 2008, 9, 530. [Google Scholar] [CrossRef]
- Saerens, S.M.; Delvaux, F.R.; Verstrepen, K.J.; Thevelein, J.M. Production and biological function of volatile esters in Saccharomyces cerevisiae. Microbial Biotechnol. 2010, 3, 165–177. [Google Scholar] [CrossRef]
- Schwartz, H.; Radler, F. Formation of l-malate by Saccharomyces cerevisiae during fermentation. Appl. Microbiol. Biotechnol. 1988, 27, 553–560. [Google Scholar] [CrossRef]
- Taing, O.; Taing, K. Production of malic and succinic acids by sugar-tolerant yeast Zygosaccharomyces rouxii. Eur. Food Res. Technol. 2007, 224, 343–347. [Google Scholar] [CrossRef]
- Takao, Y.; Takahashi, T.; Yamada, T.; Goshima, T.; Isogai, A.; Sueno, K.; Fujii, T.; Akao, T. Characteristic features of the unique house sake yeast strain Saccharomyces cerevisiae Km67 used for industrial sake brewing. J. Biosci. Bioeng. 2018, 126, 617–623. [Google Scholar] [CrossRef] [PubMed]
- Nagasawa, N.; Bogaki, T.; Iwamatsu, A.; Hamachi, M.; Kumagai, C. Cloning and nucleotide sequence of the alcohol acetyltransferase II gene (ATF2) from Saccharomyces cerevisiae Kyokai No. 7. Biosci. Biotechnol. Biochem. 1998, 62, 1852–1857. [Google Scholar] [CrossRef] [PubMed]
- Verstrepen, K.J.; Van Laere, S.D.M.; Vanderhaegen, B.M.P.; Derdelinckx, G.; Dufour, J.-P.; Pretorius, I.S.; Winderickx, J.; Thevelein, J.M.; Delvaux, F.R. Expression levels of the yeast alcohol acetyltransferase genes ATF1, Lg-ATF1, and ATF2 control the formation of a broad range of volatile esters. Appl. Environ. Microbiol. 2003, 69, 5228–5237. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Nielsen, J. Biobased organic acids production by metabolically engineered microorganisms. Curr. Opin Biotechnol. 2016, 37, 165–172. [Google Scholar] [CrossRef] [PubMed]
- Zelle, R.M.; de Hulster, E.; van Winden, W.A.; de Waard, P.; Dijkema, C.; Winkler, A.A.; Geertman, J.-M.A.; van Dijken, J.P.; Pronk, J.T.; van Maris, A.J.A. Malic Acid Production by Saccharomyces cerevisiae: Engineering of Pyruvate Carboxylation, Oxaloacetate Reduction, and Malate Export. Appl. Environ. Microbiol. 2008, 74, 2766–2777. [Google Scholar] [CrossRef]
- Ito, Y.; Hirasawa, T.; Shimizu, H. Metabolic engineering of Saccharomyces cerevisiae to improve succinic acid production based on metabolic profiling. Biosci. Biotechnol. Biochem. 2014, 78, 151–159. [Google Scholar] [CrossRef]
- Agren, R.; Otero, J.M.; Nielsen, J. Genome-scale modeling enables metabolic engineering of Saccharomyces cerevisiae for succinic acid production. J. Ind. Microbiol. Biotechnol. 2013, 40, 735–747. [Google Scholar] [CrossRef]
- Dequin, S.; Barre, P. Mixed lactic acid-alcoholic fermentation by S. cerevisiae expressing the Lactobacillus casei L(+)-LDH. Biotechnology 1994, 12, 173–177. [Google Scholar] [CrossRef]
- Novy, V.; Brunner, B.; Nidetzky, B. l-Lactic acid production from glucose and xylose with engineered strains of Saccharomyces cerevisiae: Aeration and carbon source influence yields and productivities. Microb. Cell Fact. 2018, 17, 59. [Google Scholar] [CrossRef] [PubMed]
- Bergey’s Manual of Systematic Bacteriology. Volume Three, The Firmicutes; De Vos, P., Garrity, G., Jones, D., Krieg, N.R., Ludwig, W., Rainey, F.A., Schleifer, K.-H., Whitman, W., Eds.; Springer: Dordrecht, The Netherlands; New York, NY, USA, 2009. [Google Scholar]
- König, H.; Fröhlich, J. Lactic Acid Bacteria. In Biology of Microorganisms on Grapes, in Must and in Wine; König, H., Unden, G., Fröhlich, J., Eds.; Springer: Berlin/Heidelberg, Germany, 2017. [Google Scholar]
- Mora-Villalobos, J.A.; Montero-Zamora, J.; Barboza, N.; Rojas-Garbanzo, C.; Usaga, J.; Redondo-Solano, M.; Schroedter, L.; Olszewska-Widdrat, A.; José Pablo López-Gómez, J.P. Multi-Product Lactic Acid Bacteria Fermentations: A Review. Fermentation 2020, 6, 23. [Google Scholar] [CrossRef]
- Dicks, L.M.T.; Dellaglio, F.; Collins, M.D. Proposal to reclassify Leuconostoc oenos as Oenococcus oeni (corrig.) gen. nov., comb. nov. Int. J. Sys. Bacteriol. 1995, 45, 395–397. [Google Scholar] [CrossRef] [PubMed]
- Kunkee, R.E. Some roles of malic acid in the malolactic fermentation in wine making. FEMS Microbiol. Rev. 1991, 88, 55–72. [Google Scholar]
- Bartowsky, E.J.; Costello, P.J.; Chambers, P.J. Emerging trends in the application of malolactic fermentation. Aus. J. Grape Wine Res. 2015, 21, 663–669. [Google Scholar] [CrossRef]
- Gonzàlez-Arenzana, L.; Santamaría, P.; López, R.; Tenario, C.; López-Alfaro, I. Ecology of indigenous lactic acid bacteria along different winemaking processes of Tempranillo red wine from La Rioja (Spain). Sci. World J. 2012, 7, 796327. [Google Scholar] [CrossRef]
- Betteridge, A.; Grbin, P.; Jiranek, V. Improving Oenococcus oeni to overcome challenges of wine malolactic fermentation. Trends Biotechnol. 2015, 33, 547–553. [Google Scholar] [CrossRef]
- Bravo-Ferrada, B.M.; Hollmann, A.; Delfederico, L.; La Hens, D.V.; Caballerol, A.; Semorile, L. Patagonian red wines: Selection of Lactobacillus plantarum isolates as potential starter cultures for malolactic fermentation. World J. Microbiol. Biotechnol. 2013, 29, 1537–1549. [Google Scholar] [CrossRef]
- Berbegal, C.; Peña, N.; Russo, P.; Grieco, F.; Pardo, I.; Ferrer, S.; Spano, G.; Capozzi, V. Technological properties of Lactobacillus plantarum strains isolated from grape must fermentation. Food Microbiol. 2016, 57, 187–194. [Google Scholar] [CrossRef]
- Du Toit, M.; Engelbrecht, L.; Lerm, E.; Krieger-Weber, S. Lactobacillus: The Next Generation of Malolactic Fermentation Starter Cultures—An Overview. Food Bioprocess. Technol. 2011, 4, 876–906. [Google Scholar] [CrossRef]
- Krieger-Weber, S.; Heras, J.M.; Suarez, C. Lactobacillus plantarum, a New Biological Tool to Control Malolactic Fermentation: A Review and an Outlook. Beverages 2020, 6, 23. [Google Scholar] [CrossRef]
- Matthews, A.; Grimaldi, A.; Walker, M.; Bartowsky, E.; Grbin, P.; Jiranek, V. Lactic Acid Bacteria as a Potential Source of Enzymes for Use in Vinification. Appl. Environ. Microbiol. 2004, 70, 5715–5731. [Google Scholar] [CrossRef] [PubMed]
- Cappello, M.S.; Zapparoli, G.; Logrieco, A.; Bartowsky, E.J. Linking wine lactic acid bacteria diversity with wine aroma and flavour. Int. J. Food Microbiol. 2017, 243, 16–27. [Google Scholar] [CrossRef] [PubMed]
- Brizuela, N.S.; Bravo-Ferrada, B.M.; Pozo-Bayón, M.A.; Semorile, L.; Tymczyszyna, E.E. Changes in the volatile profile of Pinot noir wines caused by Patagonian Lactobacillus plantarum and Oenococcus oeni strains. Food Res. Int. 2018, 106, 22–28. [Google Scholar] [CrossRef] [PubMed]
- Brizuela, N.S.; Tymczyszyn, E.; Semorile, L.C.; La Hens, D.; Delfederico, L.; Hollmann, A.; Bravo-Ferrada, B. Lactobacillus plantarum as a malolactic starter culture in winemaking: A new (old) player? Electron. J. Biotechnol. 2019, 38. [Google Scholar] [CrossRef]
- Milioni, C.; Martínez, B.; Degl’Innocenti, S.; Turchi, B.; Fratini, F.; Cerri, D.; Fischetti, R. A novel bacteriocin produced by Lactobacillus plantarum LpU4 as a valuable candidate for biopreservation in artisanal raw milk cheese. Dairy Sci. Technol. 2015, 95, 479–494. [Google Scholar] [CrossRef]
- Capozzi, V.; Russo, P.; Ladero, V.; Fernández, M.; Fiocco, D.; Alvarez, M.A.; Grieco, F.; Spano, G. Biogenic Amines Degradation by Lactobacillus plantarum: Toward a Potential Application in Wine. Front. Microbiol. 2012, 3, 122. [Google Scholar] [CrossRef]
- Alvarez, M.A.; Moreno-Arribas, M.V. The problem of biogenic amines in fermented foods and the use of potential biogenic amine-degrading microorganisms as a solution. Trends Food Sci. Technol. 2014, 39, 146–155. [Google Scholar] [CrossRef]
- Pilone, G.J.; Kunkee, R.E. Carbonic acid from decarboxylation by “malic” enzyme in lactic acid bacteria. J. Bacteriol. 1970, 103, 404–409. [Google Scholar] [CrossRef][Green Version]
- Schütz, M.; Radler, F. Das Vorkommen von Malatenzym und MaloLactat-Enzym bei verschiedenen Milchsäurebakterien. Arch. Microbiol. 1974, 96, 329–339. [Google Scholar] [CrossRef]
- London, J.; Meyer, E.Y. Malate utilization by a group d-Streptococcus: Physiological properties and purification of an inducible malic enzyme. J. Bacteriol. 1969, 98, 705–711. [Google Scholar] [CrossRef] [PubMed]
- Groisillier, A.; Lonvaud-Funel, A. Comparison of partial malolactic enzyme gene sequences for phylogenetic analysis of some lactic acid bacteria species and relationships with the malic enzyme. Int. J. Syst. Bacteriol. 1999, 49, 1417–1428. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Landete, J.M.; García-Haro, L.; Blasco, A.; Manzanares, P.; Berbegal, C.; Monedero, V.; Zúñiga, M. Requirement of the Lactobacillus casei MaeKR Two-Component System for l-Malic Acid Utilization via a Malic Enzyme Pathway. Appl. Environ. Microbiol. 2010, 76, 84–95. [Google Scholar] [CrossRef] [PubMed]
- Landete, J.M.; Ferrer, S.; Monedero, V.; Zúñiga, M. Malic enzyme and malolactic enzyme pathways are functionally linked but independently regulated in Lactobacillus casei BL23. Appl. Environ. Microbiol. 2013, 79, 5509–5518. [Google Scholar] [CrossRef] [PubMed]
- Cox, D.J.; Henick-Kling, T. Proton-motive force and ATP generation during malolactic fermentation. Am. J. Enol. Viticult. 1989, 46, 319–323. [Google Scholar]
- Salema, M.; Poolman, B.; Lolkema, J.S.; Loureiro Dias, M.C.; Konings, W.N. Uniport of monoanionic l-malate in membrane vesicles from Leuconostoc oenos. FEBS Eur. J. Biochem. 1994, 124, 1–7. [Google Scholar] [CrossRef]
- Labarre, C.; Diviès, C.; Guzzo, J. Genetic organization of the mle locus and identification of a mleR-like gene from Leuconostoc oenos. Appl. Environ. Microbiol. 1996, 62, 4493–4498. [Google Scholar] [CrossRef]
- García-Quintáns, N.; Repizo, G.; Martín, M.; Magni, C.; López, P. Activation of the Diacetyl/Acetoin Pathway in Lactococcus lactis subsp. lactis bv. diacetylactis CRL264 by Acidic Growth. Appl. Environ. Microbiol. 2008, 74, 1988–1996. [Google Scholar] [CrossRef]
- Pimentel, M.; Silva, M.; Cortês, I.; Mendes-Faia, A. Growth and metabolism of sugar and acids of Leuconostoc oenos under different conditions of temperature and pH. J. Appl. Bacteriol. 1994, 76, 42–48. [Google Scholar] [CrossRef]
- Hugenholtz, J. Citrate metabolism in lactic acid bacteria. FEMS Microbiol. Rev. 1993, 12, 165–178. [Google Scholar] [CrossRef]
- Kennes, C.; Dubourguier, H.C.; Albagnac, G.; Nyns, E.-J. Citrate metabolism by Lactobacillus plantarum isolated from orange juice. J. Appl. Bacteriol. 1991, 70, 380–384. [Google Scholar] [CrossRef]
- Ramos, A.; Lolkema, J.S.; Konings, W.N.; Santos, H. Enzyme basis for pH regulation of citrate and pyruvate metabolism by Leuconostoc oenos. Appl. Environ. Microbiol. 1995, 61, 1303–1310. [Google Scholar] [CrossRef] [PubMed]
- Kleerebezem, M.; Boekhorst, J.; Van Kranenburg, R.; Molenaar, D.; Kuipers, O.P.; Leer, R.; Tarchini, R.; Peters, S.A.; Sandbrink, H.M.; Fiers, M.W.E.J.; et al. Complete genome sequence of Lactobacillus plantarum WCFS1. Proc. Natl. Acad. Sci. USA 2003, 100, 1990–1995. [Google Scholar] [CrossRef] [PubMed]
- Lorquet, F.; Goffin, P.; Muscariello, L.; Baudry, J.-B.; Ladero, V.; Sacco, M.; Kleerebezem, M.; Hols, P. Characterization and Functional Analysis of the poxB Gene, Which Encodes Pyruvate Oxidase in Lactobacillus plantarum. J. Bact. 2004, 186, 3749–3759. [Google Scholar] [CrossRef]
- Lonvaud-Funel, A. Biogenic amines in wines: Role of lactic acid bacteria. FEMS Microbiol. Lett. 2001, 199, 9–13. [Google Scholar] [CrossRef]
- Ruiz-Capillas, C.; Herrero, A.M. Impact of Biogenic Amines on Food Quality and Safety. A review. Foods 2019, 8, 62. [Google Scholar] [CrossRef]
- Marcobal, A.; Martínez-Alvarez, P.J.; Polo, M.C.; Munõz, R.; Moreno-Arribas, M.V. Formation of Biogenic Amines throughout the Industrial Manufacture of Red Wine. J. Food Prot. 2006, 69, 397–404. [Google Scholar] [CrossRef]
- Radler, F.; Bröhl, K. The metabolism of several carboxylic acids by lactic acid bacteria. Z. Lebensm. Unters. Forsch. 1984, 179, 228–231. [Google Scholar] [CrossRef]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mendes Ferreira, A.; Mendes-Faia, A. The Role of Yeasts and Lactic Acid Bacteria on the Metabolism of Organic Acids during Winemaking. Foods 2020, 9, 1231. https://doi.org/10.3390/foods9091231
Mendes Ferreira A, Mendes-Faia A. The Role of Yeasts and Lactic Acid Bacteria on the Metabolism of Organic Acids during Winemaking. Foods. 2020; 9(9):1231. https://doi.org/10.3390/foods9091231
Chicago/Turabian StyleMendes Ferreira, Ana, and Arlete Mendes-Faia. 2020. "The Role of Yeasts and Lactic Acid Bacteria on the Metabolism of Organic Acids during Winemaking" Foods 9, no. 9: 1231. https://doi.org/10.3390/foods9091231
APA StyleMendes Ferreira, A., & Mendes-Faia, A. (2020). The Role of Yeasts and Lactic Acid Bacteria on the Metabolism of Organic Acids during Winemaking. Foods, 9(9), 1231. https://doi.org/10.3390/foods9091231




