A Preliminary Report for the Design of MoS (Micro-Olive-Spreadsheet), a User-Friendly Spreadsheet for the Evaluation of the Microbiological Quality of Spanish-Style Bella di Cerignola Olives from Apulia (Southern Italy)
Abstract
1. Introduction
2. Materials and Methods
2.1. Samples
2.2. Microbiological Analyses
2.3. pH and NaCl Amount
2.4. Sensory Score
2.5. Data Analysis
2.6. Spreadsheet Design
- LAB < 1% TAC. The function was evaluated through the exponential values of cell count, rather than with a logarithm;
- Yeast > 5;
- pH > 4.5.
- Staphylococci > 4.
- salt<8%.
3. Results
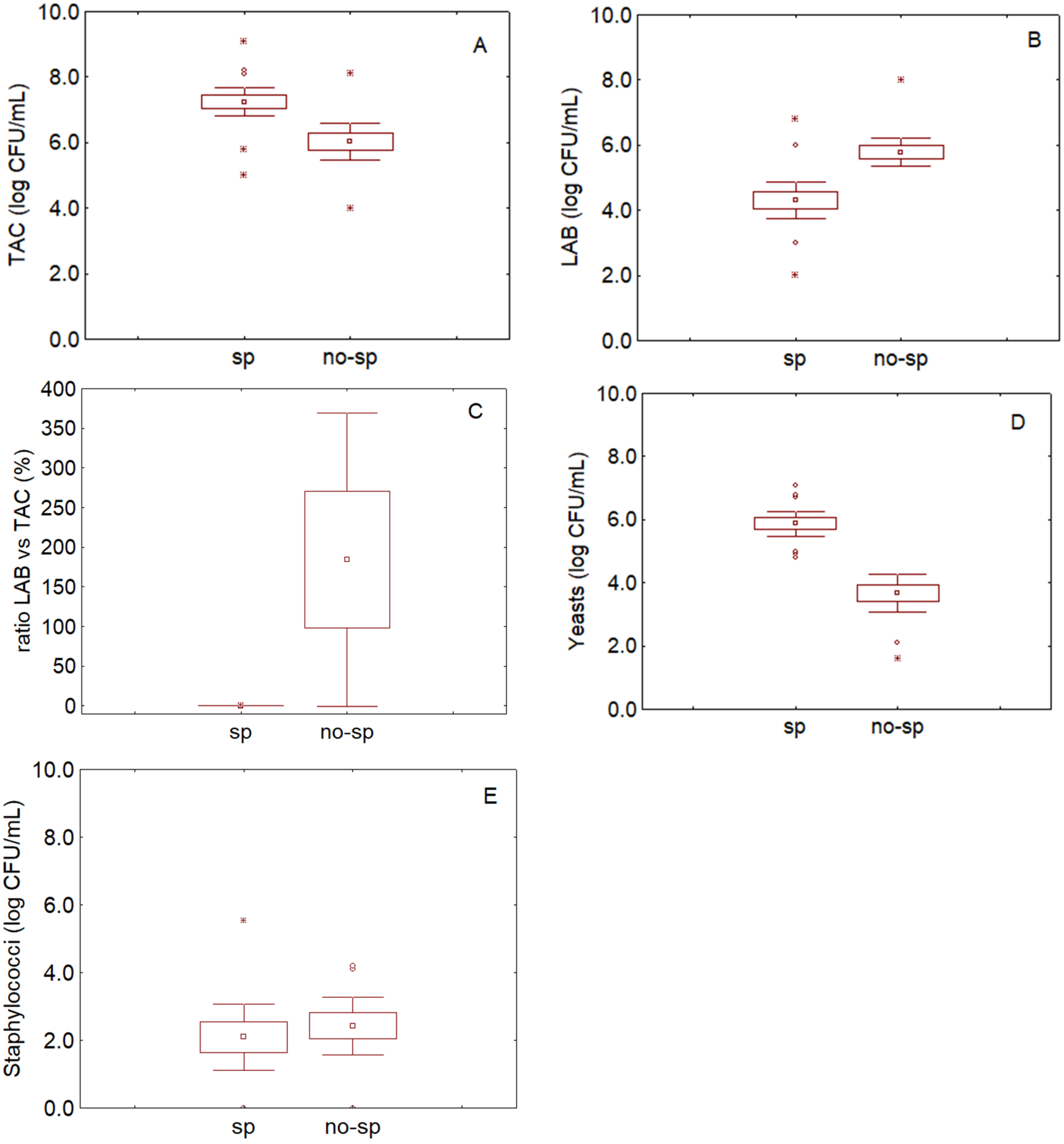
3.1. Data
3.2. MoS
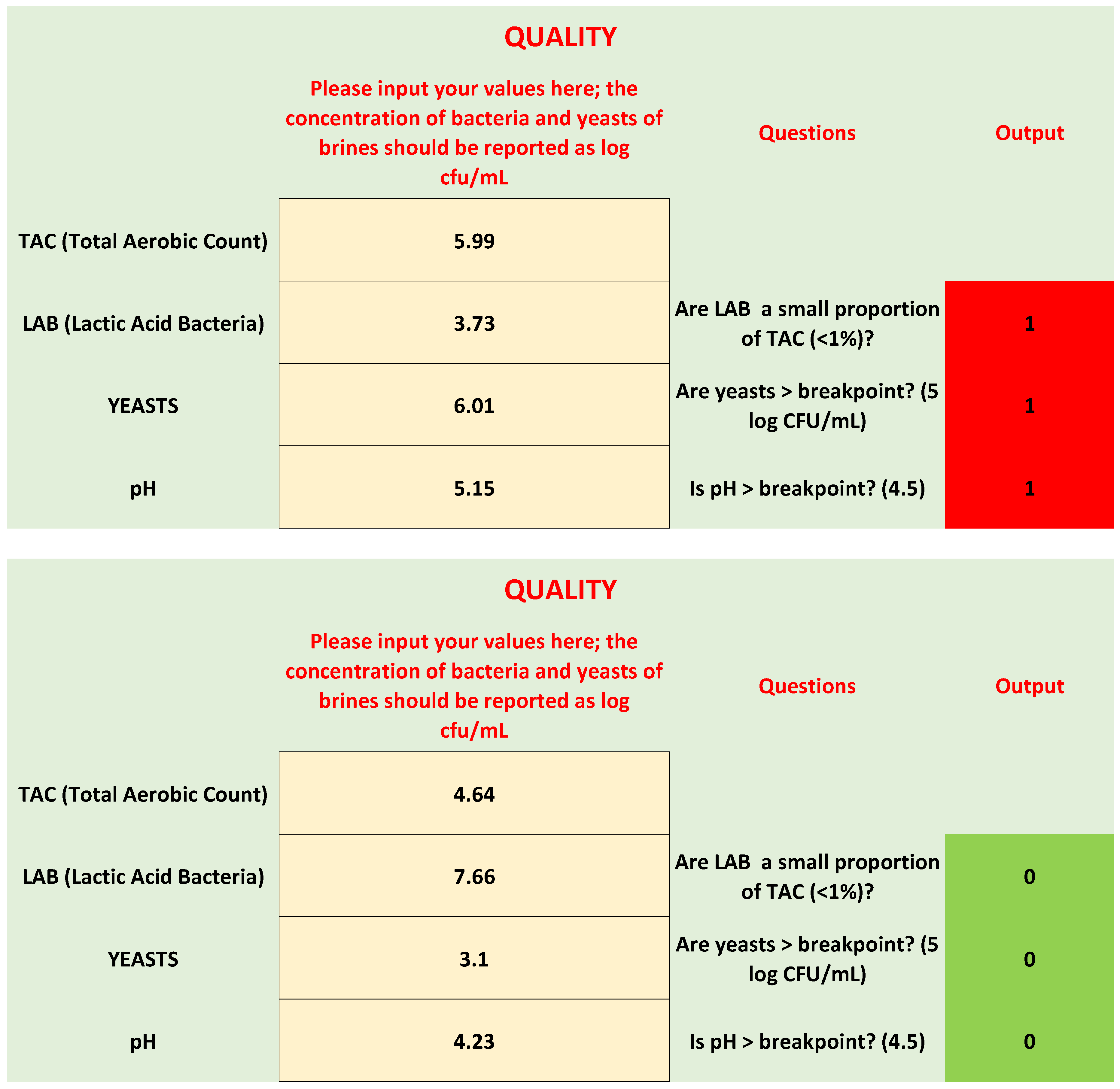
- Are LAB a small proportion of TAC in brines? This criterion was written as follows: LAB/TAC < 1%; both TAC and LAB in the equation were used as exponential values. However, user does not convert his/her values, because there is a function set in the protected cell of the “If” function, as shown in Table 1.
- Are yeasts higher than the break-point? The threshold for yeasts in brines was set to 5 log CFU/mL.
- The last criterion was on pH; although the results of the first phase did not show a clear difference between the spoiled and non-spoiled samples, a criterion on pH was added because of its role in the beginning of microbiological spoilage in the post-fermentation phase.
- Three red cells: There is probably microbiological spoilage.
- Two red cells: A correction strategy is required, because a spoilage could start or have started.
- One red cell: No spoilage and no action required; however, advice was added for pH. If pH is >breakpoint, reduce it to the break-point or, better, to 4.3.
3.3. Validation
4. Discussion
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Benítez-Cabello, A.; Romero-Gil, V.; Medinaa, E.; Sánchez, B.; Calero-Delgado, B.; Bautista-Gallego, J.; Jiménez-Díaz, R.; Arroyo-López, F.N. Metataxonomic analysis of the bacterial diversity in table olive dressing components. Food Control 2019, 105, 190–197. [Google Scholar] [CrossRef]
- Mastralexi, A.; Mantzouridou, F.T.; Tsimidou, M.Z. Evolution of safety and other quality parameters of the Greek PDO table olives “Prasines Elies Chalkidikis” during industrial scale processing and storage. Eur. J. Lipid Sci. Technol. 2019, 121, 1800171. [Google Scholar] [CrossRef]
- Perpetuini, G.; Prete, R.; Garcia-Gonzalez, N.; Alam, M.K.; Corsetti, A. Table olives more than a fermented food. Foods 2020, 9, 178. [Google Scholar] [CrossRef]
- Randazzo, C.L.; Todaro, A.; Pino, A.; Pitino, I.; Corona, O.; Caggia, C. Microbiota and metabolome during controlled and spontaneous fermentation of Nocellara Etnea table olives. Food Microbiol. 2017, 65, 136–148. [Google Scholar] [CrossRef] [PubMed]
- Fuccio, F.; Bevilacqua, A.; Sinigaglia, M.; Corbo, M.R. Using a polynomial model for fungi from table olives. Int. J. Food Sci. Technol. 2016, 51, 1276–1283. [Google Scholar] [CrossRef]
- Nychas, G.J.E.; Panagou, E.Z.; Parker, M.L.; Waldron, K.W.; Tassou, C.C. Microbial colonization of naturally black olives during fermentation and associated biochemical activities in the cover brine. Lett. Appl. Microbiol. 2002, 34, 173–177. [Google Scholar] [CrossRef]
- Romeo, F.V. Microbiological aspects of table olives. In Olive Germplasm. The Olive Cultivation, Table Olive and Olive Oil Industry in Italy; Muzzalupo, I., Ed.; InTech Publisher: Rijeka, Croatia, 2012; pp. 321–341. [Google Scholar]
- Perricone, M.; Bevilacqua, A.; Corbo, M.R.; Sinigaglia, M. Use of Lactobacillus plantarum and glucose to control the fermentation of “Bella di Cerignola” table olives, a traditional variety of Apulian region (Southern Italy). J. Food Sci. 2010, 75, M430–M436. [Google Scholar] [CrossRef]
- Bevilacqua, A.; Cannarsi, M.; Gallo, M.; Sinigaglia, M.; Corbo, M.R. Characterization and implications of Enterobacter cloacae strains, isolated from Italian table olives “Bella di Cerignola”. J. Food Sci. 2010, 75, M53–M60. [Google Scholar] [CrossRef]
- Panagou, E.Z.; Nychas, G.J.E.; Sofos, J.N. Types of traditional Greek foods and their safety. Food Control 2013, 29, 32–41. [Google Scholar] [CrossRef]
- Medina-Pradas, E.; Arroyo-López, F.N. Presence of toxic microbial metabolites in table olives. Front. Microbiol. 2015, 6, 873. [Google Scholar] [CrossRef]
- Tataridou, M.; Kotzekidou, P. Fermentation of table olives by oleuropeinolytic starter culture in reduced salt brines and inactivation of Escherichia coli O157:H7 and Listeria monocytogenes. Int. J. Food Microbiol. 2015, 208, 122–130. [Google Scholar] [CrossRef] [PubMed]
- Bevilacqua, A.; Campaniello, D.; Speranza, B.; Sinigaglia, M.; Corbo, M.R. Survival of Listeria monoctytogenes and Staphylococcus aureus in synthetic brines. Studying the effects of salt, temperature and sugar through the approach of the Design of the Experiments. Front. Microbiol. 2018, 9, 240. [Google Scholar] [CrossRef] [PubMed]
- Abriouel, H.; Benomar, N.; Lucas, R.; Gálvez, A. Culture-independent study of the diversity of microbial populations in brines during fermentation of naturally fermented Aloreña green table olives. Int. J. Food Microbiol. 2011, 144, 487–496. [Google Scholar] [CrossRef] [PubMed]
- Lavermicocca, P.; Angiolillo, L.; Lonigro, S.L.; Valerio, F.; Bevilacqua, A.; Perricone, M.; Del Nobile, M.A.; Corbo, M.R.; Conte, A. Lactobacillus plantarum 5BG survives during the refrigerated storage bio-preserving packaged Spanish-style table olives (cv. Bella di Cerignola). Front. Microbiol. 2018, 9, 889. [Google Scholar] [CrossRef] [PubMed]
- Comunian, R.; Ferrocino, I.; Paba, A.; Daga, E.; Campus, M.; Di Salvo, R.; Cauli, E.; Piras, F.; Zurru, R.; Cocolin, L. Evolution of microbiota during spontaneous and inoculated Tonda diCagliari table olives fermentation and impact on sensory characteristics. LWT-Food Sci. Technol. 2017, 84, 64–72. [Google Scholar] [CrossRef]
- Randazzo, C.L.; Todaro, A.; Pino, A.; Pitino, I.; Corona, O.; Mazzaglia, A.; Caggia, C. Giarraffa and Grossa di Spagna naturally fermented table olives: Effect of starter and probiotic cultures on chemical, microbiological and sensory traits. Food Res. Int. 2014, 62, 1154–1164. [Google Scholar] [CrossRef]
- D’Antuono, I.; Bruno, A.; Linsalata, V.; Minervini, F.; Garbetta, A.; Tufariello, M.; Mita, G.; Logrieco, A.F.; Bleve, G.; Cardinali, A. Fermented Apulian table olives: Effect of selected microbial starters on polyphenols composition, antioxidant activities and bioaccessibility. Food Chem. 2018, 248, 137–145. [Google Scholar] [CrossRef]
- Bleve, G.; Tufariello, M.; Durante, M.; Perbellini, E.; Ramires, F.A.; Grieco, F.; Cappello, M.S.; De Domenico, S.; Mita, G.; Tasioula-Margari, M.; et al. Physico-chemical and microbiological characterization of spontaneous fermentation of Cellina di Nardò and Leccino table olives. Front. Microbiol. 2014, 5, 570. [Google Scholar] [CrossRef] [PubMed]
- Lucena-Padrós, H.; Ruiz-Barba, J.L. Microbial biogeography of Spanish-style green olive fermentations in the province of Seville, Spain. Food Microbiol. 2019, 82, 259–268. [Google Scholar] [CrossRef]
- Lanza, B. Abnormal fermentations in table olive processing: microbial origin and sensory evaluation. Front. Microbiol. 2013, 4, 91. [Google Scholar] [CrossRef] [PubMed]
- Bevilacqua, A.; de Stefano, F.; Augello, S.; Pignatiello, S.; Sinigaglia, M.; Corbo, M.R. Biotechnological innovations for table olives. Int. J. Food Sci. Nutr. 2015, 66, 127–131. [Google Scholar] [CrossRef] [PubMed]
- Campaniello, D.; Bevilacqua, A.; D’Amato, D.; Corbo, M.R.; Altieri, C.; Sinigaglia, M. Microbial characterization of table olives processed according to Spanish and Natural styles. Food Technol. Biotechnol. 2005, 43, 289–294. [Google Scholar]
- International Olive Oil Council (IOC). Method. Sensory Analyses of Table Olives. COI/OT/MO No 1/Rev.2, November 2011. Available online: https://www.internationaloliveoil.org/what-we-do/chemistry-standardisation-unit/standards-and-methods/ (accessed on 28 June 2020).
- Ruiz-Bellido, M.Á.; Valero, A.; Medina Pradas, A.; Romero Gil, V.; Rodríguez-Gómez, F.; Posada-Izquierdo, G.D.; Rincón, F.; Possas, A.; García-Gimeno, R.M.; Arroyo-López, F.N. A probabilistic decision-making scoring system for quality and safety management in Aloreña de Málaga table olive processing. Front. Microbiol. 2017, 8, 2326. [Google Scholar] [CrossRef] [PubMed]
- Panagou, E.Z.; Kodogiannis, V.; Nychas, G.J.-E. Modelling fungal growth using radial basis function neural networks: The case of the ascomycetous fungus Monascus ruber van Tieghem. Int. J. Food Microbiol. 2007, 117, 276–286. [Google Scholar] [CrossRef]
- Risk Ranger. Available online: https://www.cbpremium.org/RiskRanger (accessed on 15 June 2020).
- Bevilacqua, A.; Beneduce, L.; Sinigaglia, M.; Corbo, M.R. Selection of yeasts as starter cultures for table olives. J. Food Sci. 2013, 78, M742–M751. [Google Scholar] [CrossRef]
- Bevilacqua, A.; Corbo, M.R.; Sinigaglia, M. Selection of yeasts as starter cultures for table olives: A step-by-step procedure. Front. Microbiol. 2012, 3, 194. [Google Scholar] [CrossRef]
- Fleming, H.P.; Mcfeeters, R.F.; Breidt, F. Staphylococcus aureus and staphyolococcal enterotoxins. In Compendium of methods for the Microbiological examination of Foods, 4th ed.; Downes, F.P., Ito, K., Eds.; American Public Health Association: Washington, DC, USA, 2001; pp. 387–403. [Google Scholar]
- Leventdurur, S.; Sert-Aydın, S.; Boyaci-Gunduz, C.P.; Agirman, B.; Ben Ghorbal, A.; Francesca, N.; Martorana, A.; Erten, H. Yeast biota of naturally fermented black olives in different brines made from cv. Gemlik grown in various districts of the Cukurova region of Turkey. Yeasts 2016, 33, 289–301. [Google Scholar] [CrossRef]
- Tassou, C.C.; Nychas, G.J.E. Inhibition of Staphylococcus aureus by olive phenolics in broth and in a model food system. J. Food Prot. 1994, 57, 120–124. [Google Scholar] [CrossRef]
- Perricone, M.; Gallo, M.; Corbo, M.R.; Sinigaglia, M.; Bevilacqua, A. Yeasts. In The microbiological quality of food. Foodborne spoilers; Bevilacqua, A., Corbo, M.R., Sinigaglia, M., Eds.; Elsevier: Amsterdam, The Netherlands, 2017; pp. 121–132. [Google Scholar]
- Morton, R.D. Aerobic plate count. In Compendium of Methods for the Microbiological Examination of Foods, 4th ed.; Downes, F.P., Ito, K., Eds.; American Public Health Association: Washington, DC, USA, 2001; pp. 63–67. [Google Scholar]
- Golomb, B.L.; Morales, V.; Jung, A.; Yau, B.; Boundy-Mills, K.L.; Marco, M.L. Effects of pectinolytic yeast on the microbial composition and spoilage of olive fermentations. Food Microbiol. 2013, 33, 97–106. [Google Scholar] [CrossRef]
- CODEX/COI Codex Standard for Table Olives. CODEX STAN 66-1881. Revision 1987; FAO: Rome, Italy, 2013.
- Garrido-Fernández, A.; Fernández-Díez, M.J.; Adams, R.M. Table Olives: Production and Processing, 1st ed.; Chapman and Hall: London, UK, 1997. [Google Scholar]
- Lancette, G.A.; Bennett, R.W. Fermented and acidified vegetables. In Compendium of Methods for the Microbiological Examination of Foods, 4th ed.; Downes, F.P., Ito, K., Eds.; American Public Health Association: Washington, DC, USA, 2001; pp. 521–532. [Google Scholar]
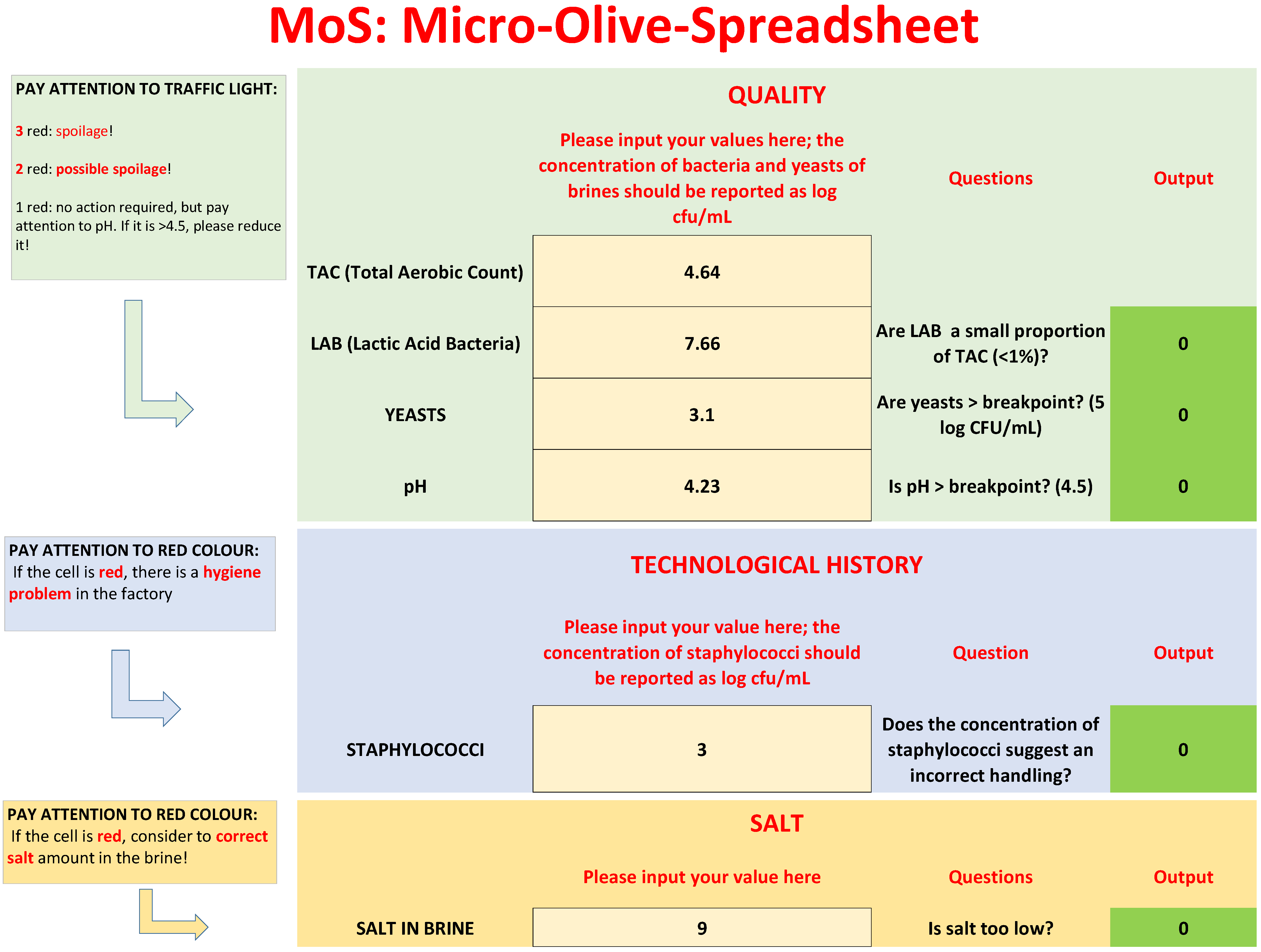
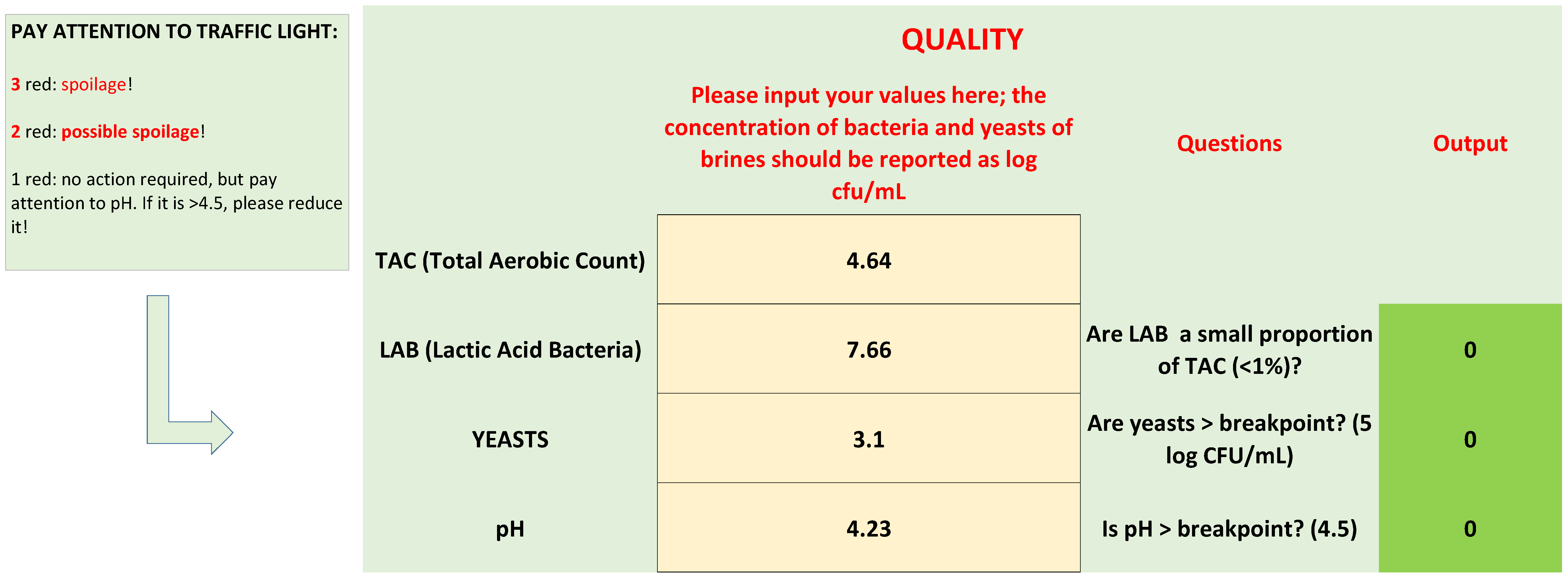
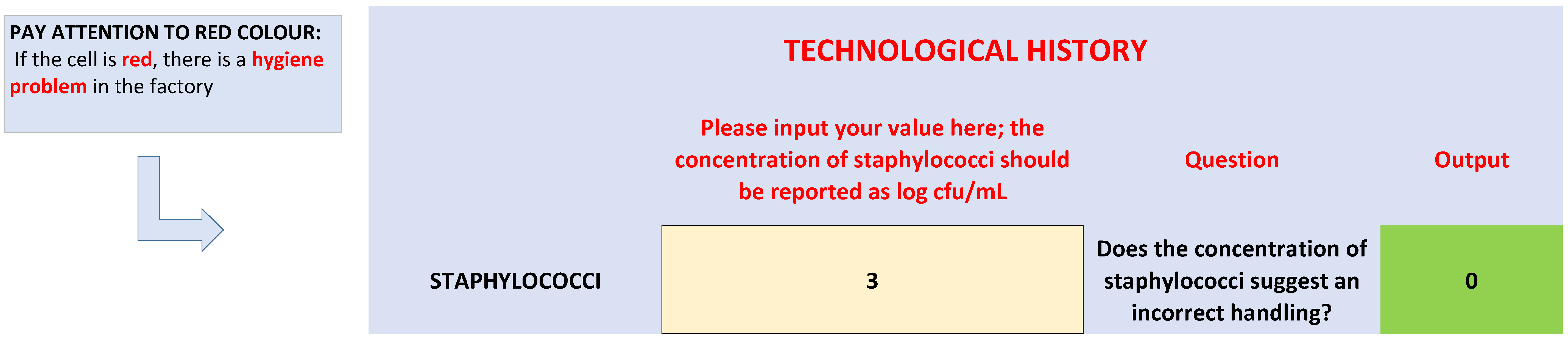

| Decision Criteria (Questions) | Equations | Yes | No |
|---|---|---|---|
| Quality | |||
| Are LAB a small proportion of bacterial microbiota? | LAB < 1% TAC | Not acceptable | Acceptable |
| Is yeast concentration > breakpoint? | Yeast > 5 | Not acceptable | Acceptable |
| Is pH > breakpoint? | pH > 4.5 | Not acceptable | Acceptable |
| Technological Process | |||
| Is handling incorrect? | Staph > 4 | Incorrect handling | - |
| Is salt too low? | Salt < 8% | Possible corrective measures | - |
| Sample | Decision by Panel | Comments | TAC | LAB | Yeasts | pH | Spreadsheet |
|---|---|---|---|---|---|---|---|
| 1 | Acceptable | Typical odor of Spanish-style olives | 2.01 | 6.65 | 4.18 | 4.05 | Acceptable |
| 2 | Not acceptable | Off-odors | 5.99 | 3.73 | 6.01 | 5.15 | Not acceptable |
| 3 | Not acceptable | Off-odors | 6.14 | 5.93 | 6.14 | 5.27 | Attention |
| 4 | Acceptable, but the sample has some problems | Films on the surface | 5.45 | 5.08 | 7.26 | 5.35 | Attention |
| 5 | Acceptable, but the sample has some problems | Strong odor | 5.53 | 2.23 | 3.46 | 4.67 | Attention |
| 6 | Acceptable | - | 4.64 | 7.66 | 3.10 | 4.23 | Acceptable |
| 7 | Acceptable, but the sample has some problems | Fruity odor is too strong | 4.82 | 7.63 | 5.87 | 4.85 | Attention |
| 8 | Acceptable | - | 4.56 | 6.12 | 4.53 | 4.43 | Acceptable |
| 9 | Acceptable | - | 4.31 | 5.99 | 3.56 | 4.36 | Acceptable |
| 10 | Not acceptable | Films on the surface | 7.80 | 5.79 | 6.01 | 4.67 | Not acceptable |
| 11 | Not acceptable | Off-odors | 5.99 | 2.34 | 5.31 | 4.89 | Not acceptable |
| 12 | Acceptable | - | 5.53 | 6.48 | 2.83 | 4.43 | Acceptable |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bevilacqua, A.; Speranza, B.; Campaniello, D.; Sinigaglia, M.; Corbo, M.R. A Preliminary Report for the Design of MoS (Micro-Olive-Spreadsheet), a User-Friendly Spreadsheet for the Evaluation of the Microbiological Quality of Spanish-Style Bella di Cerignola Olives from Apulia (Southern Italy). Foods 2020, 9, 848. https://doi.org/10.3390/foods9070848
Bevilacqua A, Speranza B, Campaniello D, Sinigaglia M, Corbo MR. A Preliminary Report for the Design of MoS (Micro-Olive-Spreadsheet), a User-Friendly Spreadsheet for the Evaluation of the Microbiological Quality of Spanish-Style Bella di Cerignola Olives from Apulia (Southern Italy). Foods. 2020; 9(7):848. https://doi.org/10.3390/foods9070848
Chicago/Turabian StyleBevilacqua, Antonio, Barbara Speranza, Daniela Campaniello, Milena Sinigaglia, and Maria Rosaria Corbo. 2020. "A Preliminary Report for the Design of MoS (Micro-Olive-Spreadsheet), a User-Friendly Spreadsheet for the Evaluation of the Microbiological Quality of Spanish-Style Bella di Cerignola Olives from Apulia (Southern Italy)" Foods 9, no. 7: 848. https://doi.org/10.3390/foods9070848
APA StyleBevilacqua, A., Speranza, B., Campaniello, D., Sinigaglia, M., & Corbo, M. R. (2020). A Preliminary Report for the Design of MoS (Micro-Olive-Spreadsheet), a User-Friendly Spreadsheet for the Evaluation of the Microbiological Quality of Spanish-Style Bella di Cerignola Olives from Apulia (Southern Italy). Foods, 9(7), 848. https://doi.org/10.3390/foods9070848







