Spectroscopic and Molecular Modeling Investigation on the Interaction between Folic Acid and Bovine Lactoferrin from Encapsulation Perspectives
Abstract
1. Introduction
2. Materials and Methods
2.1. Chemicals
2.2. Preparation of Stock Solutions
2.3. Heat Treatment
2.4. Fluorescence Spectroscopy
2.5. Molecular Modeling Investigations
2.6. Statistical Analysis
3. Results
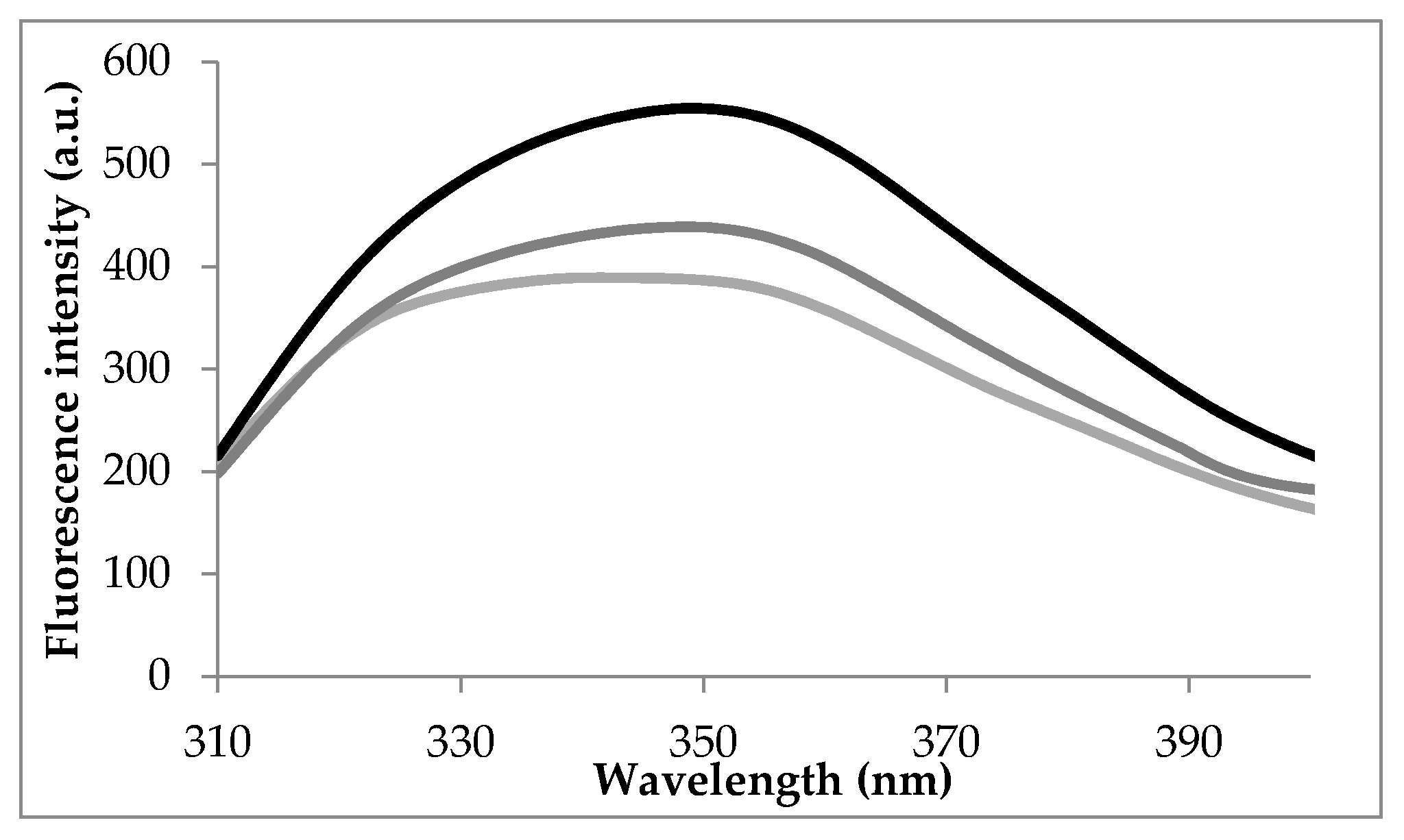
3.1. Intrinsic Fluorescence Spectra
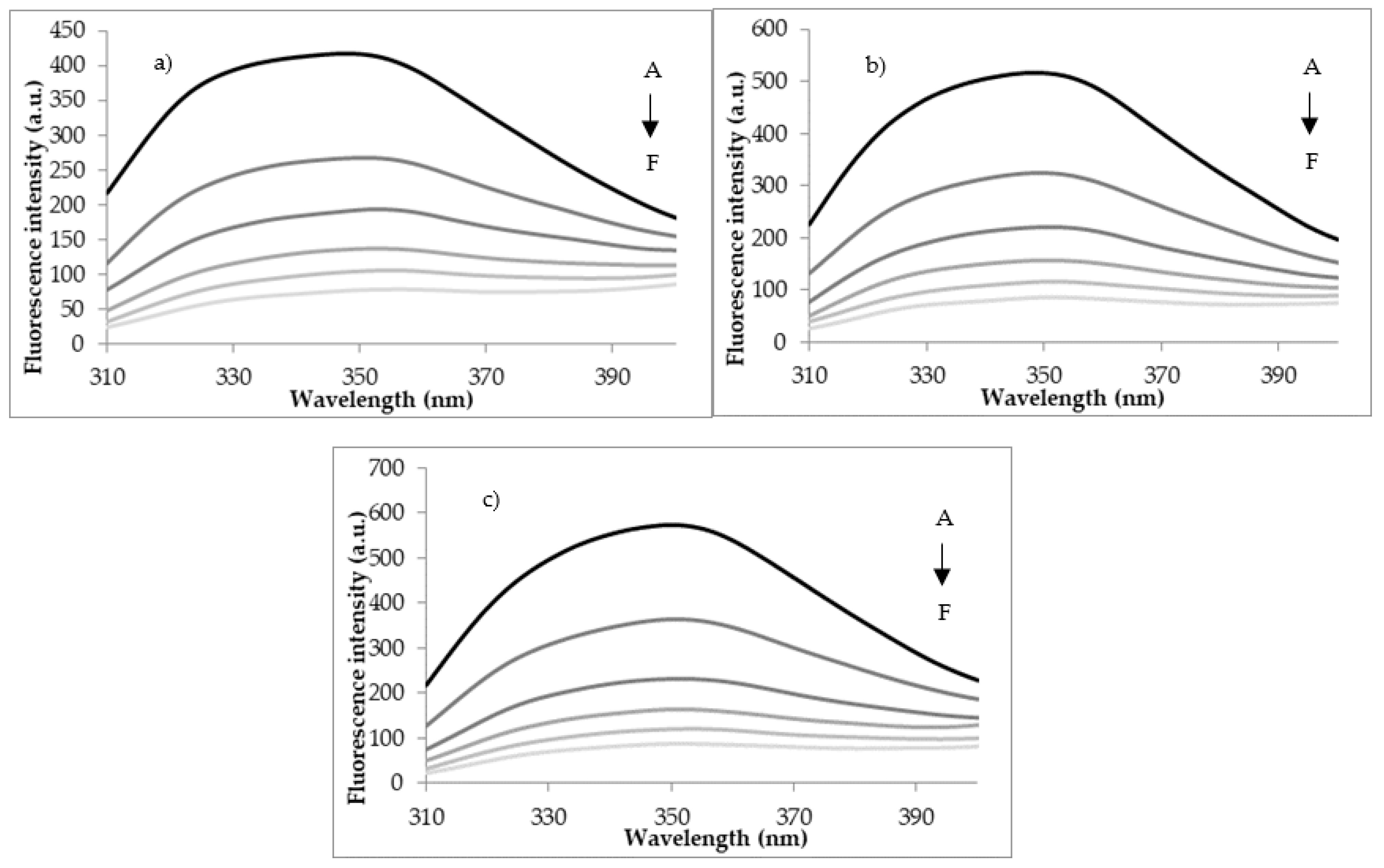
3.2. Quenching Experiments
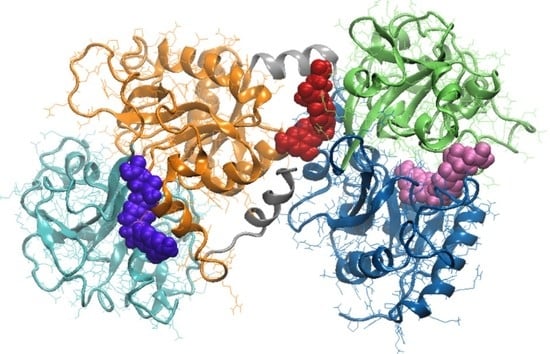
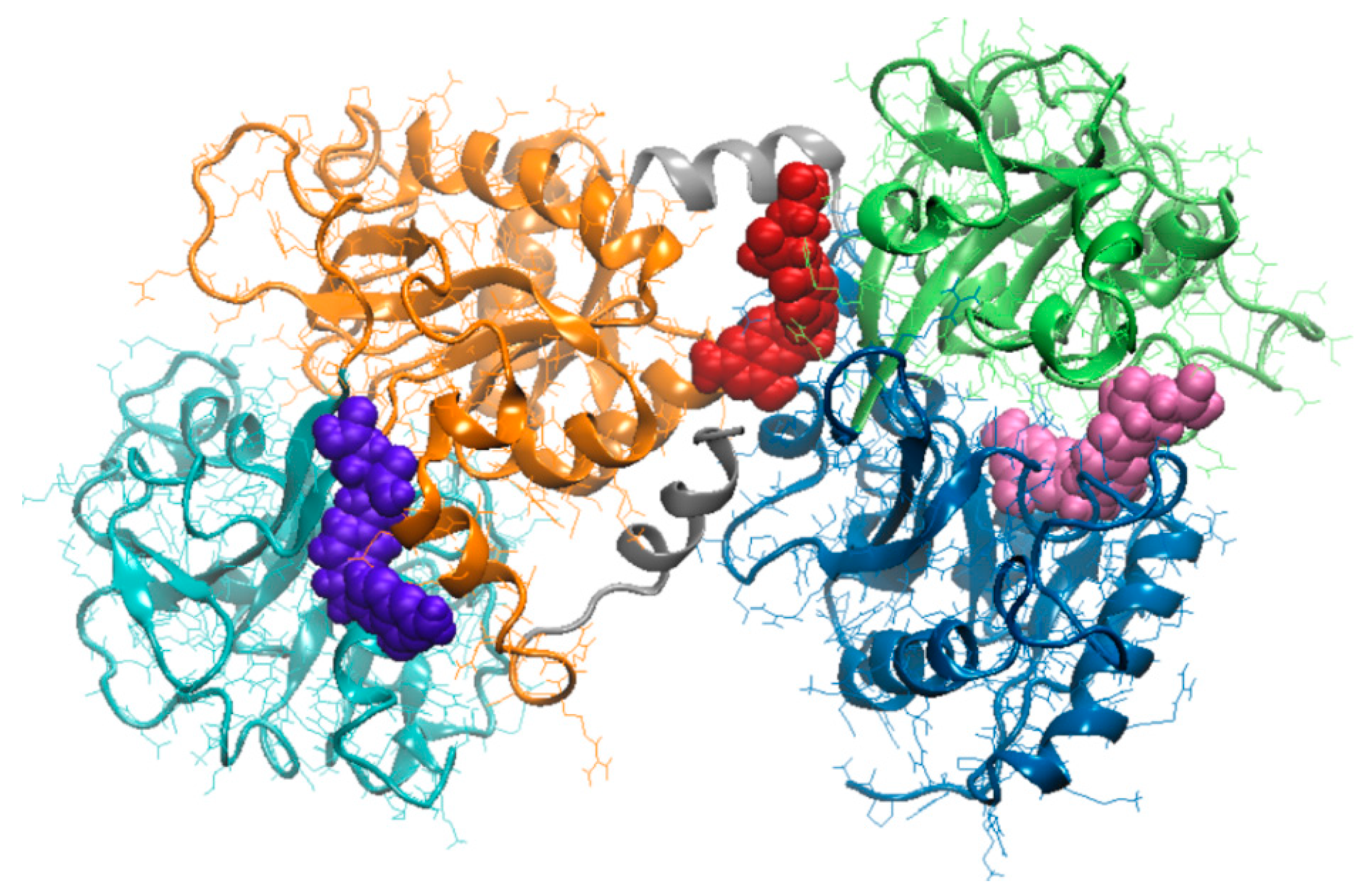
3.3. Molecular Modeling Investigation of FA Binding to LF
4. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Liu, F.; Zhang, S.; Li, J.; McClements, D.J.; Liu, X. Recent development of lactoferrin-based vehicles for the delivery of bioactive compounds: Complexes, emulsions, and nanoparticles. Trends Food Sci. Technol. 2018, 79, 67–77. [Google Scholar] [CrossRef]
- Zema, P.; Pilosof, A.M. On the binding of folic acid to food proteins performing as vitamin micro/nanocarriers. Food Hydrocoll. 2018, 79, 509–517. [Google Scholar] [CrossRef]
- Refsum, H. Folate, vitamin B12 and homocysteine in relation to birth defects and pregnancy outcome. Br. J. Nutr. 2001, 85, S109–S113. [Google Scholar] [CrossRef]
- Iyer, R.; Tomar, S. Folate: A functional food constituent. J. Food Sci. 2009, 74, R114–R122. [Google Scholar] [CrossRef]
- Off, M.K.; Steindal, A.E.; Porojnicu, A.C.; Juzeniene, A.; Vorobey, A.; Johnsson, A.; Moan, J. Ultraviolet photodegradation of folic acid. J. Photochem. Photobiol. B Biol. 2005, 80, 47–55. [Google Scholar] [CrossRef]
- Vora, A.; Riga, A.T.; Dollimore, D.; Alexander, K.S. Thermal stability of folic acid. Thermochim. Acta 2002, 392, 209–220. [Google Scholar] [CrossRef]
- Papastoyiannidis, G.; Polychroniadou, A.; Michaelidou, A.-M.; Alichanidis, E. Fermented milks fortified with B-group Vitamins: Vitamin stability and effect on resulting products. Food Sci. Technol. Int. 2006, 12, 521–529. [Google Scholar] [CrossRef]
- Coelho, Y.L.; De Paula, H.M.C.; Agudelo, A.J.P.; De Castro, A.S.; Hudson, E.A.; Pires, A.C.S.; Da Silva, L.H.M.; Benhame, A.S. Lactoferrin-phenothiazine dye interactions: Thermodynamic and kinetic approach. Int. J. Biol. Macromol. 2019, 136, 559–569. [Google Scholar] [CrossRef]
- Kühnle, A.; Veelken, R.; Galuska, C.E.; Saftenberger, M.; Verleih, M.; Schuppe, H.-C.; Rudloff, S.; Kunz, C.; Galuska, S.P. Polysialic acid interacts with lactoferrin and supports its activity to inhibit the release of neutrophil extracellular traps. Carbohydr. Polym. 2019, 208, 32–41. [Google Scholar] [CrossRef]
- Nazir, S.; Nasir, M.; Yasmeen, A.; Usman, S. Review study on lactoferrin: A multifunctional protein. Sky J. Food Sci. 2017, 6, 14–20. [Google Scholar]
- Da Silva, M.A.C.; Darr, C.R.; Moraes, L.E.; Forshey, B. Lactoferrin modulates uterine inflammation postbreeding in the Mare. J. Equine Veter-Sci. 2017, 56, 63–67. [Google Scholar] [CrossRef]
- Padrão, J.; Machado, R.; Casal, M.; Lanceros-Mendez, S.; Rodrigues, L.R.; Dourado, F.; Sencadas, V. Antibacterial performance of bovine lactoferrin-fish gelatine electrospun membranes. Int. J. Biol. Macromol. 2015, 81, 608–614. [Google Scholar] [CrossRef]
- Gifford, J.L.; Hunter, H.N.; Vogel, H.J. Lactoferricin: A lactoferrin-derived peptide with antimicrobial, antiviral, antitumor and immunological properties. Cell. Mol. Life Sci. 2015, 62, 2588–2598. [Google Scholar] [CrossRef]
- Okubo, K.; Kamiya, M.; Urano, Y.; Nishi, H.; Herter, J.M.; Mayadas, T.; Hirohama, D.; Suzuki, K.; Kawakami, H.; Tanaka, M.; et al. Lactoferrin suppresses neutrophil extracellular traps release in inflammation. EBioMedicine 2016, 10, 204–215. [Google Scholar] [CrossRef]
- Lakowicz, J.R. Principles of Fluorescence Spectroscopy, 3rd ed.; Springer: Baltimore, MD, USA, 2006. [Google Scholar]
- Horincar, G.; Aprodu, I.; Barbu, V.; Râpeanu, G.; Bahrim, G.-E.; Stănciuc, N. Interactions of flavonoids from yellow onion skins with whey proteins: Mechanisms of binding and microencapsulation with different combinations of polymers. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2019, 215, 158–167. [Google Scholar] [CrossRef]
- Fasman, G.D. Practical Handbook of Biochemistry and Molecular Biology; CRC Press: Boca Raton, FL, USA, 1989; p. 275. [Google Scholar]
- Ghisaidoobe, A.B.T.; Chung, S.J. Intrinsic tryptophan fluorescence in the detection and analysis of proteins: A focus on Förster resonance energy transfer techniques. Int. J. Mol. Sci. 2014, 15, 22518–22538. [Google Scholar] [CrossRef]
- Moore, S.A.; Anderson, B.F.; Groom, C.; Haridas, M.; Baker, E.N. Three-dimensional structure of diferric bovine lactoferrin at 2.8 Å resolution. J. Mol. Biol. 1997, 274, 222–236. [Google Scholar] [CrossRef]
- Stănciuc, N.; Aprodu, I.; Râpeanu, G.; Van Der Plancken, I.; Bahrim, G.; Hendrickx, M. Analysis of the thermally induced structural changes of Bovine Lactoferrin. J. Agric. Food Chem. 2013, 61, 2234–2243. [Google Scholar] [CrossRef]
- Abraham, M.; Murtola, T.; Schulz, R.; Páll, S.; Smith, J.C.; Hess, B.; Lindahl, E. GROMACS: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX 2015, 19–25. [Google Scholar] [CrossRef]
- Schneidman-Duhovny, D.; Inbar, Y.; Nussinov, R.; Wolfson, H.J. PatchDock and SymmDock: Servers for rigid and symmetric docking. Nucleic Acids Res. 2005, 33, W363–W367. [Google Scholar] [CrossRef]
- Laskowski, R.A. PDBsum new things. Nucleic Acids Res. 2008, 37, D355–D359. [Google Scholar] [CrossRef]
- Krissinel, E. Crystal contacts as nature’s docking solutions. J. Comput. Chem. 2010, 31, 133–143. [Google Scholar] [CrossRef]
- Laskowski, R.A.; Swindells, M.B. LigPlot+: Multiple ligand–protein interaction diagrams for drug discovery. J. Chem. Inf. Model. 2011, 51, 2778–2786. [Google Scholar] [CrossRef]
- Guo, M.; Zhang, L.-Y.; Lu, W.-J.; Cao, H.-R. Analysis of the spectroscopic characteristics on the binding interaction between tosufloxacin and bovine lactoferrin. J. Lumin. 2011, 131, 768–775. [Google Scholar] [CrossRef]
- Yang, W.; Liu, F.; Xu, C.; Yuan, F.; Gao, Y. Molecular interaction between (−)-epigallocatechin-3-gallate and bovine lactoferrin using multi-spectroscopic method and isothermal titration calorimetry. Food Res. Int. 2014, 64, 141–149. [Google Scholar] [CrossRef]
- Jayabharathi, J.; Thanikachalam, V.; Perumal, M.V. A study on the binding interaction between the imidazole derivative and bovine serum albumin by fluorescence spectroscopy. J. Lumin. 2012, 132, 707–712. [Google Scholar] [CrossRef]
- Schanbacher, F.; Goodman, R.; Talhouk, R. Bovine mammary Lactoferrin: Implications from Messenger Ribonucleic Acid (mRNA) sequence and regulation contrary to other milk proteins. J. Dairy Sci. 1993, 76, 3812–3831. [Google Scholar] [CrossRef]
- Bokkhim, H.; Bansal, N.; Grøndahl, L.; Bhandari, B. Physico-chemical properties of different forms of bovine lactoferrin. Food Chem. 2013, 141, 3007–3013. [Google Scholar] [CrossRef]
- Ward, P.P.; Paz, E.; Conneely, O.M. Multifunctional roles of lactoferrin: A critical overview. Cell. Mol. Life Sci. 2005, 62, 2540–2548. [Google Scholar] [CrossRef]
- Matsuura, J.E.; Manning, M.C. Heat-induced gel formation of β-lactoglobulin: A study on the secondary and tertiary structure as followed by circular dichroism spectroscopy. J. Agric. Food Chem. 1994, 42, 1650–1656. [Google Scholar] [CrossRef]
- Van De Weert, M.; Stella, L. Fluorescence quenching and ligand binding: A critical discussion of a popular methodology. J. Mol. Struct. 2011, 998, 144–150. [Google Scholar] [CrossRef]
- Callis, P.R. Binding phenomena and fluorescence quenching. I: Descriptive quantum principles of fluorescence quenching using a supermolecule approach. J. Mol. Struct. 2014, 1077, 14–21. [Google Scholar] [CrossRef]
- Ross, P.D.; Subramanian, S. Thermodynamics of protein association reactions: Forces contributing to stability. Biochemistry 1981, 20, 3096–3102. [Google Scholar] [CrossRef]
- Nhan, C.; Rix, C.J.; May, B.K.; Hung, A. Temperature-induced structural changes of apo-lactoferrin and their functional implications: A molecular dynamics simulation study. Mol. Simul. 2018, 45, 533–548. [Google Scholar] [CrossRef]
- Chen, D.; Oezguen, N.; Urvil, P.; Ferguson, C.; Dann, S.; Savidge, T.C. Regulation of protein-ligand binding affinity by hydrogen bond pairing. Sci. Adv. 2016, 2, e1501240. [Google Scholar] [CrossRef]
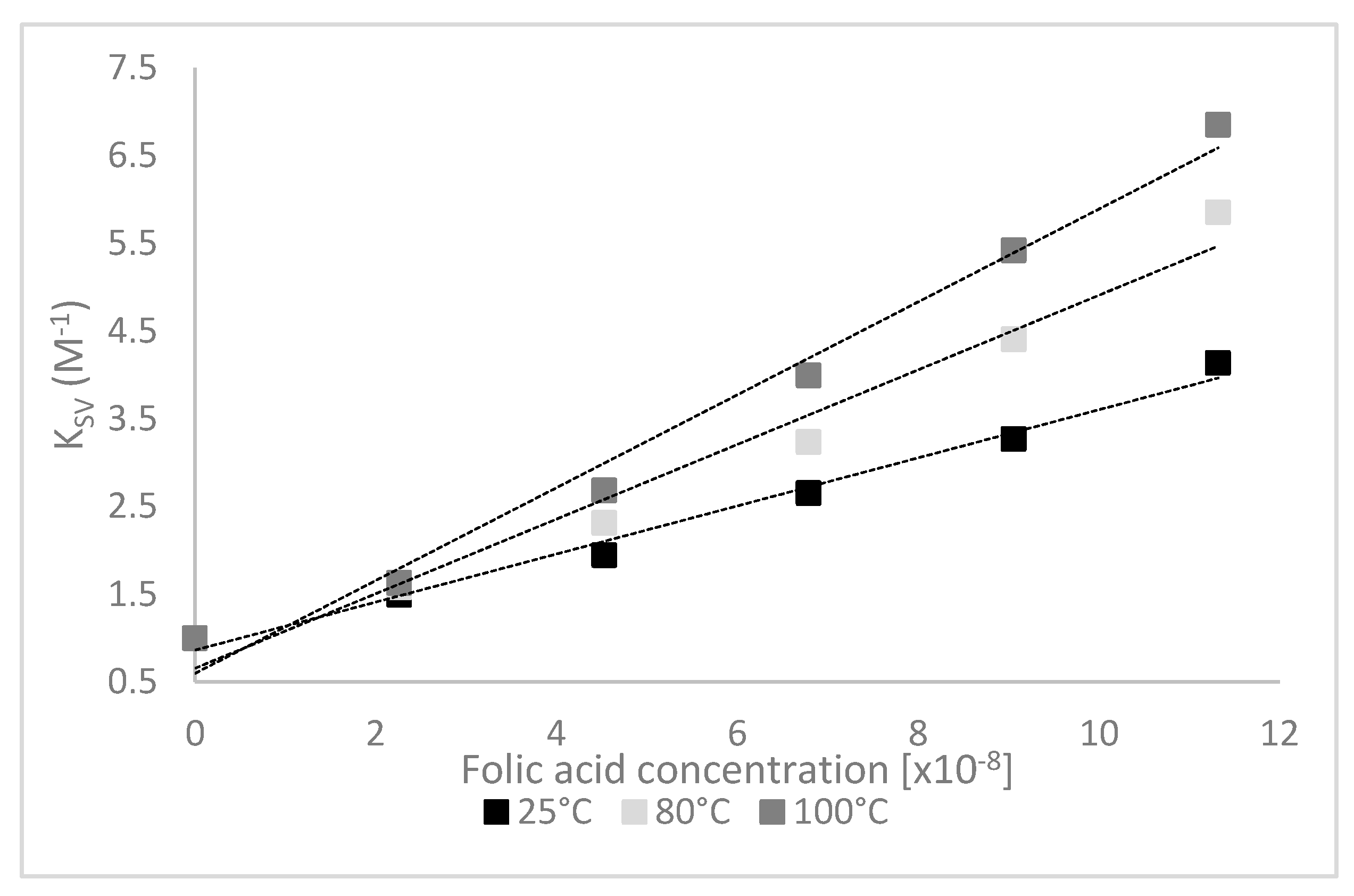
| T (°C) | KSV (108 M−1) | Kq (1018 mol−1s−1) | Kb (108 M−1) | n |
|---|---|---|---|---|
| 25 | 0.28 ± 0.01 b | 0.28 ± 0.01 b | 1.68 ± 0.01 a | 1.24 ± 0.13 a |
| 80 | 0.48 ± 0.03 a | 0.48 ± 0.03 a | 1.67 ± 0.23 a | 1.30 ± 0.02 a |
| 100 | 0.52 ± 0.01 a | 0.52 ± 0.01 a | 1.66 ± 0.11 a | 1.32 ± 0.08 a |
| T (K) | ΔHo (J·mol−1) | ΔSo (J·mol−1K−1) | ΔGo (J·mol−1) | R a |
|---|---|---|---|---|
| 298 | 15.29 ± 1.25 a | 0.47 ± 0.04 | −124.77 ± 21.03 | 0.97 |
| 353 | −150.62 ± 11.32 | |||
| 373 | −160.02 ± 16.98 |
| Descriptors | Temperature, °C | ||
|---|---|---|---|
| 25 | 80 | 100 | |
| Surface descriptors | |||
| Total protein surface, Å2 | 30,974.0 | 31,226.3 | 33,964.8 |
| Total LF–FA interface area, Å2 | 324.1 | 341.1 | 345.1 |
| Amino acids interacting with FA | Ile11, Ser12, Glu15, Ser42, Val57, Thr58, Leu59, Asp60, Arg121, Ser185, Phe190, His253, Gln296, Asp297, Leu298, Leu299 | Ile469, Gly472, Leu473, Phe475, Asn476, Leu589, Ala590, Lys654, Leu661, Glu664, Tyr665, Thr667, Ser668, Asn671 | Arg91, Ile129, Gly130, Thr131, Pro232, His246, Leu247, Ala248, Arg249, Leu320, Gly321, Asn323, Cys405, Gly406 |
| Energy descriptors | |||
| LF–FA binding energy, kcal/mol | −57.20 | −46.03 | −47.19 |
| aVdW, kcal/mol | −24.97 | −20.47 | −22.45 |
| rVdW, kcal/mol | 5.59 | 4.24 | 4.86 |
| Thermodynamic descriptors | |||
| ΔGf, kcal/mol | −604.7 | −602.9 | −594.0 |
| ΔGint, kcal/mol | −0.49 | −2.36 | −1.26 |
| TΔSdiss, kcal/mol | 0.5 | 0.5 | 0.5 |
| ΔGdiss, kcal/mol | −0.9 | 0.1 | −1.1 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Aprodu, I.; Dumitrașcu, L.; Râpeanu, G.; Bahrim, G.-E.; Stănciuc, N. Spectroscopic and Molecular Modeling Investigation on the Interaction between Folic Acid and Bovine Lactoferrin from Encapsulation Perspectives. Foods 2020, 9, 744. https://doi.org/10.3390/foods9060744
Aprodu I, Dumitrașcu L, Râpeanu G, Bahrim G-E, Stănciuc N. Spectroscopic and Molecular Modeling Investigation on the Interaction between Folic Acid and Bovine Lactoferrin from Encapsulation Perspectives. Foods. 2020; 9(6):744. https://doi.org/10.3390/foods9060744
Chicago/Turabian StyleAprodu, Iuliana, Loredana Dumitrașcu, Gabriela Râpeanu, Gabriela-Elena Bahrim, and Nicoleta Stănciuc. 2020. "Spectroscopic and Molecular Modeling Investigation on the Interaction between Folic Acid and Bovine Lactoferrin from Encapsulation Perspectives" Foods 9, no. 6: 744. https://doi.org/10.3390/foods9060744
APA StyleAprodu, I., Dumitrașcu, L., Râpeanu, G., Bahrim, G.-E., & Stănciuc, N. (2020). Spectroscopic and Molecular Modeling Investigation on the Interaction between Folic Acid and Bovine Lactoferrin from Encapsulation Perspectives. Foods, 9(6), 744. https://doi.org/10.3390/foods9060744