Effect of Casein Hydrolysates on Intestinal Cell Migration and Their Peptide Profiles by LC-ESI/MS/MS
Abstract
1. Introduction
2. Materials and Methods
2.1. Casein Hydrolysates
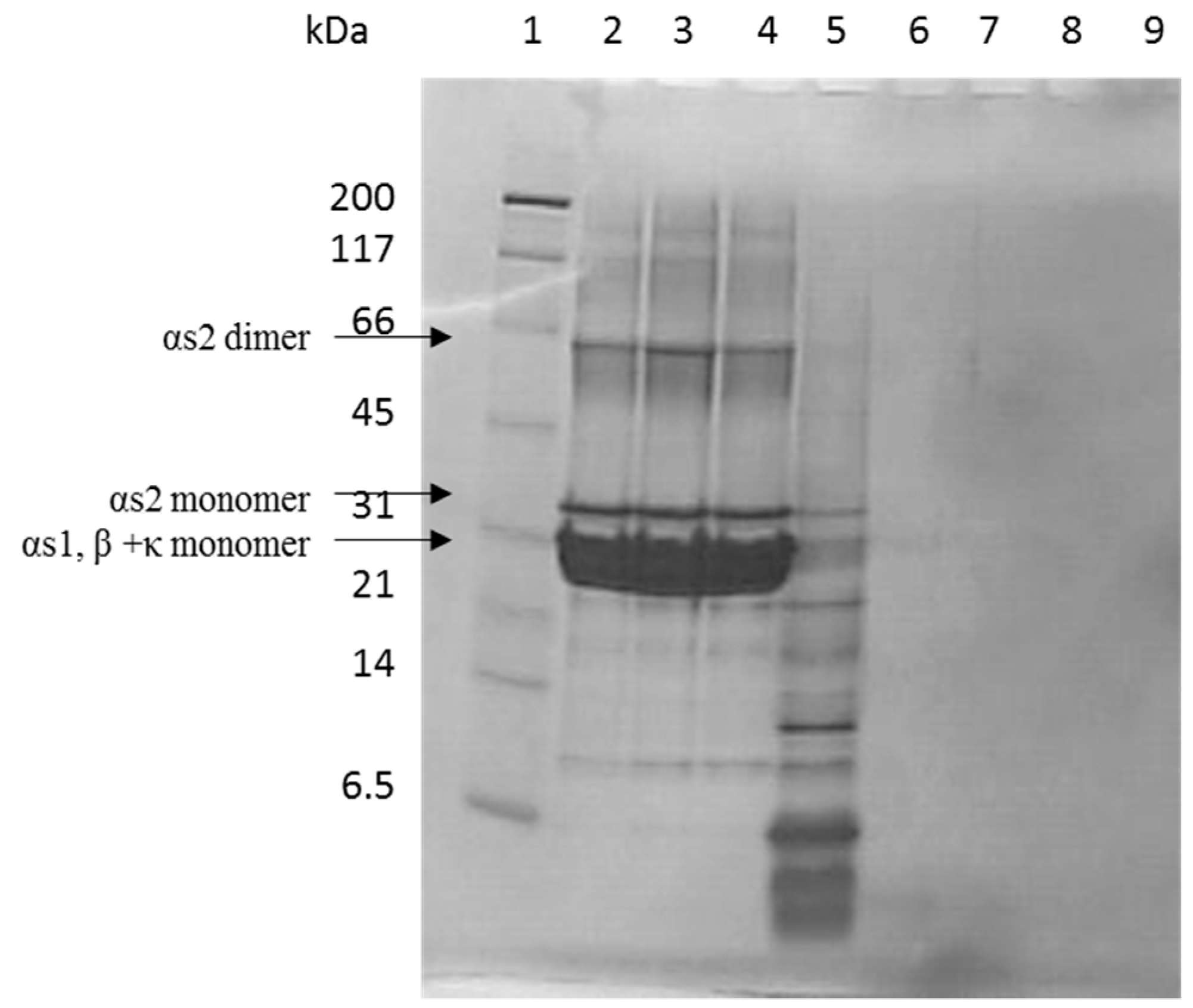
2.2. Tris-Tricine SDS-PAGE
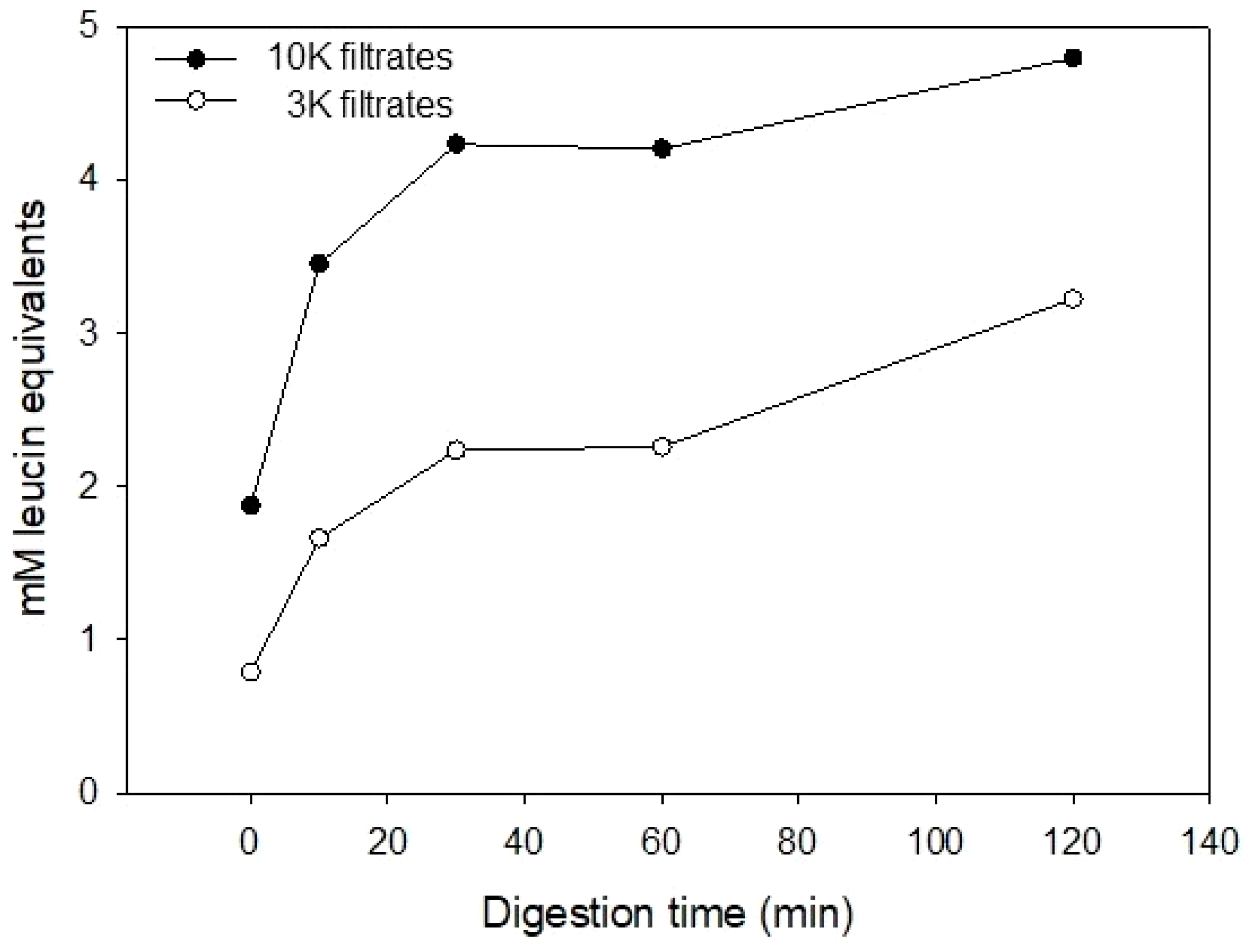
2.3. Free Amino Terminal Determination
2.4. Cell Culture
2.5. Cell Migration Assay
2.6. Cell Proliferation Assay
2.7. Liquid Chromatography–Electrospray Ionization Tandem Mass Spectrometry Ion Trap Ion Trap Peptide Profiling
2.8. Statistical Analysis
3. Results
3.1. Casein Hydrolysate Analysis
3.2. Effects of Casein Hydrolysates on Intestinal Cell Migration and Proliferation
3.3. Identification of Peptides in Casein Hydrolysates by LC-ESI/MS/MS Ion Trap
3.4. Literature-Identified Bioactive Peptides
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Silva, S.V.; Malcata, F.X. Caseins as source of bioactive peptides. Int. Dairy J. 2005, 15, 1–15. [Google Scholar] [CrossRef]
- Playford, R.J.; Macdonald, C.E.; Johnson, W.S. Colostrum and milk-derived peptide growth factors for the treatment of gastrointestinal disorders. Am. J. Clin. Nutr. 2000, 72, 5–14. [Google Scholar] [CrossRef] [PubMed]
- Nielsen, S.D.; Beverly, R.L.; Qu, Y.Y.; Dallas, D.C. Milk bioactive peptide database: A comprehensive database of milk protein-derived bioactive peptides and novel visualization. Food Chem. 2017, 232, 673–682. [Google Scholar] [CrossRef] [PubMed]
- Kanwar, J.R.; Kanwar, R.K.; Sun, X.Y.; Punj, V.; Matta, H.; Morley, S.M.; Parratt, A.; Puri, M.; Sehgal, R. Molecular and biotechnological advances in milk proteins in relation to human health. Curr. Protein Pept. Sci. 2009, 10, 308–338. [Google Scholar] [CrossRef] [PubMed]
- Nielsen, S.D.; Beverly, R.L.; Underwood, M.A.; Dallas, D.C. Release of functional peptides from mother’s milk and fortifier proteins in the premature infant stomach. PLoS ONE 2018, 13, e0208204. [Google Scholar] [CrossRef] [PubMed]
- Fernández-Tomé, S.; Martínez-Maqueda, D.; Girón, R.; Goicoechea, C.; Miralles, B.; Recio, I. Novel peptides derived from αs1-casein with opioid activity and mucin stimulatory effect on ht29-mtx cells. J. Funct. Foods 2016, 25, 466–476. [Google Scholar] [CrossRef]
- Yamada, A.; Sakurai, T.; Ochi, D.; Mitsuyama, E.; Yamauchi, K.; Abe, F. Antihypertensive effect of the bovine casein-derived peptide met-lys-pro. Food Chem. 2015, 172, 441–446. [Google Scholar] [CrossRef] [PubMed]
- Jorgensen, A.L.W.; Juul-Madsen, H.R.; Stagsted, J. Colostrum and bioactive, colostral peptides differentially modulate the innate immune response of intestinal epithelial cells. J. Pept. Sci. 2010, 16, 21–30. [Google Scholar] [CrossRef] [PubMed]
- Phelan, M.; Aherne, A.; FitzGerald, R.J.; O’Brien, N.M. Casein-derived bioactive peptides: Biological effects, industrial uses, safety aspects and regulatory status. Int. Dairy J. 2009, 19, 643–654. [Google Scholar] [CrossRef]
- Purup, S.; Nielsen, S.D.; Le, T.T.; Bertelsen, H.; Sørensen, J.; Larsen, L.B. Wound healing properties of commercial milk hydrolysates in intestinal cells. Int. J. Pept. Res. Ther. 2018. [Google Scholar] [CrossRef]
- Ebaid, H.; Abdel-Salam, B.; Hassan, I.; Al-Tamimi, J.; Metwalli, A.; Alhazza, I. Camel milk peptide improves wound healing in diabetic rats by orchestrating the redox status and immune response. Lipids Health Dis. 2015, 14, 132. [Google Scholar] [CrossRef] [PubMed]
- Melgar, S.; Shanahan, F. Inflammatory bowel disease-from mechanisms to treatment strategies. Autoimmunity 2010, 43, 463–477. [Google Scholar] [CrossRef] [PubMed]
- Dignass, A.U. Mechanisms and modulation of intestinal epithelial repair. Inflamm. Bowel Dis. 2001, 7, 68–77. [Google Scholar] [CrossRef] [PubMed]
- Karrasch, T.; Jobin, C. Wound healing responses at the gastrointestinal epithelium: A close look at novel regulatory factors and investigative approaches. Z. Gastroenterol. 2009, 47, 1221–1229. [Google Scholar] [CrossRef] [PubMed]
- Larsen, L.B.; Rasmussen, M.D.; Bjerring, M.; Nielsen, J.H. Proteases and protein degradation in milk from cows infected with streptococcus uberis. Int. Dairy J. 2004, 14, 899–907. [Google Scholar] [CrossRef]
- McCormack, S.A.; Viar, M.J.; Johnson, L.R. Migration of iec-6 cells—A model for mucosal healing. Am. J. Physiol. 1992, 263, G426–G435. [Google Scholar] [CrossRef] [PubMed]
- Purup, S.; Nielsen, T.S. Cell-based models to test the effects of milk-derived bioactives. Animal 2012, 6, 423–432. [Google Scholar] [CrossRef] [PubMed]
- Purup, S.; Larsen, E.; Christensen, L.P. Differential effects of falcarinol and related aliphatic c(17)-polyacetylenes on intestinal cell proliferation. J. Agric. Food Chem. 2009, 57, 8290–8296. [Google Scholar] [CrossRef] [PubMed]
- Nielsen, S.D.; Jansson, T.; Le, T.T.; Jensen, S.; Eggers, N.; Rauh, V.; Sundekilde, U.K.; Sørensen, J.; Andersen, H.J.; Bertram, H.C.; et al. Correlation between sensory properties and peptides derived from hydrolysed-lactose UHT milk during storage. Int. Dairy J. 2017, 68, 23–31. [Google Scholar] [CrossRef]
- Read, N.W.; Aljanabi, M.N.; Holgate, A.M.; Barber, D.C.; Edwards, C.A. Simultaneous measurement of gastric-emptying, small-bowel residence and colonic filling of a solid meal by the use of the gamma-camera. Gut 1986, 27, 300–308. [Google Scholar] [CrossRef] [PubMed]
- Piper, D.W.; Fenton, B.H. Ph stability and activity curves of pepsin with special reference to their clinical importance. Gut 1965, 6, 506. [Google Scholar] [CrossRef] [PubMed]
- McClements, D.J.; Li, Y. Review of in vitro digestion models for rapid screening of emulsion-based systems. Food Funct. 2010, 1, 32–59. [Google Scholar] [CrossRef] [PubMed]
- Hur, S.J.; Lim, B.O.; Decker, E.A.; McClements, D.J. In vitro human digestion models for food applications. Food Chem. 2011, 125, 1–12. [Google Scholar] [CrossRef]
- Hagiwara, T.; Shinoda, I.; Fukuwatari, Y.; Shimamura, S. Effects of lactoferrin and its peptides on proliferation of rat intestinal epithelial-cell line, iec-18, in the presence of epidermal growth-factor. Biosci. Biotechnol. Biochem. 1995, 59, 1875–1881. [Google Scholar] [CrossRef] [PubMed]
- Hayes, M.; Ross, R.P.; Fitzgerald, G.F.; Hill, C.; Stanton, C. Casein-derived antimicrobial peptides generated by lactobacillus acidophilus dpc6026. Appl. Environ. Microbiol. 2006, 72, 2260–2264. [Google Scholar] [CrossRef] [PubMed]
- Kohmura, M.; Nio, N.; Kubo, K.; Minoshima, Y.; Munekata, E.; Ariyoshi, Y. Inhibition of angiotensin-converting enzyme by synthetic peptides of human beta-casein. Agric. Biol. Chem. 1989, 53, 2107–2114. [Google Scholar] [CrossRef]
- Yamamoto, N.; Akino, A.; Takano, T. Antihypertensive effect of the peptides derived from casein by an extracellular proteinase from lactobacillus-helveticus cp790. J. Dairy Sci. 1994, 77, 917–922. [Google Scholar] [CrossRef]
- Almaas, H.; Eriksen, E.; Sekse, C.; Comi, I.; Flengsrud, R.; Holm, H.; Jensen, E.; Jacobsen, M.; Langsrud, T.; Vegarud, G.E. Antibacterial peptides derived from caprine whey proteins, by digestion with human gastrointestinal juice. Br. J. Nutr. 2011, 106, 896–905. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Chen, R.; Ma, H.; Chen, S. Isolation and identification of dipeptidyl peptidase iv-inhibitory peptides from trypsin/chymotrypsin-treated goat milk casein hydrolysates by 2d-tlc and lc-ms/ms. J. Agric. Food Chem. 2015, 63, 8819–8828. [Google Scholar] [CrossRef] [PubMed]
| Casein Hydrolysate a | |||||
|---|---|---|---|---|---|
| Time Point (min) | 0.0% | 0.1% | 1.0% | 2.5% | |
| 10 K | 0 | 1.00 ± 0.13 | 1.20 ± 0.13 | 1.12 ± 0.13 | 1.01 ± 0.14 |
| 10 b | 1.00 ± 0.16 | 1.42 ± 0.15 ** | 1.36 ± 0.15 * | 1.45 ± 0.17 ** | |
| 30 b | 1.00 ± 0.20 | 1.36 ± 0.18 * | 1.55 ± 0.17 ** | 1.15 ± 0.17 | |
| 60 | 1.00 ± 0.14 | 0.91 ± 0.14 | 1.13 ± 0.13 | 0.96 ± 0.14 | |
| 120 | 1.00 ± 0.12 | 1.18 ± 0.12 | 1.10 ± 0.12 | 0.77 ± 0.18 | |
| 3 K | 0 | 1.00 ± 0.13 | 1.01 ± 0.14 | 0.97 ± 0.15 | 0.98 ± 0.13 |
| 10 b | 1.00 ± 0.12 | 1.08 ± 0.10 | 1.09 ± 0.12 | 1.09 ± 0.12 | |
| 30 b | 1.00 ± 0.10 | 1.14 ± 0.10 | 1.22 ± 0.10 ** | 1.05 ± 0.10 | |
| 60 | 1.00 ± 0.14 | 1.19 ± 0.15 | 1.23 ± 0.05 | 1.13 ± 0.19 | |
| 120 | 1.00 ± 0.11 | 1.10 ± 0.11 | 1.00 ± 0.1 | 0.94 ± 0.10 | |
| Protein | Mass [M + H]+ | Peptide Sequence |
|---|---|---|
| αs1-casein (148–158) | 1200.6 | EPMIGVNQELA |
| αs1-casein (189–208) | 2105.0 | TDAPSFSDIPNPIGSENSEK |
| β-casein (139–154) | 1775.0 | SLTLTDVENLHLPLPL |
| β-casein (158–171) | 1698.8 | WMHQPHQPLPPTVM |
| β-casein (158–177) | 2354.2 | WMHQPHQPLPPTVMFPPQSV |
| β-casein (158–178) | 2467.2 | WMHQPHQPLPPTVMFPPQSVL |
| β-casein (160–171) | 1381.7 | HQPHQPLPPTVM |
| β-casein (84–108) | 2741.5 | SLPQNIPPLTQTPVVVPPFLQPEVM |
| β-casein (96–107) | 1320.8 | PVVVPPFLQPEV |
| κ-casein (134–145) | 1373.7 | NQDKTEIPTINT |
| κ-casein (70–81) | 1452.8 | ALINNQFLPYPY |
| Protein | Sequence | Function |
|---|---|---|
| αs1-casein (39–48) | FVAPFPEVFG | ACE-inhibitory a |
| αs1-casein (119–124) | YKVPQL | ACE-inhibitory |
| αs1-casein (195–208) | SDIPNPIGSENSEK | Antimicrobial |
| αs2-casein (96–104) | ALNEINQFY | ACE-inhibitory |
| αs2-casein (204–212) | AMKPWIQPK | ACE-inhibitory |
| β-casein (74–83) | VYPFPGPIPN | ACE-inhibitory, antioxidant |
| β-casein (113–120) | VKEAMAPK | Antimicrobial, antioxidant |
| β-casein (115–120) | EAMAPK | Antimicrobial |
| β-casein (121–128) | HKEMPFPK | Antimicrobial |
| β-casein (185–191) | VLPVPQK | Antimicrobial, ACE-inhibitory, antioxidant |
| β-casein (208–224) | YQEPVLGPVRGPFPIIV | ACE-inhibitory, immunoregulatory |
| β-casein (209–224) | QEPVLGPVRGPFPIIV | ACE-inhibitory |
| β-casein (208–222) | YQEPVLGPVRGPFPI | Antimicrobial |
| κ-casein (72–81) | INNQFLPYPY | ACE-inhibitory |
| κ-casein (46–51) | YIPIQY | ACE-inhibitory |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nielsen, S.D.; Purup, S.; Larsen, L.B. Effect of Casein Hydrolysates on Intestinal Cell Migration and Their Peptide Profiles by LC-ESI/MS/MS. Foods 2019, 8, 91. https://doi.org/10.3390/foods8030091
Nielsen SD, Purup S, Larsen LB. Effect of Casein Hydrolysates on Intestinal Cell Migration and Their Peptide Profiles by LC-ESI/MS/MS. Foods. 2019; 8(3):91. https://doi.org/10.3390/foods8030091
Chicago/Turabian StyleNielsen, Søren D., Stig Purup, and Lotte B. Larsen. 2019. "Effect of Casein Hydrolysates on Intestinal Cell Migration and Their Peptide Profiles by LC-ESI/MS/MS" Foods 8, no. 3: 91. https://doi.org/10.3390/foods8030091
APA StyleNielsen, S. D., Purup, S., & Larsen, L. B. (2019). Effect of Casein Hydrolysates on Intestinal Cell Migration and Their Peptide Profiles by LC-ESI/MS/MS. Foods, 8(3), 91. https://doi.org/10.3390/foods8030091






