The Biological Properties of the FAS and TACR3 Genes and the Association of Single-Nucleotide Polymorphisms with Milk Quality Traits in Gannan Yak
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethics Statement
2.2. Experimental Animal Selection
2.3. Sample Collection and DNA Extraction
2.4. Genotyping
2.5. Analysis of Milk Composition of Gannan Yak
2.6. Association Analysis Between Different SNP Loci and Dairy Quality Traits
2.7. Bioinformatics Analysis
2.8. Data Statistical Analysis
3. Results
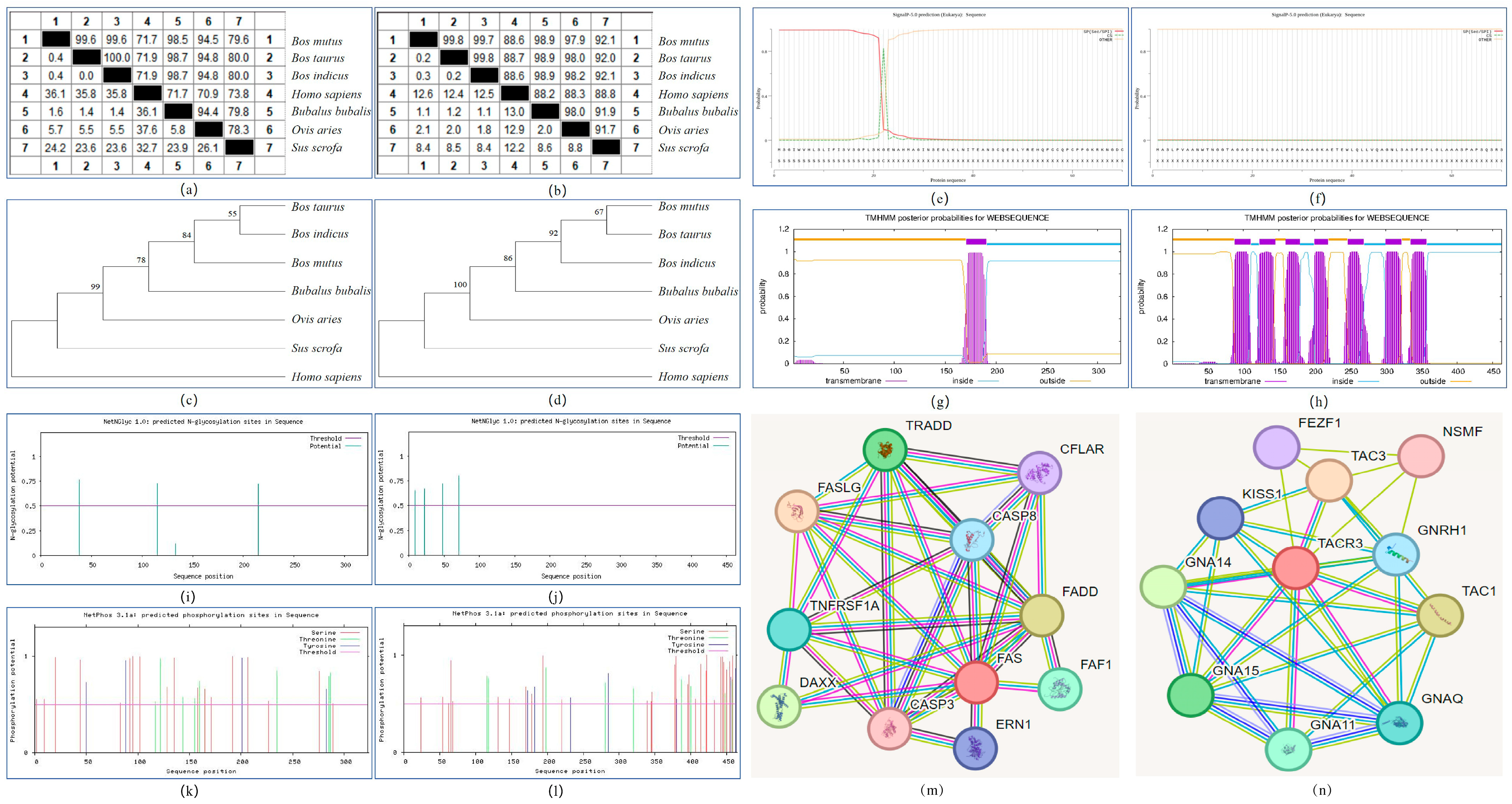
3.1. Homology Analysis and Phylogenetic Tree Construction of FAS and TACR3 Genes
3.2. Physicochemical Properties, Hydrophilicity/Hydrophobicity, Signal Peptide, Transmembrane Domain, and Advanced Structure Analysis of FAS and TACR3 Proteins
3.3. Genotype and Allele Frequencies and Polymorphism Information Content of FAS and TACR3 Genes at Different Loci
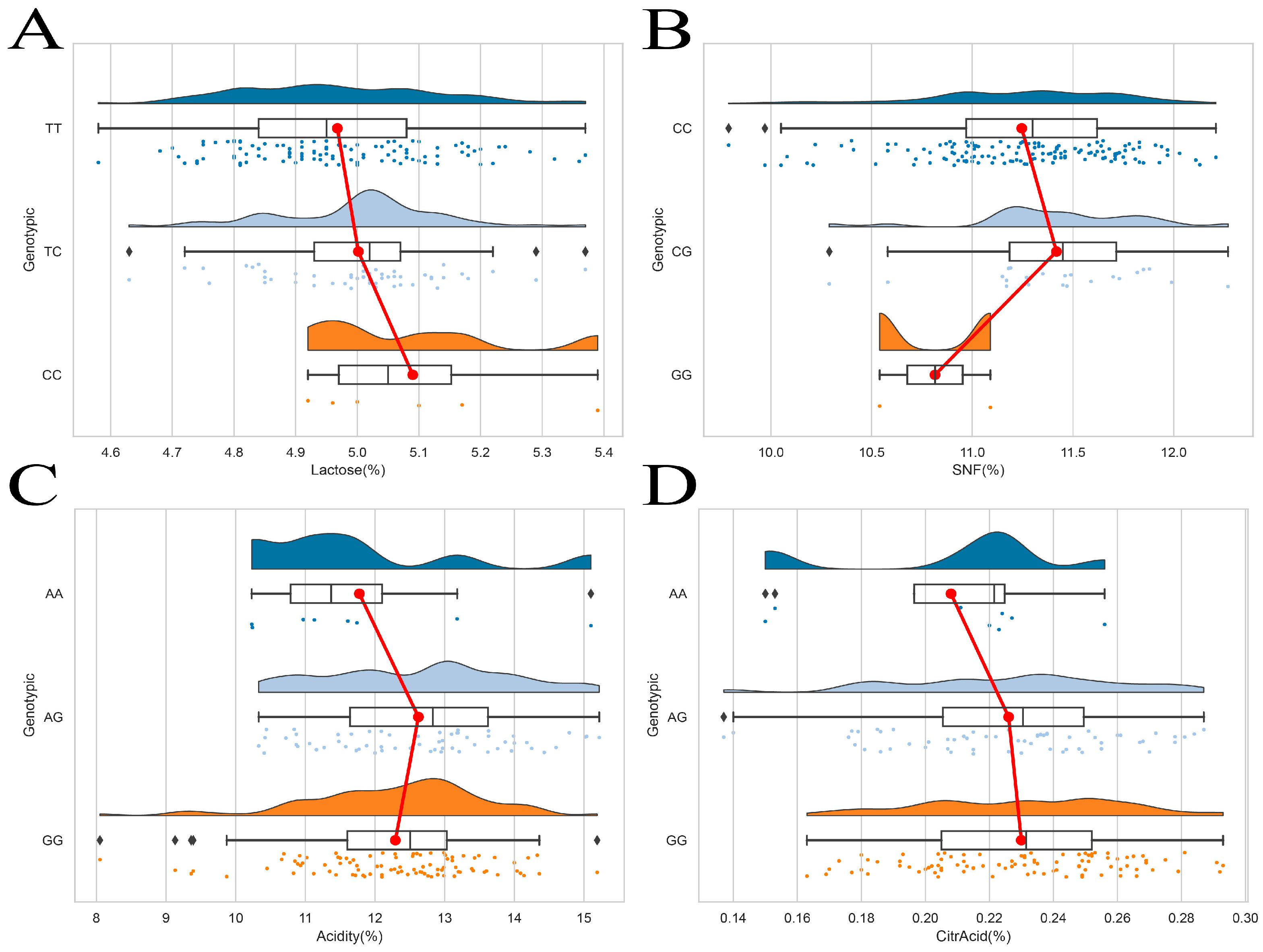
3.4. Association Analysis of FAS and TACR3 Gene Polymorphisms with Milk Quality Traits in Gannan Yaks
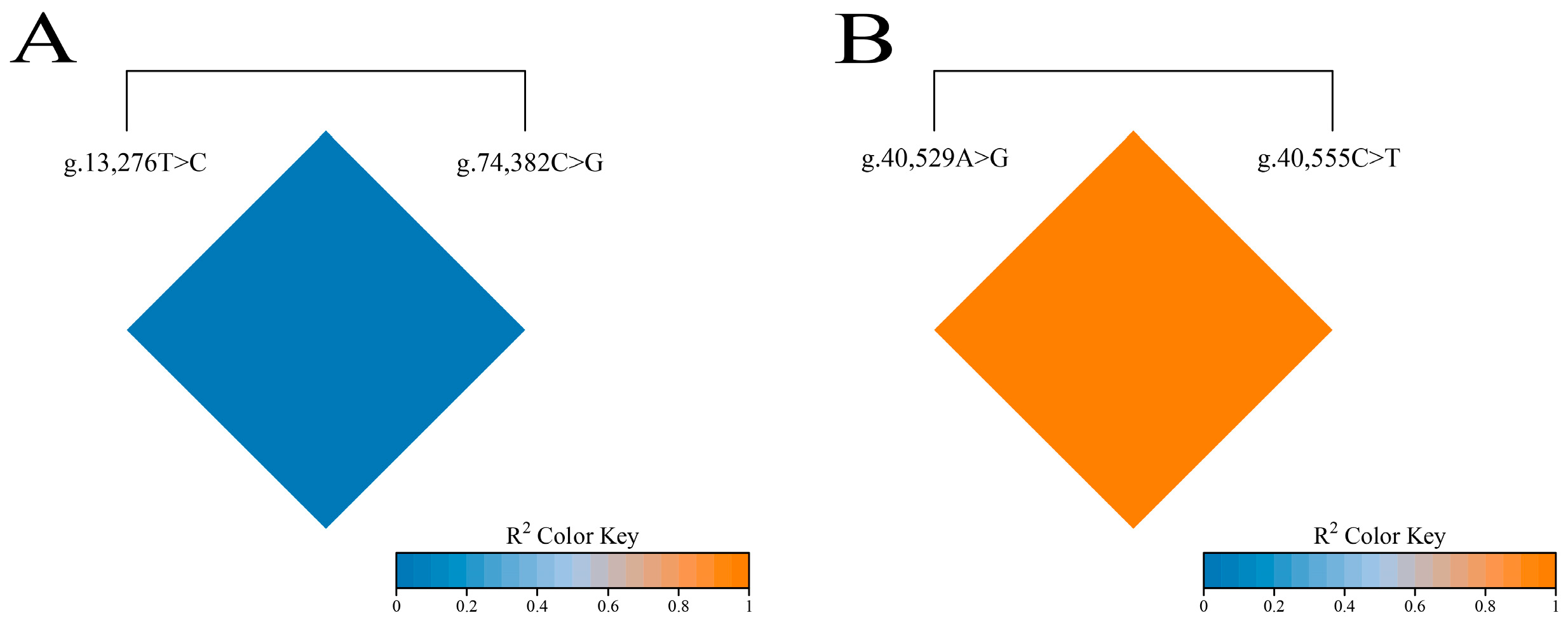
3.5. Linkage Disequilibrium Analysis of SNPs in FAS and TACR3 Genes
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Guo, X.; Long, R.; Kreuzer, M.; Ding, L.; Shang, Z.; Zhang, Y.; Yang, Y.; Cui, G. Importance of functional ingredients in yak milk-derived food on health of Tibetan nomads living under high-altitude stress: A review. Crit. Rev. Food Sci. Nutr. 2014, 54, 292–302. [Google Scholar] [CrossRef] [PubMed]
- Cui, G.; Yuan, F.; Degen, A.; Liu, S.; Zhou, J.; Shang, Z.; Ding, L.; Mi, J.; Wei, X.; Long, R. Composition of the milk of yaks raised at different altitudes on the Qinghai–Tibetan Plateau. Int. Dairy J. 2016, 59, 29–35. [Google Scholar] [CrossRef]
- Li, H.; Ma, Y.; Li, Q.; Wang, J.; Cheng, J.; Xue, J.; Shi, J. The chemical composition and nitrogen distribution of Chinese yak (Maiwa) milk. Int. J. Mol. Sci. 2011, 12, 4885–4895. [Google Scholar] [CrossRef]
- Qu, S.; Barrett-Wilt, G.; Fonseca, L.; Rankin, S. A profile of sphingolipids and related compounds tentatively identified in yak milk. J. Dairy Sci. 2016, 99, 5083–5092. [Google Scholar] [CrossRef]
- Li, H.; Xi, B.; Yang, X.; Wang, H.; He, X.; Li, W.; Gao, Y. Evaluation of change in quality indices and volatile flavor components in raw milk during refrigerated storage. LWT 2022, 165, 113674. [Google Scholar] [CrossRef]
- Wang, P.; Liu, H.; Wen, P.; Zhang, H.; Guo, H.; Ren, F. The composition, size and hydration of yak casein micelles. Int. Dairy J. 2013, 31, 107–110. [Google Scholar] [CrossRef]
- Liu, Q.; Yang, M.; Zhao, B.; Yang, F. Isolation of antioxidant peptides from yak casein hydrolysate. RSC Adv. 2020, 10, 19844–19851. [Google Scholar] [CrossRef] [PubMed]
- Li, A.; Liu, C.; Han, X.; Zheng, J.; Zhang, G.; Qi, X.; Du, P.; Liu, L. Tibetan Plateau yak milk: A comprehensive review of nutritional values, health benefits, and processing technology. Food Chem. X 2023, 20, 100919. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Jiang, Y.; Wang, S.; Fu, L.; Liang, Z.; Zhang, Y.; Huang, X.; Li, X.; Feng, M.; Long, D. Yak milk protects against alcohol-induced liver injury in rats. Food Funct. 2023, 14, 9857–9871. [Google Scholar] [CrossRef]
- Chen, Y.; Ren, Y.; Wang, L.; Huang, Z. Analysis of A1-type and A2-type β-casein in Maiwa Yak and Pien-niu milk by HPLC-high-resolution MS and tandem MS. J. Sep. Sci. 2021, 44, 1913–1922. [Google Scholar] [CrossRef]
- Chen, Y.; Qu, S.; Huang, Z.; Ren, Y.; Wang, L.; Rankin, S. Analysis and comparison of key proteins in Maiwa yak and bovine milk using high-performance liquid chromatography mass spectrometry. J. Dairy Sci. 2021, 104, 8661–8672. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.J.; Liang, A.X.; Campanile, G.; Plastow, G.; Zhang, C.; Wang, Z.; Salzano, A.; Gasparrini, B.; Cassandro, M.; Yang, L. Genome-wide association studies to identify quantitative trait loci affecting milk production traits in water buffalo. J. Dairy Sci. 2018, 101, 433–444. [Google Scholar] [CrossRef] [PubMed]
- Shahid, R.; Sahrish, F.; Hira, M. Role of Molecular Markers and their Significance. Am. J. Pharm. Health Res. 2015, 3, 2321–3647. [Google Scholar]
- LaFramboise, T. Single nucleotide polymorphism arrays: A decade of biological, computational and technological advances. Nucleic Acids Res. 2009, 37, 4181–4193. [Google Scholar] [CrossRef] [PubMed]
- Marian, A.J. Molecular genetic studies of complex phenotypes. Transl. Res. J. Lab. Clin. Med. 2012, 159, 64–79. [Google Scholar] [CrossRef] [PubMed]
- Göttgens, B.; Barton, L.M.; Gilbert, J.G.; Bench, A.J.; Sanchez, M.-J.; Bahn, S.; Mistry, S.; Grafham, D.; McMurray, A.; Vaudin, M.; et al. Analysis of vertebrate SCL loci identifies conserved enhancers. Nat. Biotechnol. 2000, 18, 181–186. [Google Scholar] [CrossRef]
- Loots, G.G.; Locksley, R.M.; Blankespoor, C.M.; Wang, Z.E.; Miller, W.; Rubin, E.M.; Frazer, K.A. Identification of a coordinate regulator of interleukins 4, 13, and 5 by cross-species sequence comparisons. Science 2000, 288, 136–140. [Google Scholar] [CrossRef] [PubMed]
- Viale, E.; Tiezzi, F.; Maretto, F.; De Marchi, M.; Penasa, M.; Cassandro, M. Association of candidate gene polymorphisms with milk technological traits, yield, composition, and somatic cell score in Italian Holstein-Friesian sires. J. Dairy Sci. 2017, 100, 7271–7281. [Google Scholar] [CrossRef]
- Liu, Y.; Han, B.; Zheng, W.; Peng, P.; Yang, C.; Jiang, G.; Ma, Y.; Li, J.; Ni, J.; Sun, D. Identification of genetic associations and functional SNPs of bovine KLF6 gene on milk production traits in Chinese holstein. BMC Genom. Data 2023, 24, 72. [Google Scholar] [CrossRef]
- Selionova, M.; Trukhachev, V.; Aibazov, M.; Sermyagin, A.; Belous, A.; Gladkikh, M.; Zinovieva, N. Genome-Wide Association Study of Milk Composition in Karachai Goats. Animals 2024, 14, 327. [Google Scholar] [CrossRef] [PubMed]
- Zwierzchowski, L.; Ostrowska, M.; Żelazowska, B.; Bagnicka, E. Single nucleotide polymorphisms in the bovine SLC2A12 and SLC5A1 glucose transporter genes—The effect on gene expression and milk traits of Holstein Friesian cows. Anim. Biotechnol. 2023, 34, 225–235. [Google Scholar] [CrossRef]
- Luo, J.-X.; Li, C.-F.; Chu, X.-H.; Gu, D.-H.; Li, X.-Q.; Xu, Z.-Q. Research on the Fatty Acid Synthase (FAS) Gene. China Anim. Husb. Vet. Med. 2011, 38, 118–123. [Google Scholar]
- Menendez, J.A.; Lupu, R. Fatty acid synthase-catalyzed de novo fatty acid biosynthesis: From anabolic-energy-storage pathway in normal tissues to jack-of-all-trades in cancer cells. Arch. Immunol. Ther. Exp. 2004, 52, 414–426. [Google Scholar]
- Lacasa, D.; Le Liepvre, X.; Ferre, P.; Dugail, I. Progesterone stimulates adipocyte determination and differentiation 1/sterol regulatory element-binding protein 1c gene expression. potential mechanism for the lipogenic effect of progesterone in adipose tissue. J. Biol. Chem. 2001, 276, 11512–11516. [Google Scholar] [CrossRef] [PubMed]
- Migliaccio, A.; Castoria, G.; DI Domenico, M.; DE Falco, A.; Bilancio, A.; Auricchio, F. Src is an initial target of sex steroid hormone action. Ann. N. Y. Acad. Sci. 2002, 963, 185–190. [Google Scholar] [CrossRef] [PubMed]
- Chirala, S.S.; Chang, H.; Matzuk, M.; Abu-Elheiga, L.; Mao, J.; Mahon, K.; Finegold, M.; Wakil, S.J. Fatty acid synthesis is essential in embryonic development: Fatty acid synthase null mutants and most of the heterozygotes die in utero. Proc. Natl. Acad. Sci. USA 2003, 100, 6358–6363. [Google Scholar] [CrossRef] [PubMed]
- Kalkhoven, E.; Kwakkenbos-Isbrücker, L.; de Laat, S.W.; van der Saag, P.T.; van der Burg, B. Synthetic progestins induce proliferation of breast tumor cell lines via the progesterone or estrogen receptor. Mol. Cell. Endocrinol. 1994, 102, 45–52. [Google Scholar] [CrossRef]
- Morris, C.A.; Cullen, N.G.; Glass, B.C.; Hyndman, D.L.; Manley, T.R.; Hickey, S.M.; McEwan, J.C.; Pitchford, W.S.; Bottema, C.D.; Lee, M.A. Fatty acid synthase effects on bovine adipose fat and milk fat. Mamm. Genome 2007, 18, 64–74. [Google Scholar] [CrossRef]
- Zhang, W.; Wang, Y.; Chu, Y. Tacr3/NK3R: Beyond Their Roles in Reproduction. ACS Chem. Neurosci. 2020, 11, 2935–2943. [Google Scholar] [CrossRef]
- Chen, W.H.; Tian, Z.L.; Sun, W.; Chu, M.X. Polymorphism of Tacr3 Gene and lts Association with Litter Size in Sheep. China Anim. Husb. Vet. Med. 2019, 46, 1405–1412. [Google Scholar] [CrossRef]
- Young, J.; Bouligand, J.; Francou, B.; Raffin-Sanson, M.-L.; Gaillez, S.; Jeanpierre, M.; Grynberg, M.; Kamenicky, P.; Chanson, P.; Brailly-Tabard, S.; et al. TAC3 and TACR3 defects cause hypothalamic congenital hypogonadotropic hypogonadism in humans. J. Clin. Endocrinol. Metab. 2010, 95, 2287–2295. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.J.; Caligioni, C.S.; Chan, Y.-M.; Seminara, S.B. Uncovering novel reproductive defects in neurokinin B receptor null mice: Closing the gap between mice and men. Endocrinology 2012, 153, 1498–1508. [Google Scholar] [CrossRef] [PubMed]
- Lasaga, M.; Debeljuk, L. Tachykinins and the hypothalamo-pituitary-gonadal axis: An update. Peptides 2011, 32, 1972–1978. [Google Scholar] [CrossRef] [PubMed]
- Corander, M.P.; Challis, B.G.; Thompson, E.L.; Jovanovic, Z.; Tung, Y.C.L.; Rimmington, D.; Huhtaniemi, I.T.; Murphy, K.G.; Topaloglu, A.K.; Yeo, G.S.H.; et al. The effects of neurokinin B upon gonadotrophin release in male rodents. J. Neuroendocrinol. 2010, 22, 181–187. [Google Scholar] [CrossRef]
- Yang, G.; Zhang, J.; Ma, X.; Ma, R.; Shen, J.; Liu, M.; Yu, D.; Feng, F.; Huang, C.; Ma, X.; et al. Polymorphisms of CCSER1 Gene and Their Correlation with Milk Quality Traits in Gannan Yak (Bos grunniens). Foods 2023, 12, 4318. [Google Scholar] [CrossRef]
- Yang, B.G.; Hao, H.S.; Du, W.H.; Zhu, H.B.; Zhao, X.M. Advances in research on plateau adaptation of yak. Acta Vet. Zootech. Sin. 2023, 54, 12–23. [Google Scholar]
- Du, M.; Yang, C.; Liang, Z.; Zhang, J.; Yang, Y.; Ahmad, A.A.; Yan, P.; Ding, X. Dietary Energy Levels Affect Carbohydrate Metabolism-Related Bacteria and Improve Meat Quality in the Longissimus Thoracis Muscle of Yak (Bos grunniens). Front. Vet. Sci. 2021, 8, 718036. [Google Scholar] [CrossRef]
- Dai, A.; Yang, J.T.; Ding, B.; Liu, H.N. Research progress on components and functional characteristics of yak milk. Chin. J. Anim. Nutr. 2022, 34, 3443–3453. [Google Scholar]
- Dong, J. The future development direction of Chinese dairy products. Agric. Prod. Process. 2009, 55–56. [Google Scholar]
- Li, Y.; Yang, X.; Zhao, S.; Zhang, Z.; Bai, L.; Zhaxi, P.; Qu, S.; Zhao, Y. Effects of sampling time and location on the geographical origin traceability of protected geographical indication (PGI) Hongyuan yak milk: Based on stable isotope ratios. Food Chem. 2024, 441, 138283. [Google Scholar] [CrossRef]
- Fang, D. Regulation of Duck Preadoipocyte Differentiation and the Cloneand Expression Patterns of FAS and SCDI Gene; Sichuan Agricultural University: Ya’an, China, 2015. [Google Scholar]
- Wang, T.; Ma, X.; Feng, F.; Zheng, F.; Zheng, Q.; Zhang, J.; Zhang, M.; Ma, C.; Deng, J.; Guo, X.; et al. Study on Single Nucleotide Polymorphism of LAP3 Gene and Its Correlation with Dairy Quality Traits of Gannan Yak. Foods 2024, 13, 2953. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Wang, J.; Zhang, X.; Shi, Q.; Xin, L.; Fu, H.; Wang, Y. Effects of radio frequency assisted blanching on polyphenol oxidase, weight loss, texture, color and microstructure of potato. Food Chem. 2018, 248, 173–182. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Knight, T.J.; Reecy, J.M.; Beitz, D.C. DNA polymorphisms in bovine fatty acid synthase are associated with beef fatty acid composition. Anim. Genet. 2008, 39, 62–70. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Sun, D.; Zhang, S.; Yang, S.; Alim, M.A.; Zhang, Q.; Li, Y.; Liu, L. Genetic effects of FASN, PPARGC1A, ABCG2 and IGF1 revealing the association with milk fatty acids in a Chinese Holstein cattle population based on a post genome-wide association study. BMC Genet. 2016, 17, 110. [Google Scholar] [CrossRef]
- Roy, R.; Gautier, M.; Hayes, H.; Laurent, P.; Osta, R.; Zaragoza, P.; Eggen, A.; Rodellar, C. Assignment of the fatty acid synthase (FASN) gene to bovine chromosome 19 (19q22) by in situ hybridization and confirmation by somatic cell hybrid mapping. Cytogenet. Cell Genet. 2001, 93, 141–142. [Google Scholar] [CrossRef]
- Schennink, A.; Bovenhuis, H.; Léon-Kloosterziel, K.M.; Van Arendonk, J.A.M.; Visker, M.H.P.W. Effect of polymorphisms in the FASN, OLR1, PPARGC1A, PRL and STAT5A genes on bovine milk-fat composition. Anim. Genet. 2009, 40, 909–916. [Google Scholar] [CrossRef]
- Ordovás, L.; Roy, R.; Pampín, S.; Zaragoza, P.; Osta, R.; Rodríguez-Rey, J.C.; Rodellar, C. The g.763G>C SNP of the bovine FASN gene affects its promoter activity via Sp-mediated regulation: Implications for the bovine lactating mammary gland. Physiol. Genom. 2008, 34, 144–148. [Google Scholar] [CrossRef]
- Noel, S.D.; Abreu, A.P.; Xu, S.; Muyide, T.; Gianetti, E.; Tusset, C.; Carroll, J.; Latronico, A.C.; Seminara, S.B.; Carroll, R.S.; et al. TACR3 mutations disrupt NK3R function through distinct mechanisms in GnRH-deficient patients. FASEB J. 2014, 28, 1924–1937. [Google Scholar] [CrossRef]
- Gazave, E.; Marqués-Bonet, T.; Fernando, O.; Charlesworth, B.; Navarro, A. Patterns and rates of intron divergence between humans and chimpanzees. Genome Biol. 2007, 8, R21. [Google Scholar] [CrossRef]
- Nott, A.; Meislin, S.H.; Moore, M.J. A quantitative analysis of intron effects on mammalian gene expression. RNA 2003, 9, 607–617. [Google Scholar] [CrossRef]
| Gene | SNPs | Position | Genotypic Frequencies | Allelic Frequencies | He | Ho | X2 | PIC | HW p Value | |||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| FAS | g.13,276T>C | Intron | TT | TC | CC | T | C | 0.321 | 0.679 | 0.065 | 0.269 | 0.799 |
| 0.636 | 0.327 | 0.037 | 0.799 | 0.201 | ||||||||
| g.74,382C>G | Intron | CC | CG | GG | C | G | 0.153 | 0.847 | 0.810 | 0.141 | 0.368 | |
| 0.846 | 0.142 | 0.012 | 0.917 | 0.083 | ||||||||
| TACR3 | g.40,529A>G | Intron | AA | AG | GG | A | G | 0.359 | 0.641 | 0.160 | 0.295 | 0.689 |
| 0.049 | 0.370 | 0.580 | 0.235 | 0.765 | ||||||||
| g.40,555C>T | Intron | CC | CT | TT | C | T | 0.362 | 0.638 | 0.004 | 0.297 | 0.948 | |
| 0.580 | 0.364 | 0.056 | 0.762 | 0.238 | ||||||||
| FAS g.13,276T>C | ||||||||
| Genotype | Casein/% | Protein/% | Fat/% | TS/% | SNF/% | Lactose/% | Citric Acid/% | Acidity/°T |
| TT | 4.11 ± 0.28 | 4.92 ± 0.36 | 5.24 ± 2.20 | 16.66 ± 2.54 | 11.25 ± 0.46 | 4.97 ± 0.16 | 0.23 ± 0.34 | 12.44 ± 1.19 |
| TC | 4.12 ± 0.24 | 4.95 ± 0.33 | 5.30 ± 1.76 | 16.48 ± 1.67 | 11.39 ± 0.36 | 5.00 ± 0.12 | 0.23 ± 0.30 | 12.53 ± 1.26 |
| CC | 3.93 ± 0.35 | 4.75 ± 0.48 | 3.41 ± 0.37 | 14.12 ± 1.22 | 11.20 ± 0.51 | 5.09 ± 0.17 | 0.22 ± 0.28 | 13.03 ± 0.93 |
| p value | p = 0.274 | p = 0.414 | p = 0.202 | p = 0.051 | p = 0.144 | p = 0.104 | p = 0.424 | p = 0.627 |
| FAS g.74,382C>G | ||||||||
| Genotype | Casein/% | Protein/% | Fat/% | TS/% | SNF/% | Lactose/% | Citric Acid/% | Acidity/°T |
| CC | 4.08 ± 0.26 a | 4.90 ± 0.36 a | 5.21 ± 2.10 | 16.35 ± 2.09 | 11.27 ± 0.45 ab | 5.00 ± 0.16 | 0.23 ± 0.33 | 12.34 ± 1.22 a |
| CG | 4.24 ± 0.27 a | 5.10 ± 0.33 a | 5.39 ± 1.94 | 16.73 ± 1.94 | 11.47 ± 0.37 a | 4.93 ± 0.15 | 0.24 ± 0.22 | 13.01 ± 1.22 a |
| GG | 3.58 ± 0.24 b | 4.35 ± 0.38 b | 3.34 ± 0.78 | 13.94 ± 0.40 | 10.82 ± 0.39 b | 5.03 ± 0.57 | 0.25 ± 0.24 | 10.82 ± 0.14 b |
| p value | p = 0.001 | p = 0.004 | p = 0.412 | p = 0.184 | p = 0.043 | p = 0.162 | p = 0.170 | p = 0.010 |
| TACR3 g.40,529A>G | ||||||||
| Genotype | Casein/% | Protein/% | Fat/% | TS/% | SNF/% | Lactose/% | Citric Acid/% | Acidity/°T |
| AA | 4.04 ± 0.41 | 4.72 ± 0.53 b | 6.96 ± 3.21 | 17.96 ± 3.28 | 10.99 ± 0.60 b | 5.02 ± 0.21 | 0.21 ± 0.37 | 11.78 ± 1.64 |
| AG | 4.18 ± 0.27 | 5.02 ± 0.37 a | 5.79 ± 2.78 | 17.05 ± 2.72 | 11.38 ± 0.43 a | 4.98 ± 0.16 | 0.23 ± 0.34 | 12.62 ± 1.31 |
| GG | 4.05 ± 0.26 | 4.84 ± 0.39 ab | 5.30 ± 2.49 | 16.39 ± 2.41 | 11.21 ± 0.48 ab | 4.99 ± 0.15 | 0.23 ± 0.31 | 12.29 ± 1.23 |
| p value | p = 0.127 | p = 0.008 | p = 0.169 | p = 0.113 | p = 0.026 | p = 0.714 | p = 0.175 | p = 0.147 |
| TACR3 g.40,555C>T | ||||||||
| Genotype | Casein/% | Protein/% | Fat/% | TS/% | SNF/% | Lactose/% | Citric Acid/% | Acidity/°T |
| CC | 4.03 ± 0.28 | 4.84 ± 0.39 | 5.19 ± 2.25 | 16.39 ± 2.41 | 11.21 ± 0.48 | 4.99 ± 0.15 | 0.23 ± 0.31 | 12.29 ± 1.23 |
| CT | 4.17 ± 0.26 | 5.01 ± 0.36 | 5.62 ± 2.50 | 17.04 ± 2.74 | 11.37 ± 0.42 | 4.98 ± 0.16 | 0.23 ± 0.35 | 12.58 ± 1.27 |
| TT | 4.11 ± 0.43 | 4.84 ± 0.61 | 6.79 ± 3.05 | 17.90 ± 3.07 | 11.13 ± 0.69 | 5.00 ± 0.21 | 0.21 ± 0.38 | 12.16 ± 1.92 |
| p value | p = 0.136 | p = 0.170 | p = 0.124 | p = 0.114 | p = 0.105 | p = 0.934 | p = 0.281 | p = 0.351 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, T.; Ma, X.; Ma, C.; Yu, Q.; Liang, C.; Yan, P. The Biological Properties of the FAS and TACR3 Genes and the Association of Single-Nucleotide Polymorphisms with Milk Quality Traits in Gannan Yak. Foods 2025, 14, 1575. https://doi.org/10.3390/foods14091575
Wang T, Ma X, Ma C, Yu Q, Liang C, Yan P. The Biological Properties of the FAS and TACR3 Genes and the Association of Single-Nucleotide Polymorphisms with Milk Quality Traits in Gannan Yak. Foods. 2025; 14(9):1575. https://doi.org/10.3390/foods14091575
Chicago/Turabian StyleWang, Tong, Xiaoming Ma, Chaofan Ma, Qinran Yu, Chunnian Liang, and Ping Yan. 2025. "The Biological Properties of the FAS and TACR3 Genes and the Association of Single-Nucleotide Polymorphisms with Milk Quality Traits in Gannan Yak" Foods 14, no. 9: 1575. https://doi.org/10.3390/foods14091575
APA StyleWang, T., Ma, X., Ma, C., Yu, Q., Liang, C., & Yan, P. (2025). The Biological Properties of the FAS and TACR3 Genes and the Association of Single-Nucleotide Polymorphisms with Milk Quality Traits in Gannan Yak. Foods, 14(9), 1575. https://doi.org/10.3390/foods14091575






