Metaproteomic Analysis of Fermented Vegetable Formulations with Lactic Acid Bacteria: A Comparative Study from Initial Stage to 15 Days of Production
Abstract
1. Introduction
2. Materials and Methods
2.1. Process of Fermented Vegetable Production
2.2. Measurement of the Antioxidant Activity of Fermented Vegetables
2.2.1. Total Phenolic Content (TPC)
2.2.2. Ferric-Reducing Antioxidant Power (FRAP)
2.2.3. Oxygen Radical Absorbance Capacity (ORAC)
2.2.4. DPPH Radical Scavenging Activity (DPPH)
2.3. DNA Extraction and Sequencing
2.4. Microbiome Bioinformatic Analysis
2.5. Proteomic Analysis of All Formulas of Fermented Vegetables by LC-MS/MS
3. Results
3.1. Microbial Diversity of Fermented Foods Classified by Formula
3.2. Metaproteomic Analysis of Individual LAB Genera and Species
3.2.1. Genus Lactobacillus
3.2.2. Genus Weissella
3.2.3. Species Lacticaseibacillus rhamnosus
3.2.4. Species Levilactobacillus brevis
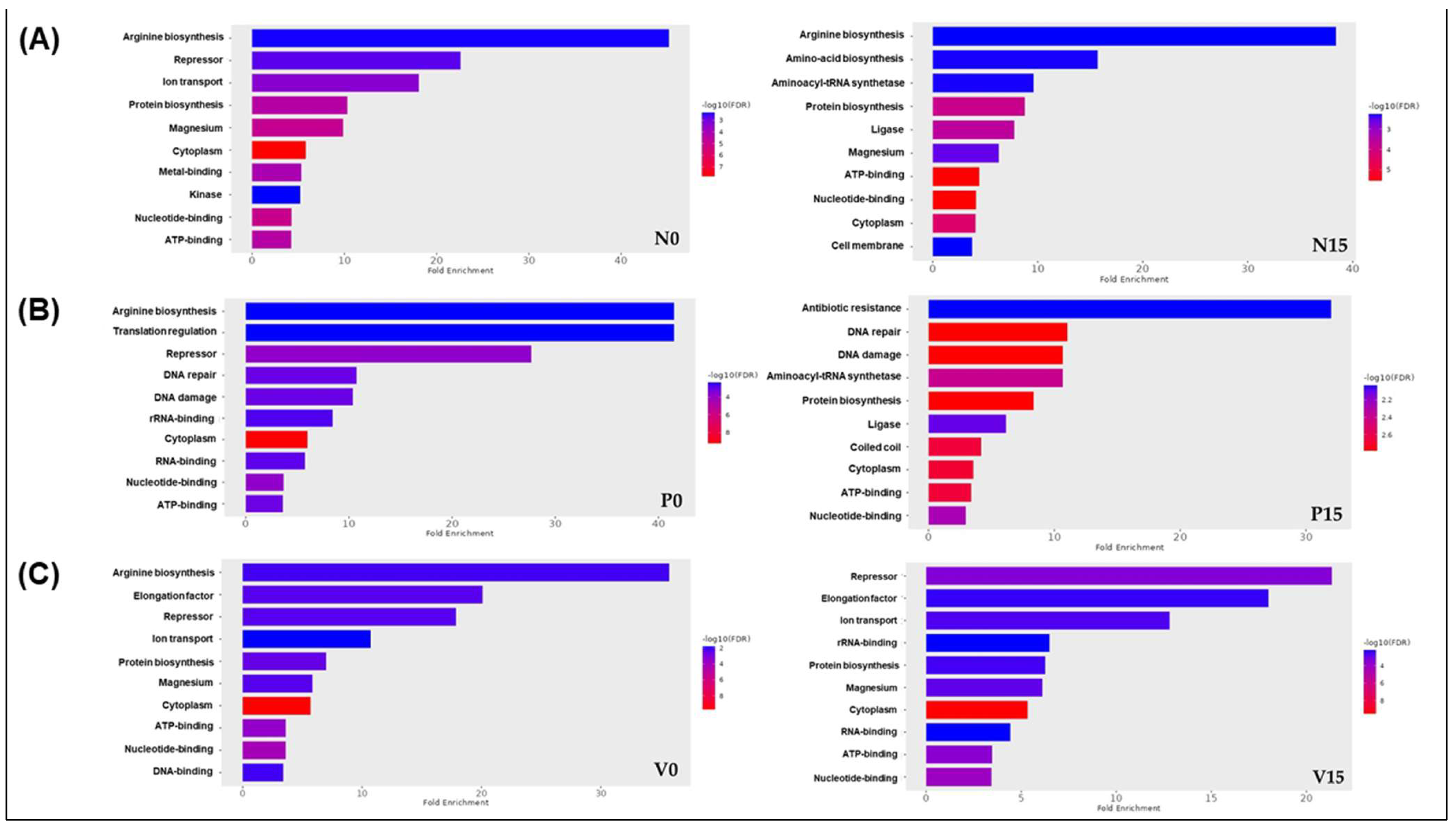
3.2.5. Assessment of Differences in Proteins Profiles Between Six Groups of Fermented Vegetables at Two Time Points (Day 0 and Day 15)
3.2.6. The Correlation Between Significant Proteins Identified Through ANOVA and the genera Lactobacillus, Weissella, Lacticaseibacillus rhamnosus, and Levilactobacillus brevis with Key Antioxidant Parameters
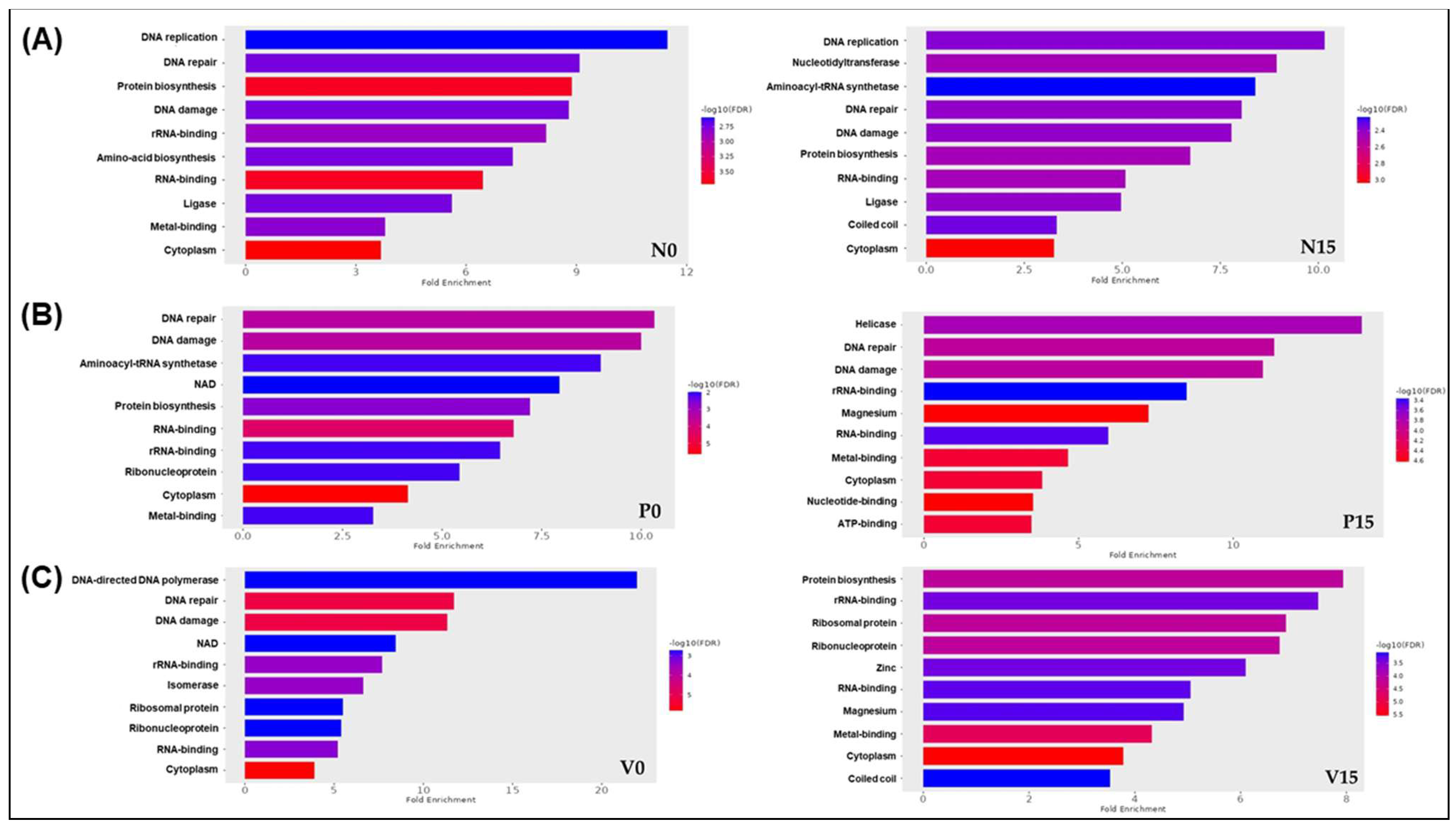
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Gul, K.; Singh, A.K.; Jabeen, R. Nutraceuticals and Functional Foods: The Foods for the Future World. Crit. Rev. Food Sci. Nutr. 2016, 56, 2617–2627. [Google Scholar] [CrossRef]
- Chareonrungrueangchai, K.; Wongkawinwoot, K.; Anothaisintawee, T.; Reutrakul, S. Dietary Factors and Risks of Cardiovascular Diseases: An Umbrella Review. Nutrients 2020, 12, 1088. [Google Scholar] [CrossRef] [PubMed]
- Knez, E.; Kadac-Czapska, K.; Grembecka, M. Effect of Fermentation on the Nutritional Quality of the Selected Vegetables and Legumes and Their Health Effects. Life 2023, 13, 655. [Google Scholar] [CrossRef] [PubMed]
- Yu, H.S.; Lee, N.K.; Choi, A.J.; Choe, J.S.; Bae, C.H.; Paik, H.D. Anti-Inflammatory Potential of Probiotic Strain Weissella cibaria JW15 Isolated from Kimchi through Regulation of NF-κB and MAPKs Pathways in LPS-Induced RAW 264.7 Cells. J. Microbiol. Biotechnol. 2019, 29, 1022–1032. [Google Scholar] [CrossRef] [PubMed]
- Woo, M.; Kim, M.; Noh, J.S.; Park, C.H.; Song, Y.O. Preventative activity of kimchi on high cholesterol diet-induced hepatic damage through regulation of lipid metabolism in LDL receptor knockout mice. Food Sci. Biotechnol. 2018, 27, 211–218. [Google Scholar] [CrossRef]
- Ngamsamer, C.; Muangnoi, C.; Tongkhao, K.; Sae-Tan, S.; Treesuwan, K.; Sirivarasai, J. Potential Health Benefits of Fermented Vegetables with Additions of Lacticaseibacillus rhamnosus GG and Polyphenol Vitexin Based on Their Antioxidant Properties and Prohealth Profiles. Foods 2024, 13, 982. [Google Scholar] [CrossRef]
- Rizo, J.; Guillén, D.; Farrés, A.; Díaz-Ruiz, G.; Sánchez, S.; Wacher, C.; Rodríguez-Sanoja, R. Omics in traditional vegetable fermented foods and beverages. Crit. Rev. Food Sci. Nutr. 2020, 60, 791–809. [Google Scholar] [CrossRef]
- Heyer, R.; Schallert, K.; Büdel, A.; Zoun, R.; Dorl, S.; Behne, A.; Kohrs, F.; Püttker, S.; Siewert, C.; Muth, T.; et al. A Robust and Universal Metaproteomics Workflow for Research Studies and Routine Diagnostics Within 24 h Using Phenol Extraction, FASP Digest, and the MetaProteomeAnalyzer. Front. Microbiol. 2019, 10, 1883. [Google Scholar] [CrossRef]
- Xie, M.; An, F.; Yue, X.; Liu, Y.; Shi, H.; Yang, M.; Cao, X.; Wu, J.; Wu, R. Characterization and comparison of metaproteomes in traditional and commercial dajiang, a fermented soybean paste in northeast China. Food Chem. 2019, 301, 125270. [Google Scholar] [CrossRef]
- Lee, H.W.; Yoon, S.R.; Yang, J.S.; Lee, H.M.; Kim, S.J.; Lee, J.Y.; Hwang, I.M.; You, S.Y.; Ha, J.H. Proteomic evaluation of kimchi, a traditional Korean fermented vegetable, and comparison of kimchi manufactured in China and Korea. J. Food Sci. Technol. 2021, 58, 389–396. [Google Scholar] [CrossRef]
- Keser, G.; Ozcan, T. Cross-over fermentation dynamics and proteomic properties of acid gels with indigenous Lactobacillus spp. isolated from cheeses. Food Microbiol. 2025, 128, 104700. [Google Scholar] [CrossRef]
- Park, J.-M.; Shin, J.-H.; Gu, J.-G.; Yoon, S.-J.; Song, J.-C.; Jeon, W.-M.; Suh, H.-J.; Chang, U.-J.; Yang, C.-Y.; Kim, J.-M. Effect of Antioxidant Activity in Kimchi during a Short-term and Over-ripening Fermentation Period. J. Biosci. Bioeng. 2011, 112, 356–359. [Google Scholar] [PubMed]
- Johnson, J.B.; Mani, J.S.; Naiker, M. Microplate Methods for Measuring Phenolic Content and Antioxidant Capacity in Chick-pea: Impact of Shaking. Eng. Proc. 2023, 48, 57. [Google Scholar] [CrossRef]
- Halvorsen, B.; Holte, K.; Myhrstad, M.; Barikmo, I.; Hvattum, E.; Remberg, S.; Wold, A.-B.; Haffner, K.; Baugerød, H.; Ander-sen, L.; et al. A Systematic Screening of Total Antioxidants in Dietary Plants. J. Nutr. 2002, 132, 461–471. [Google Scholar]
- Huang, D.; Ou, B.; Hampsch-Woodill, M.; Flanagan, J.A.; Prior, R.L. High-throughput Assay of Oxygen Radical Absorbance Capacity (ORAC) using a Multichannel Liquid Handling System Coupled with a Microplate Fluorescence Reader in 96-well Format. Food Chem. 2002, 50, 4437–4444. [Google Scholar]
- Amarowicz, R.; Pegg, R.B.; Rahimi-Moghaddam, P.; Barl, B.; Weil, J.A. Free-radical Scavenging Capacity and Antioxidant Activity of Selected Plant Species from the Canadian Prairies. Food Chem. 2004, 84, 551–562. [Google Scholar]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat Biotechnol 2019, 37, 852–857. [Google Scholar] [CrossRef]
- Lowry, O.H.; Rosebrough, N.J.; Farr, A.L.; Randall, R.J. Protein measurement with the Folin phenol reagent. J. Biol. Chem. 1951, 193, 265–275. [Google Scholar]
- Tyanova, S.; Temu, T.; Cox, J. The MaxQuant computational platform for mass spectrometry-based shotgun proteomics. Nat. Protoc. 2016, 11, 2301–2319. [Google Scholar] [CrossRef]
- Pang, Z.; Lu, Y.; Zhou, G.; Hui, F.; Xu, L.; Viau, C.; Aliya, F.S.; Patrick, E.M.; David, S.W.; Li, S.; et al. MetaboAnalyst 6.0: Towards a unified platform for metabolomics data processing, analysis and interpretation. Nucleic Acids Res. 2024, 52, W398–W406. [Google Scholar] [CrossRef]
- Bardou, P.; Mariette, J.; Escudié, F.; Djemiel, C.; Klopp, C. jvenn: An interactive Venn diagram viewer. BMC Bioinform. 2014, 15, 293. [Google Scholar] [CrossRef]
- Ge, S.X.; Jung, D.; Yao, R. ShinyGO: A graphical gene-set enrichment tool for animals and plants. Bioinformatics 2020, 36, 2628–2629. [Google Scholar] [CrossRef] [PubMed]
- Szklarczyk, D.; Santos, A.; von Mering, C.; Jensen, L.J.; Bork, P.; Kuhn, M. STITCH 5: Augmenting protein-chemical interaction networks with tissue and affinity data. Nucleic Acids Res. 2016, 44, D380–D384. [Google Scholar] [CrossRef]
- Mir, S.A.; Raja, J.; Masoodi, F.A. Fermented Vegetables, a Rich Repository of Beneficial Probiotics-A Review. Ferment. Technol. 2018, 7, 1–6. [Google Scholar]
- Park, S.; Saravanakumar, K.; Sathiyaseelan, A.; Han, K.S.; Lee, J.; Wang, M.H. Polysaccharides of Weissella cibaria Act as a Prebiotic to Enhance the Probiotic Potential of Lactobacillus rhamnosus. Appl. Biochem. Biotechnol. 2023, 195, 3928–3940. [Google Scholar]
- Feyereisen, M.; Mahony, J.; Kelleher, P.; Roberts, R.J.; O’Sullivan, T.; Geertman, J.-M.A.; van Sinderen, D. Comparative Ge-nome Analysis of the Lactobacillus brevis Species. BMC Genomics 2019, 20, 416. [Google Scholar]
- Marr, M.T.; Roberts, J.W. Function of transcription cleavage factors GreA and GreB at a regulatory pause site. Mol. Cell 2000, 6, 1275–1285. [Google Scholar] [CrossRef]
- Zhang, L.; Ma, H.; Kulyar, M.F.; Pan, H.; Li, K.; Li, A.; Mo, Q.; Wang, Y.; Dong, H.; Bao, Y.; et al. Complete genome analysis of Lactobacillus fermentum YLF016 and its probiotic characteristics. Microb. Pathog. 2022, 162, 105212. [Google Scholar] [CrossRef]
- Hu, F.; Liu, F. Mitochondrial stress: A bridge between mitochondrial dysfunction and metabolic diseases? Cell Signal. 2011, 23, 1528–1533. [Google Scholar] [CrossRef]
- Penin, F.; Geourjon, C.; Montserret, R.; Böckmann, A.; Lesage, A.; Yang, Y.S.; Bonod-Bidaud, C.; Cortay, J.C.; Nègre, D.; Cozzone, A.J.; et al. Three-dimensional structure of the DNA-binding domain of the fructose repressor from Escherichia coli by 1H and 15N NMR. J. Mol. Biol. 1997, 270, 496–510. [Google Scholar] [CrossRef]
- Ilyina, A.; Ramos-González, R.; Vargas-Segura, A.; Sánchez-Ramírez, J.; Palacios-Ponce, S.A.; Martínez-Hernández, J.L.; Segura-Ceniceros, E.P.; Contreras-Esquivel, J.C.; Aguilar-González, C.N. 3—Magnetic separation of nanobiostructured systems for innovation of biocatalytic processes in food industry. In Novel Approaches of Nanotechnology in Food; Grumezescu, A.M., Ed.; Academic Press: Cambridge, MA, USA, 2016; pp. 67–96. [Google Scholar]
- Yared, M.J.; Marcelot, A.; Barraud, P. Beyond the Anticodon: tRNA Core Modifications and Their Impact on Structure, Translation and Stress Adaptation. Genes 2024, 15, 374. [Google Scholar] [CrossRef] [PubMed]
- Musatov, A.; Sedlák, E. Role of cardiolipin in stability of integral membrane proteins. Biochimie 2017, 142, 102–111. [Google Scholar] [CrossRef] [PubMed]
- Nepper, J.F.; Lin, Y.C.; Weibel, D.B. Rcs Phosphorelay Activation in Cardiolipin-Deficient Escherichia coli Reduces Biofilm Formation. J. Bacteriol. 2019, 201, e00804-18. [Google Scholar] [CrossRef] [PubMed]
- Soumya, M.P.; Nampoothiri, K.M. An overview of functional genomics and relevance of glycosyltransferases in exopolysaccharide production by lactic acid bacteria. Int. J. Biol. Macromol. 2021, 184, 1014–1025. [Google Scholar] [CrossRef]
- Singleton, M.R.; Dillingham, M.S.; Wigley, D.B. Structure and mechanism of helicases and nucleic acid translocases. Annu. Rev. Biochem. 2007, 76, 23–50. [Google Scholar] [CrossRef]
- Owttrim, G.W. RNA helicases and abiotic stress. Nucleic Acids Res. 2006, 34, 3220–3230. [Google Scholar] [CrossRef]
- Jankowsky, E.; Fairman, M.E. RNA helicases—one fold for many functions. Curr. Opin. Struct. Biol. 2007, 17, 316–324. [Google Scholar] [CrossRef]
- Savijoki, K.; Ingmer, H.; Varmanen, P. Proteolytic systems of lactic acid bacteria. Appl. Microbiol. Biotechnol. 2006, 71, 394–406. [Google Scholar] [CrossRef]
- Fusco, V.; Quero, G.M.; Cho, G.S.; Kabisch, J.; Meske, D.; Neve, H.; Bockelmann, W.; Franz, C.M. The genus Weissella: Taxonomy, ecology and biotechnological potential. Front. Microbiol. 2015, 6, 155. [Google Scholar] [CrossRef]
- Peña-Ramos, E.A.; Xiong, Y.L. Whey and soy protein hydrolysates inhibit lipid oxidation in cooked pork patties. Meat Sci. 2003, 64, 259–263. [Google Scholar] [CrossRef]
- Devirgiliis, C.; Zinno, P.; Perozzi, G. Update on antibiotic resistance in foodborne Lactobacillus and Lactococcus species. Front. Microbiol. 2013, 4, 301. [Google Scholar] [CrossRef]
- Gänzle, M.G. Lactic metabolism revisited: Metabolism of lactic acid bacteria in food fermentations and food spoilage. Curr. Opin. Food Sci. 2015, 2, 106–117. [Google Scholar] [CrossRef]
- Wood, J.M. Bacterial responses to osmotic challenges. J. Gen. Physiol. 2015, 145, 381–388. [Google Scholar] [CrossRef]
- Parvez, S.; Malik, K.A.; Ah Kang, S.; Kim, H.Y. Probiotics and their fermented food products are beneficial for health. J. Appl. Microbiol. 2006, 100, 1171–1185. [Google Scholar] [CrossRef]
- Khodzhaieva, R.S.; Gladkov, E.S.; Kyrychenko, A.; Roshal, A.D. Progress and Achievements in Glycosylation of Flavonoids. Front. Chem. 2021, 9, 637994. [Google Scholar] [CrossRef]
- Ellis, E.M. Microbial aldo-keto reductases. FEMS Microbiol. Lett. 2002, 216, 123–131. [Google Scholar] [CrossRef]
- Wang, C.; Yan, R.; Luo, D.; Watabe, K.; Liao, D.F.; Cao, D. Aldo-keto reductase family 1 member B10 promotes cell survival by regulating lipid synthesis and eliminating carbonyls. J. Biol. Chem. 2009, 284, 26742–26748. [Google Scholar] [CrossRef]
- Adebawo, O.O.; Ruiz-Barba, J.L.; Warner, P.J.; Oguntimein, G.B. Regulation of aspartokinase in Lactobacillus plantarum. J. Appl. Microbiol. 1997, 82, 191–196. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Sybesma, W.; Starrenburg, M.; Tijsseling, L.; Hoefnagel, M.H.; Hugenholtz, J. Effects of cultivation conditions on folate production by lactic acid bacteria. Appl. Environ. Microbiol. 2003, 69, 4542–4548. [Google Scholar] [CrossRef]
- Li, S.F.; Zhu, C.S.; Wang, Y.M.; Xie, X.X.; Xiao, L.L.; Zhang, Z.C.; Tang, Q.Q.; Li, X. Downregulation of β1,4-galactosyltransferase 5 improves insulin resistance by promoting adipocyte commitment and reducing inflammation. Cell Death Dis. 2018, 9, 196. [Google Scholar] [CrossRef]
- Tian, M.; Wang, H.; Li, Y.; Wang, J.; Ren, D.; Lin, K. A comprehensive safety assessment of a novel starter Weissella confusa M1 combining with whole-genome sequencing. Food Res. Int. 2025, 202, 115748. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Mao, Y. AAA+ ATPases in Protein Degradation: Structures, Functions and Mechanisms. Biomolecules 2020, 10, 629. [Google Scholar] [CrossRef] [PubMed]
- Elsholz, A.K.W.; Birk, M.S.; Charpentier, E.; Turgay, K. Functional Diversity of AAA+ Protease Complexes in Bacillus subtilis. Front. Mol. Biosci. 2017, 4, 44. [Google Scholar] [CrossRef]
- Jurášková, D.; Ribeiro, S.C.; Silva, C.C.G. Exopolysaccharides Produced by Lactic Acid Bacteria: From Biosynthesis to Health-Promoting Properties. Foods 2022, 11, 156. [Google Scholar] [CrossRef]
- Cui, Y.; Qu, X. CRISPR-Cas systems of lactic acid bacteria and applications in food science. Biotechnol. Adv. 2024, 71, 108323. [Google Scholar] [CrossRef] [PubMed]
- Mugwanda, K.; Hamese, S.; Van Zyl, W.F.; Prinsloo, E.; Du Plessis, M.; Dicks, L.M.T.; Thimiri Govinda Raj, D.B. Recent advances in genetic tools for engineering probiotic lactic acid bacteria. Biosci. Rep. 2023, 43, 1299. [Google Scholar] [CrossRef]
- Zuo, D.; Yin, Y.; Fang, T.; Jiang, H.; Ding, J.; Hu, H.; Wang, S.; Qi, J.; Tian, M.; Yu, S. A homolog of low molecular weight protein tyrosine phosphatase isolated from Brucella melitensis displays an acidic dual specific phosphatase activity, nonessential for bacterial resistance to bactericidal factors and virulence. Comp. Immunol. Microbiol. Infect. Dis. 2022, 90–91, 101904. [Google Scholar] [CrossRef]
- Delgado, S.; Flórez, A.B.; Guadamuro, L.; Mayo, B. Genetic and biochemical characterization of an oligo-α-1,6-glucosidase from Lactobacillus plantarum. Int. J. Food Microbiol. 2017, 246, 32–39. [Google Scholar] [CrossRef]
- Illikoud, N.; do Carmo, F.L.R.; Daniel, N.; Jan, G.; Gagnaire, V. Development of innovative fermented products by exploiting the diversity of immunomodulatory properties and fermentative activity of lactic and propionic acid bacteria. Food Res. Int. 2023, 166, 112557. [Google Scholar] [CrossRef]
- Zhang, z.; Song, T.; Cai, W.; Wang, F.; Lv, G. Effects of different depolymerisation methods on the physicochemical and antioxidant properties of polysaccharides derived from Sparassis latifolia. Process Biochem. 2021, 110, 110–117. [Google Scholar] [CrossRef]
- Zhang, B.; Gu, H.; Yang, Y.; Bai, H.; Zhao, C.; Si, M.; Su, T.; Shen, X. Molecular Mechanisms of AhpC in Resistance to Oxidative Stress in Burkholderia thailandensis. Front. Microbiol. 2019, 10, 1483. [Google Scholar] [CrossRef]
- Muangrat, R.; Thipsuwan, Y. Enhancing bacterial inhibition in beetroot beverages: The potent combination of sappan heartwood extracts via accelerated solvent extraction and pulsed electric field methods. J. Agric. Food Res. 2024, 18, 101268. [Google Scholar] [CrossRef]
- Sharma, R.; Garg, P.; Kumar, P.; Bhatia, S.K.; Kulshrestha, S. Microbial Fermentation and Its Role in Quality Improvement of Fermented Foods. Fermentation 2020, 6, 106. [Google Scholar] [CrossRef]
- Wilkins, M.R.; Sanchez, J.C.; Gooley, A.A.; Appel, R.D.; Humphery-Smith, I.; Hochstrasser, D.F.; Williams, K.L. Progress with proteome projects: Why all proteins expressed by a genome should be identified and how to do it. Biotechnol. Genet. Eng. Rev. 1996, 13, 19–50. [Google Scholar] [CrossRef] [PubMed]
- Aebersold, R.; Mann, M. Mass-spectrometric exploration of proteome structure and function. Nature 2016, 537, 347–355. [Google Scholar] [CrossRef]
- Van Den Bossche, T.; Kunath, B.J.; Schallert, K.; Schäpe, S.S.; Abraham, P.E.; Armengaud, J.; Arntzen, M.; Bassignani, A.; Benndorf, D.; Fuchs, S.; et al. Critical Assessment of MetaProteome Investigation (CAMPI): A multi-laboratory comparison of established workflows. Nat. Commun. 2021, 12, 7305. [Google Scholar] [CrossRef]

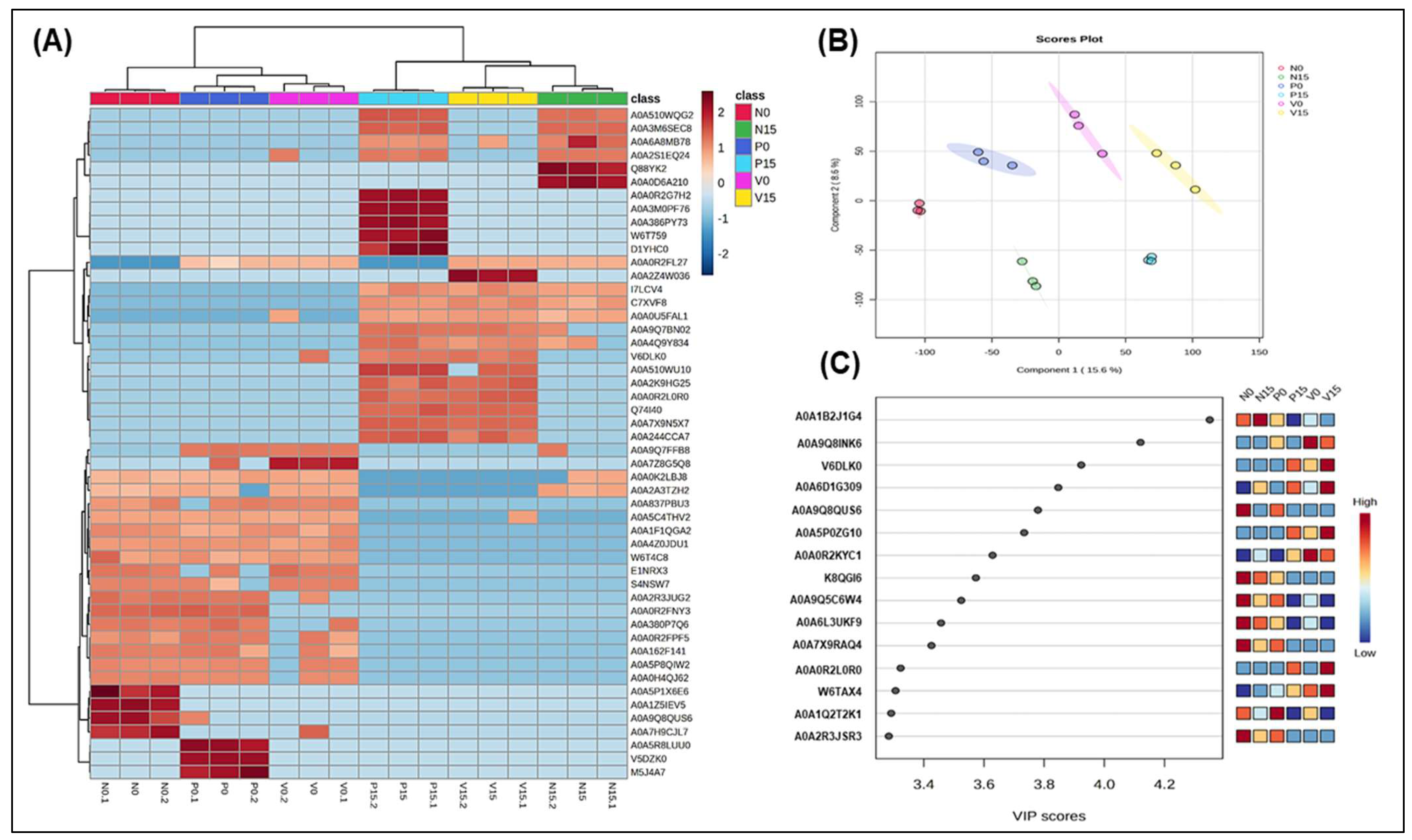
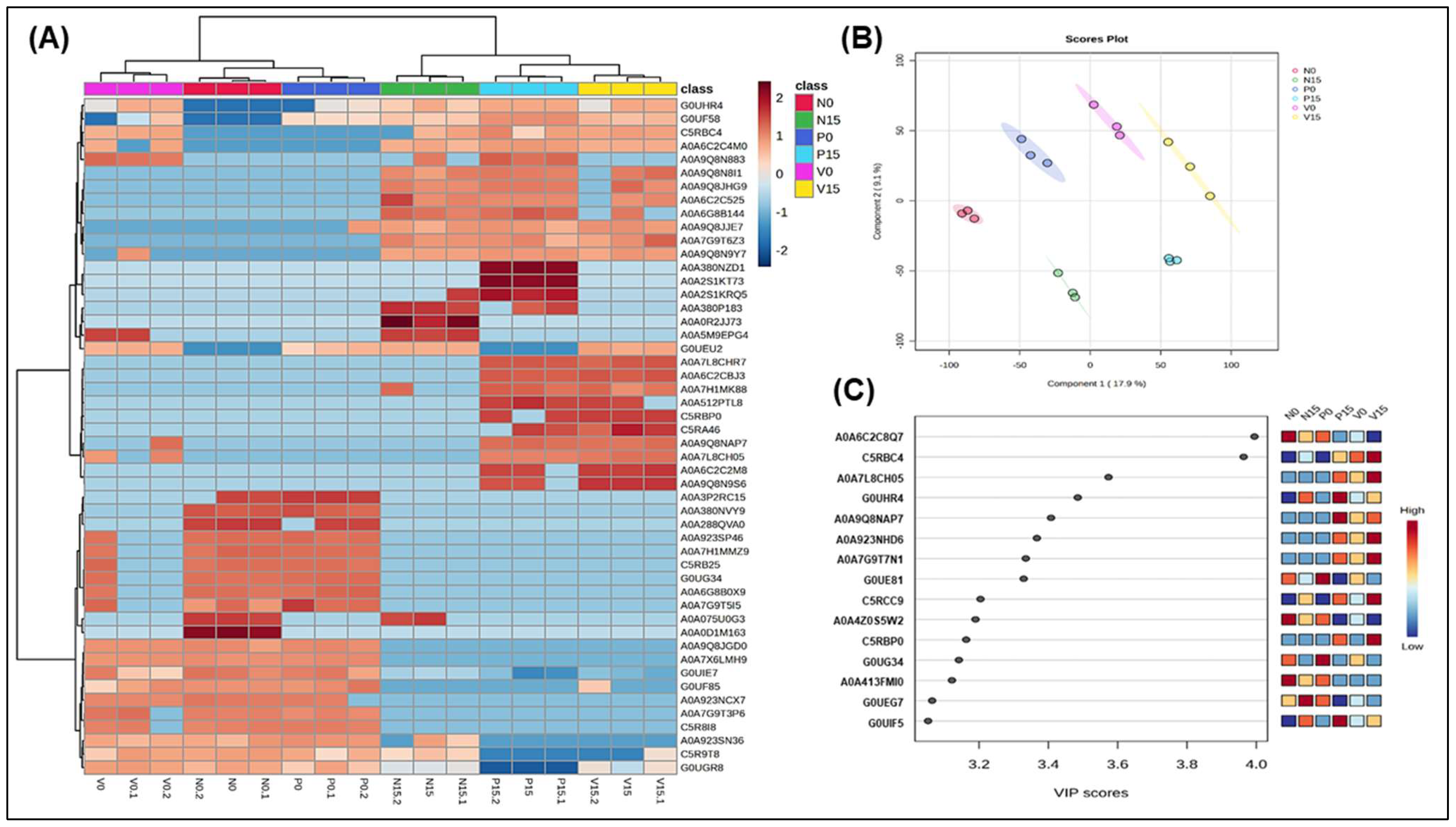
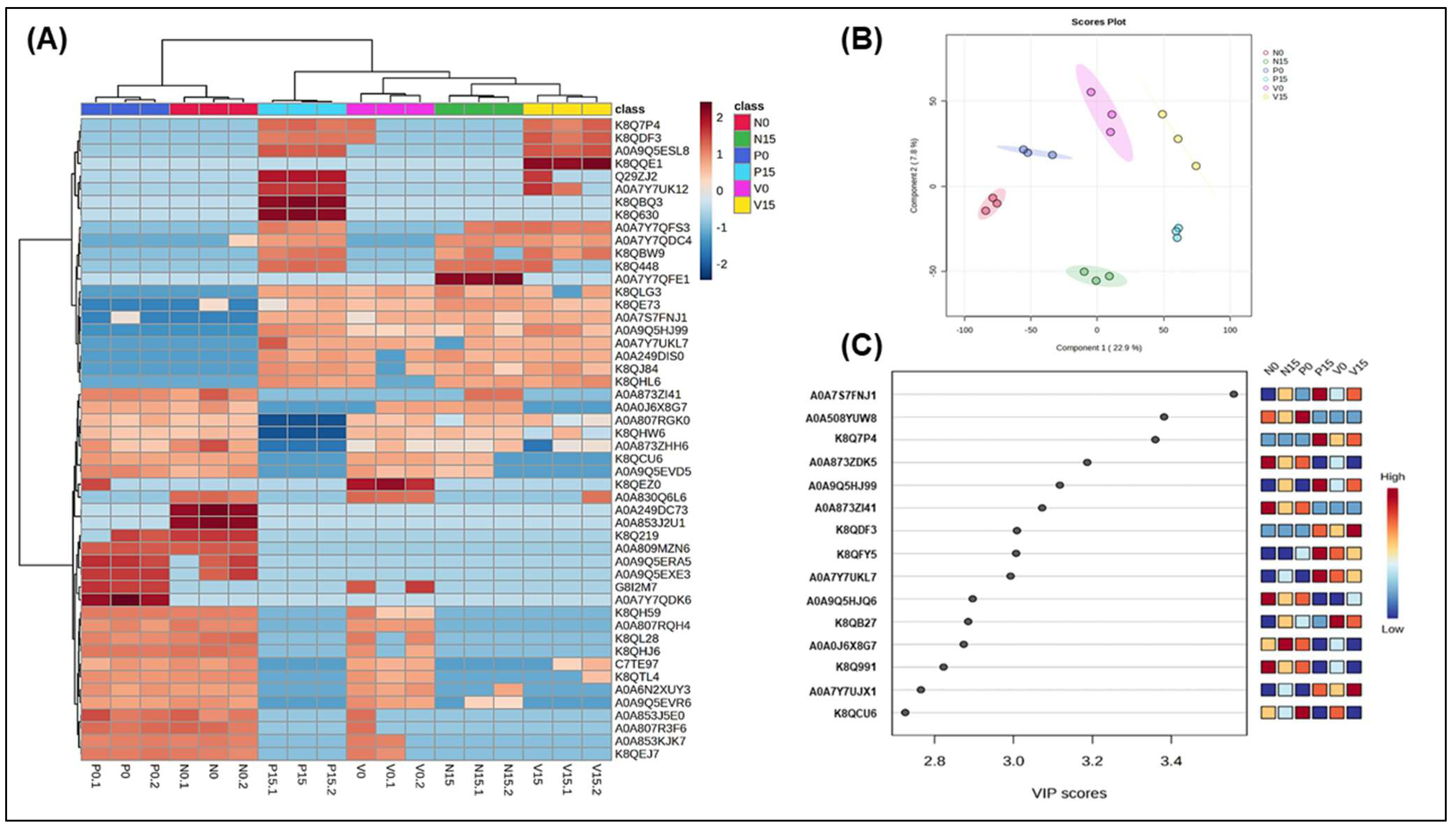

| Genus/Species | Protein Names | Tukey’s HSD (p Value < 0.05) | |
|---|---|---|---|
| Lactobacillus | |||
| 1 | A0A0R2G7H2 | Diadenosine tetraphosphatase-like protein | P15-N0; P15-N15; P15-P0; V0-P15; V15-P15 |
| 2 | A0A7X9N5X7 | GNAT family N-acetyltransferase | P15-N0; V15-N0; P15-N15; V15-N15; P15-P0; V15-P0; V0-P15; V15-P15; V15-V0 |
| 3 | A0A1Z5IEV5 | UDP-N-acetylglucosamine 1-carboxyvinyltransferase | N15-N0; P0-N0; P15-N0; V0-N0; V15-N0 |
| 4 | A0A510WQG2 | ATP-dependent Clp protease ATP-binding subunit ClpC | N15-N0; P15-N0; P0-N15; P15-N15; V0-N15; V15-N15; P15-P0; V0-P15; V15-P15 |
| 5 | A0A0R2FNY3 | Transposase | N15-N0; P15-N0; V0-N0; V15-N0; P0-N15; P15-P0; V0-P0; V15-P0 |
| Weissella | |||
| 1 | A0A7X6LMH9 | Aminoacyltransferase | N15-N0; P0-N0; P15-N0; V15-N0; P0-N15; V0-N15; P15-P0; V0-P0; V15-P0; V0-P15; V15-V0 |
| 2 | A0A6C2CBJ3 | ABC transporter permease | P15-N0; V15-N0; P15-N15; V15-N15; P15-P0; V15-P0; V0-P15; V15-V0 |
| 3 | A0A2S1KT73 | ATP-binding/permease protein CydD | P15-N0; P15-N15; P15-P0; V0-P15; V15-P15 |
| 4 | A0A0D1M163 | VWA-like domain-containing protein | N15-N0; P0-N0; P15-N0; V0-N0; V15-N0 |
| 5 | A0A7L8CHR7 | ABC transporter permease | P15-N0; V15-N0; P15-N15; V15-N15; P15-P0; V15-P0; V0-P15; V15-V0 |
| Lacticaseibacillus rhamnosus | |||
| 1 | A0A249DF14 | tRNA (guanine-N(1)-methyltransferase | P15-N0; V0-N0; P15-P0; V0-P0 |
| 2 | A0A6N2XUY3 | Capsid protein (F protein) | N15-N0; P15-N0; V15-N0; P0-N15; V0-N15; P15-P0; V15-P0; V0-P15; V15-V0 |
| 3 | A0A6N2ZUA9 | site-specific DNA-methyltransferase (adenine-specific) | N15-N0; P15-N0; V15-N0; P0-N15; P15-P0; V15-P0 |
| 4 | A0A7Y7QFE1 | Signal peptidase I | N15-N0; P0-N15; P15-N15; V0-N15; V15-N15 |
| 5 | A0A7Y7QFS3 | Aldo/keto reductase | N15-N0; P15-N0; V15-N0; P0-N15; V0-N15; P15-P0; V15-P0; V0-P15; V15-V0 |
| Levilactobacillus brevis | |||
| 1 | A0A5B7Y3K0 | RepC protein | N15-N0; P15-N0; V15-N0; P0-N15; V0-N15; P15-P0; V15-P0; V0-P15; V15-V0 |
| 2 | Q03U02 | Uronate isomerase | N15-N0; P15-N0; V15-N0; P0-N15; V0-N15; P15-P0; V15-P0; V0-P15; V15-V0 |
| 3 | A0A5B7Y1U6 | DNA helicase RecQ | P15-N0; V15-N0; P15-N15; V15-N15; P15-P0; V15-P0; V0-P15; V15-V0 |
| 4 | Q03N47 | Nickase | N15-N0; P15-N0; V0-N0; V15-N0; P0-N15; P15-P0; V0-P0; V15-P0 |
| 5 | A0A7Z6MKU9 | Protein-tyrosine-phosphatase | N15-N0; P15-N0; V15-N0; P0-N15; V0-N15; P15-P0; V15-P0; V0-P15; V15-V0 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rueangsri, N.; Roytrakul, S.; Muangnoi, C.; Tongkhao, K.; Sae-Tan, S.; Treesuwan, K.; Sirivarasai, J. Metaproteomic Analysis of Fermented Vegetable Formulations with Lactic Acid Bacteria: A Comparative Study from Initial Stage to 15 Days of Production. Foods 2025, 14, 1148. https://doi.org/10.3390/foods14071148
Rueangsri N, Roytrakul S, Muangnoi C, Tongkhao K, Sae-Tan S, Treesuwan K, Sirivarasai J. Metaproteomic Analysis of Fermented Vegetable Formulations with Lactic Acid Bacteria: A Comparative Study from Initial Stage to 15 Days of Production. Foods. 2025; 14(7):1148. https://doi.org/10.3390/foods14071148
Chicago/Turabian StyleRueangsri, Narisa, Sittiruk Roytrakul, Chawanphat Muangnoi, Kullanart Tongkhao, Sudathip Sae-Tan, Khemmapas Treesuwan, and Jintana Sirivarasai. 2025. "Metaproteomic Analysis of Fermented Vegetable Formulations with Lactic Acid Bacteria: A Comparative Study from Initial Stage to 15 Days of Production" Foods 14, no. 7: 1148. https://doi.org/10.3390/foods14071148
APA StyleRueangsri, N., Roytrakul, S., Muangnoi, C., Tongkhao, K., Sae-Tan, S., Treesuwan, K., & Sirivarasai, J. (2025). Metaproteomic Analysis of Fermented Vegetable Formulations with Lactic Acid Bacteria: A Comparative Study from Initial Stage to 15 Days of Production. Foods, 14(7), 1148. https://doi.org/10.3390/foods14071148








