Quantitative Analysis of Peanut Skin Adulterants by Fourier Transform Near-Infrared Spectroscopy Combined with Chemometrics
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Preparation
2.2. Spectral Acquisition
2.3. Chemometric Analysis
2.3.1. Spectral Data Processing
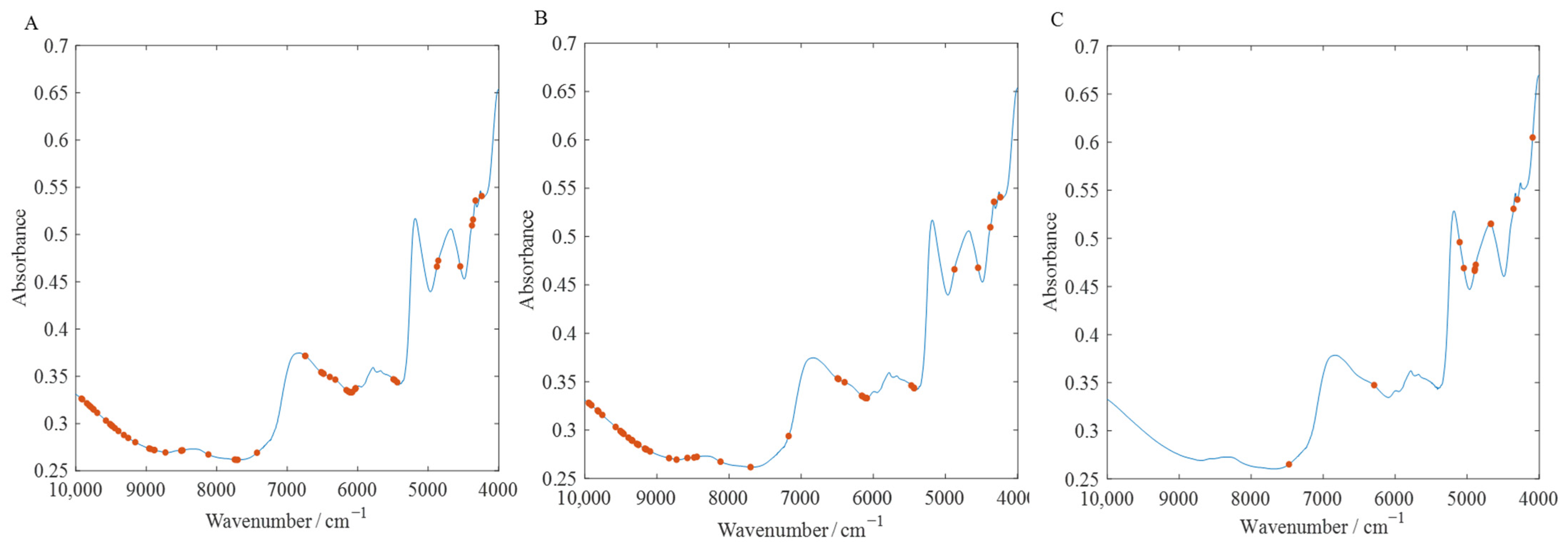
2.3.2. Feature Selection
2.3.3. Quantitative Models
2.4. Model Evaluation
3. Results and Discussion
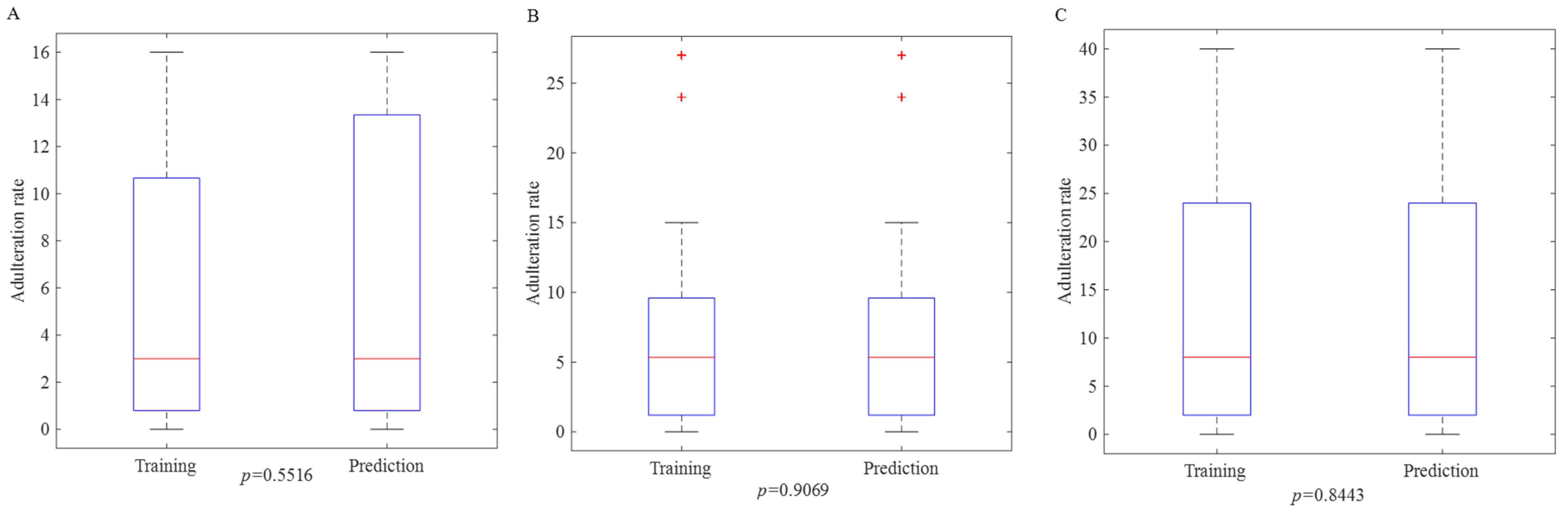
3.1. Dividing the Samples into Prediction and Calibration Sets
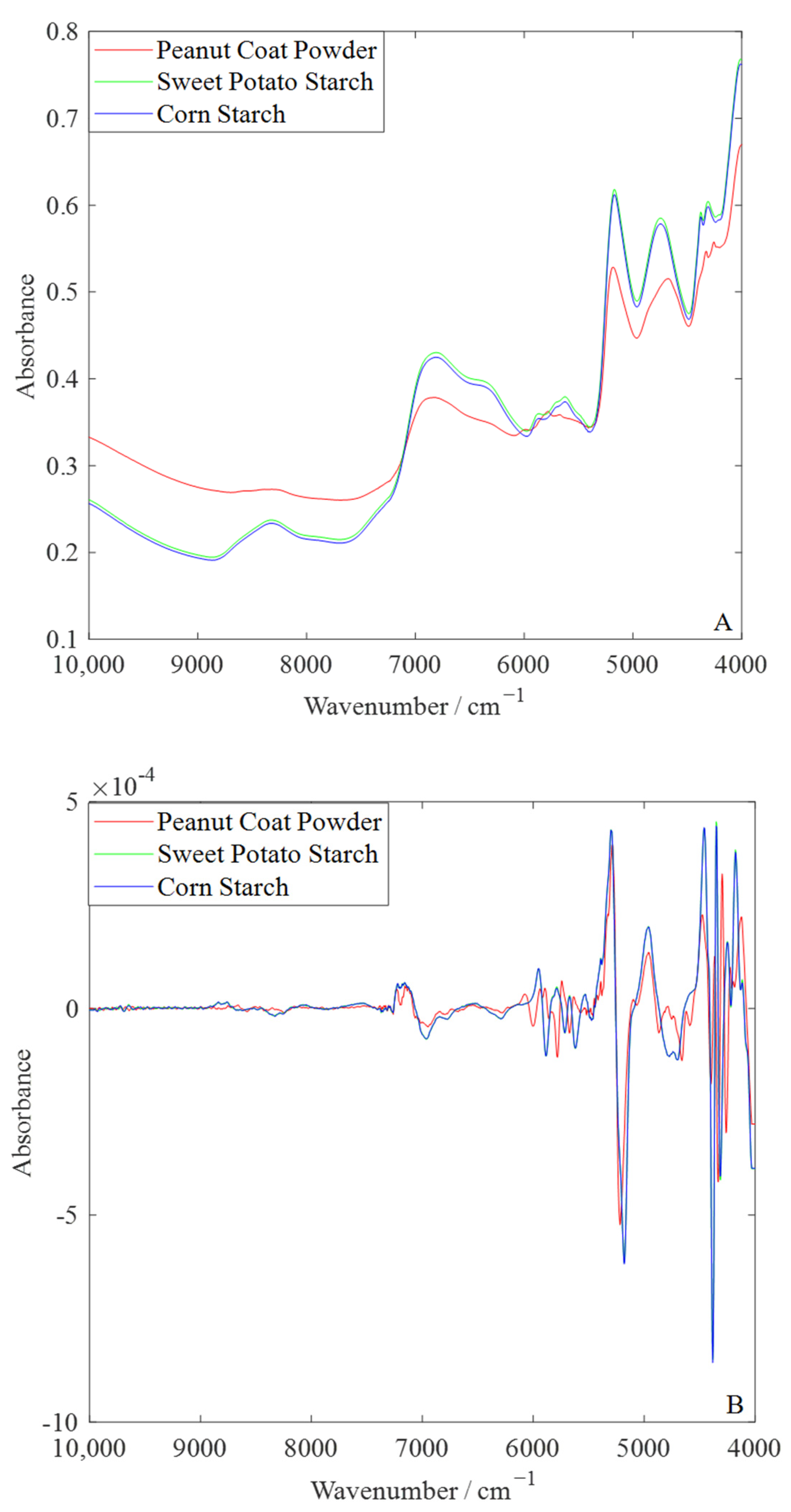
3.2. Spectral Characteristics
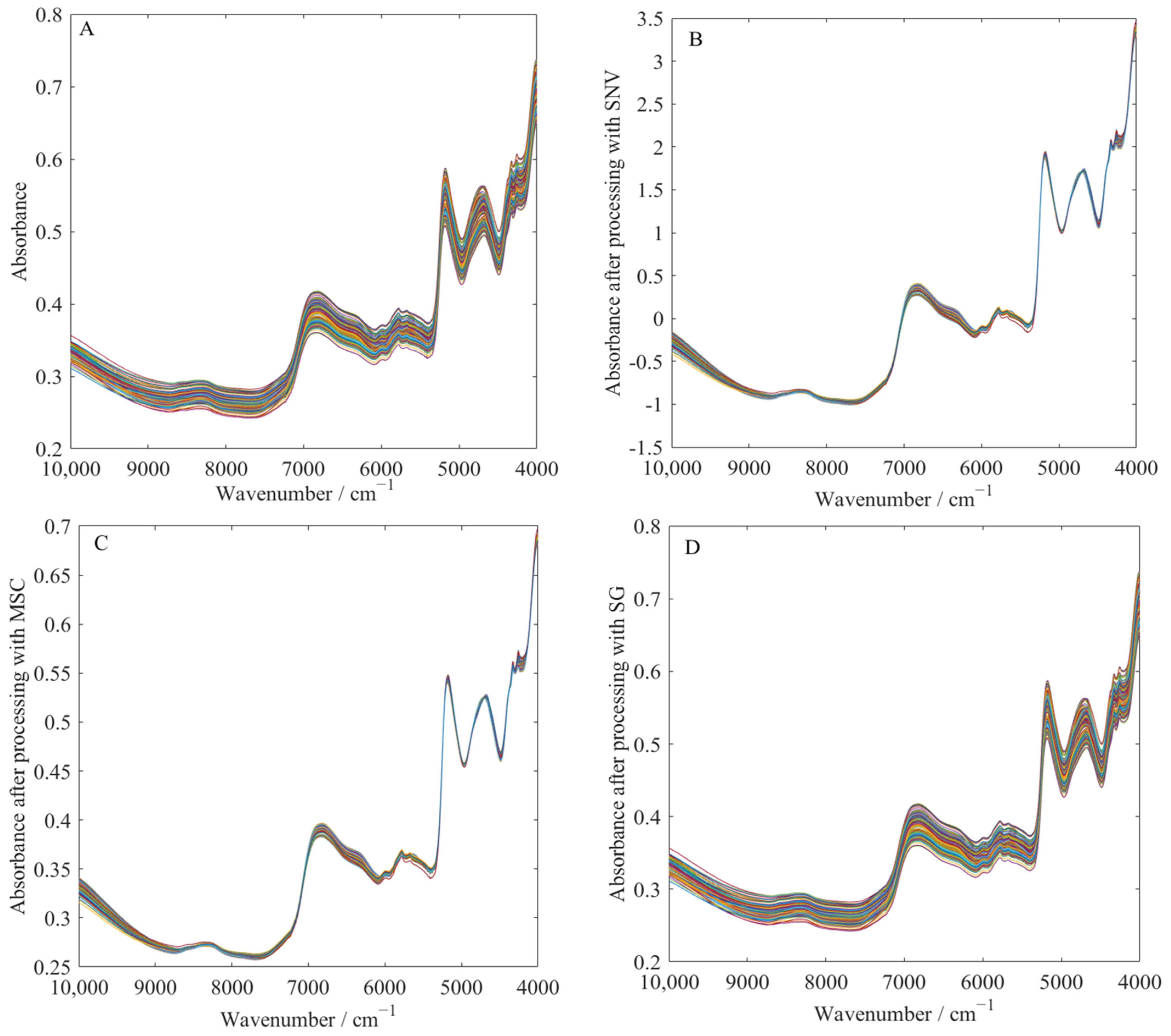
3.3. Analysis of Spectral Preprocessing Results
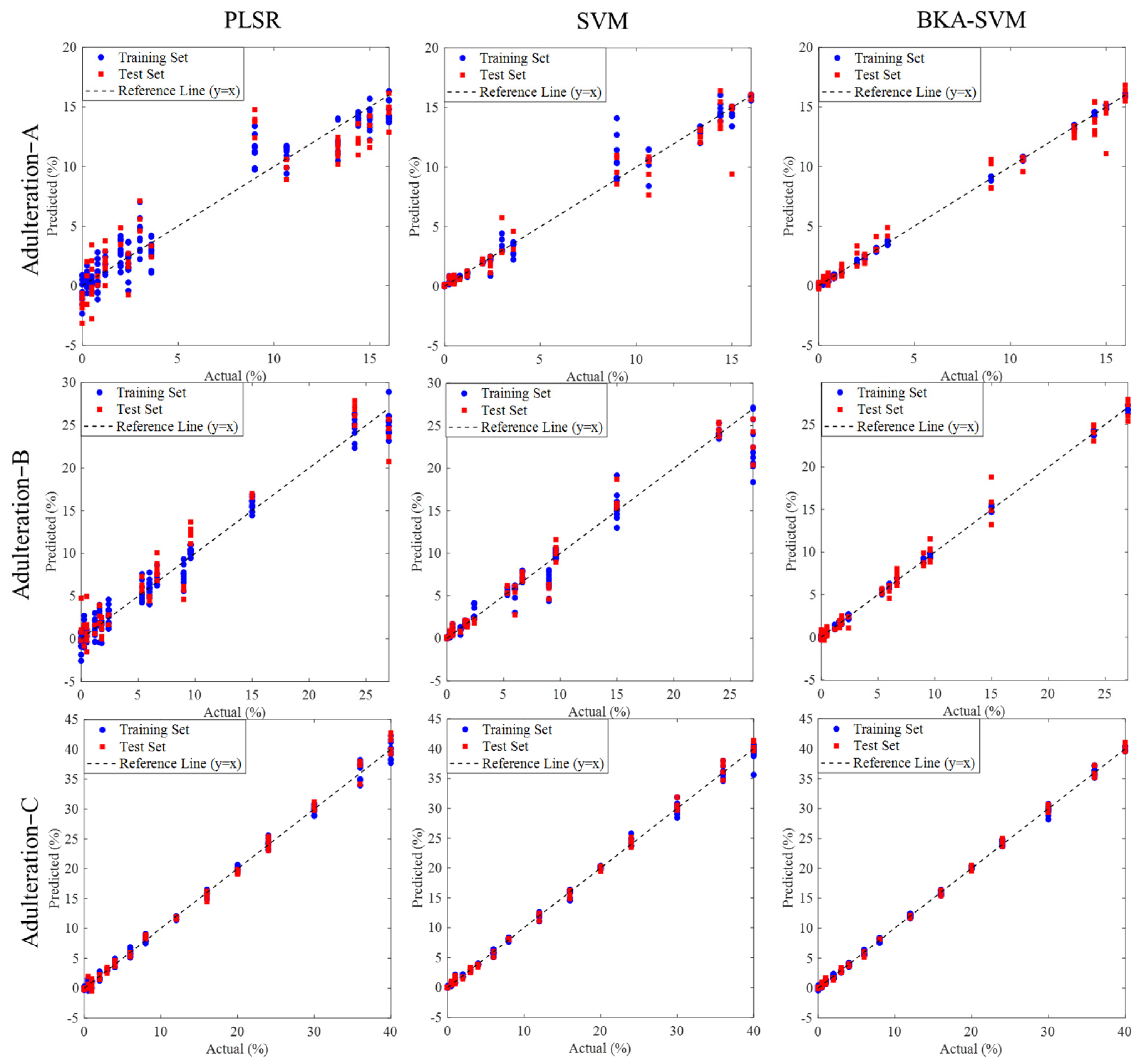
3.4. Prediction Results for Different Adulterations
3.5. Discussion
3.5.1. Discussion of Selected Variables
3.5.2. Discussion of Different Predicting Models
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Toomer, O.T.; Redhead, A.K.; Vu, T.C.; Santos, F.; Malheiros, R.; Proszkowiec-Weglarz, M. The effect of peanut skins as a natural antimicrobial feed additive on ileal and cecal microbiota in broiler chickens inoculated with Salmonella enterica Enteritidis. Poult. Sci. 2024, 103, 104159. [Google Scholar] [CrossRef] [PubMed]
- Redhead, A.K.; Azman, N.; Nasaruddin, A.I.; Vu, T.; Santos, F.; Malheiros, R.; Hussin, A.S.M.; Toomer, O.T. Peanut Skins as a Natural Antimicrobial Feed Additive to Reduce the Transmission of Salmonella in Poultry Meat Produced for Human Consumption. J. Food Prot. 2022, 85, 1479–1487. [Google Scholar] [CrossRef]
- Muñoz-Arrieta, R.; Esquivel-Alvarado, D.; Alfaro-Viquez, E.; Alvarez-Valverde, V.; Krueger, C.G.; Reed, J.D. Nutritional and bioactive composition of Spanish, Valencia, and Virginia type peanut skins. J. Food Compos. Anal. 2021, 98, 103816. [Google Scholar] [CrossRef]
- Bodoira, R.M.; Rodríguez Ruiz, A.C.; Martínez, M.L.; Velez, A.R.; Ribotta, P.D.; Maestri, D.M. From by-product to natural antioxidant: Incorporation of peanut skin extract in mayonnaise and its effect on physico-chemical and sensory properties. Food Biosci. 2024, 61, 104680. [Google Scholar] [CrossRef]
- Zhao, L.; Zhang, X.; He, L.; Li, Y.; Yu, Y.; Lu, Q.; Liu, R. Diet with high content of advanced glycation end products induces oxidative stress damage and systemic inflammation in experimental mice: Protective effect of peanut skin procyanidins. Food Sci. Hum. Wellness 2024, 13, 3570–3581. [Google Scholar] [CrossRef]
- Egido, C.; Saurina, J.; Sentellas, S.; Nunez, O. Honey fraud detection based on sugar syrup adulterations by HPLC-UV fingerprinting and chemometrics. Food Chem. 2024, 436, 137758. [Google Scholar] [CrossRef]
- Shi, T.; Wu, G.; Jin, Q.; Wang, X. Detection of camellia oil adulteration using chemometrics based on fatty acids GC fingerprints and phytosterols GC-MS fingerprints. Food Chem. 2021, 352, 129422. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.-D.; Zhang, M.; Liang, H.; Wang, P.-J.; Wang, T.; Gao, X.-L. Identification and quantification of cinnamon adulteration using non-targeted HPLC-DAD fingerprints and chemometrics. J. Food Compos. Anal. 2025, 139, 107076. [Google Scholar] [CrossRef]
- Shi, T.; Dai, T.; Wu, G.; Jin, Q.; Wang, X. Camellia oil grading adulteration detection using characteristic volatile components GC-MS fingerprints combined with chemometrics. Food Control 2025, 169, 111033. [Google Scholar] [CrossRef]
- Uncu, A.O.; Uncu, A.T. A barcode-DNA analysis method for the identification of plant oil adulteration in milk and dairy products. Food Chem. 2020, 326, 126986. [Google Scholar] [CrossRef] [PubMed]
- Dodd, S.; Kevei, Z.; Karimi, Z.; Parmar, B.; Franklin, D.; Koidis, A.; Anastasiadi, M. Detection of sugar syrup adulteration in UK honey using DNA barcoding. Food Control 2025, 167, 110772. [Google Scholar] [CrossRef]
- Chen, Y.; Li, S.; Jia, J.; Sun, C.; Cui, E.; Xu, Y.; Shi, F.; Tang, A. FT-NIR combined with machine learning was used to rapidly detect the adulteration of pericarpium citri reticulatae (chenpi) and predict the adulteration concentration. Food Chem. X 2024, 24, 101798. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.; Chen, Y.; Deng, J.; Jiang, H. Improve the accuracy of FT-NIR for determination of zearalenone content in wheat by using the characteristic wavelength optimization algorithm. Spectrochim. Acta A Mol. Biomol. Spectrosc. 2024, 313, 124169. [Google Scholar] [CrossRef] [PubMed]
- Yu, D.X.; Qu, C.; Xu, J.Y.; Lu, J.Y.; Wu, D.D.; Wu, Q.N. Rapid discrimination and quantification of chemotypes in Perillae folium using FT-NIR spectroscopy and GC-MS combined with chemometrics. Food Chem. X 2024, 24, 101881. [Google Scholar] [CrossRef] [PubMed]
- Deng, J.; Jiang, H.; Chen, Q. Qualitative and quantitative analysis of mineral oil pollution in peanut oil by Fourier transform near-infrared spectroscopy. Food Chem. 2024, 469, 142590. [Google Scholar] [CrossRef]
- Meng, X.; Yin, C.; Yuan, L.; Zhang, Y.; Ju, Y.; Xin, K.; Chen, W.; Lv, K.; Hu, L. Rapid detection of adulteration of olive oil with soybean oil combined with chemometrics by Fourier transform infrared, visible-near-infrared and excitation-emission matrix fluorescence spectroscopy: A comparative study. Food Chem. 2023, 405, 134828. [Google Scholar] [CrossRef]
- Kuang, L.; Tian, X.; Su, Y.; Chen, C.; Zhao, L.; Ma, X.; Han, L.; Chen, C.; Zhang, J. Rapid identification of horse oil adulteration based on deep learning infrared spectroscopy detection method. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2024, 330, 125604. [Google Scholar] [CrossRef]
- Chaharlangi, M.; Tashkhourian, J.; Weller, P.; Bodenbender, L.; Hemmateenejad, B. Paper-based optical nose arrays and untargeted GC-IMS for the adulteration detection of cherry seed oils. Microchem. J. 2025, 209, 112610. [Google Scholar] [CrossRef]
- Khodabakhshian, R.; Seyedalibeyk Lavasani, H.; Weller, P. A methodological approach to preprocessing FTIR spectra of adulterated sesame oil. Food Chem. 2023, 419, 136055. [Google Scholar] [CrossRef]
- Millatina, N.R.N.; Calle, J.L.P.; Barea-Sepulveda, M.; Setyaningsih, W.; Palma, M. Detection and quantification of cocoa powder adulteration using Vis-NIR spectroscopy with chemometrics approach. Food Chem. 2024, 449, 139212. [Google Scholar] [CrossRef]
- Olivieri, A.C. Handling non-linearities and pre-processing in multivariate calibration of vibrational spectra. Microchem. J. 2025, 208, 112323. [Google Scholar] [CrossRef]
- Tsagkaris, A.S.; Bechynska, K.; Ntakoulas, D.D.; Pasias, I.N.; Weller, P.; Proestos, C.; Hajslova, J. Investigating the impact of spectral data pre-processing to assess honey botanical origin through Fourier transform infrared spectroscopy (FTIR). J. Food Compos. Anal. 2023, 119, 105276. [Google Scholar] [CrossRef]
- Chen, D.; Guo, C.; Lu, W.; Zhang, C.; Xiao, C. Rapid quantification of royal jelly quality by mid-infrared spectroscopy coupled with backpropagation neural network. Food Chem. 2023, 418, 135996. [Google Scholar] [CrossRef] [PubMed]
- Li, M.X.; Shi, Y.B.; Zhang, J.B.; Wan, X.; Fang, J.; Wu, Y.; Fu, R.; Li, Y.; Li, L.; Su, L.L.; et al. Rapid evaluation of Ziziphi Spinosae Semen and its adulterants based on the combination of FT-NIR and multivariate algorithms. Food Chem. X 2023, 20, 101022. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Zareef, M.; Wang, Z.; Li, H.; Chen, Q.; Ouyang, Q. Monitoring chlorophyll changes during Tencha processing using portable near-infrared spectroscopy. Food Chem. 2023, 412, 135505. [Google Scholar] [CrossRef]
- Wen, Y.; Li, Z.; Ning, Y.; Yan, Y.; Li, Z.; Wang, N.; Wang, H. Portable Raman spectroscopy coupled with PLSR analysis for monitoring and predicting of the quality of fresh-cut Chinese yam at different storage temperatures. Spectrochim. Acta A Mol. Biomol. Spectrosc. 2024, 310, 123956. [Google Scholar] [CrossRef]
- Lomarat, P.; Phechkrajang, C.; Sunghad, P.; Anantachoke, N. Raman spectroscopy coupled with the PLSR model: A rapid method for analyzing gamma-oryzanol content in rice bran oil. Food Chem. X 2024, 24, 101923. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Lei, T.; Cheng, Y.; Wei, X.; Sun, D.W. Predicting wheat gluten concentrations in potato starch using GPR and SVM models built by terahertz time-domain spectroscopy. Food Chem. 2024, 432, 137235. [Google Scholar] [CrossRef]
- Gao, X.; Dong, W.; Ying, Z.; Li, G.; Cheng, Q.; Zhao, Z.; Li, W. Rapid discriminant analysis for the origin of specialty yam based on multispectral data fusion strategies. Food Chem. 2024, 460, 140737. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.; Chai, Y.; Yan, Y.; Li, Z.; Huang, Y.; Chen, L.; Dong, H. Near-infrared spectroscopy combined with support vector machine for the identification of Tartary buckwheat (Fagopyrum tataricum (L.) Gaertn) adulteration using wavelength selection algorithms. Food Chem. 2025, 463, 141548. [Google Scholar] [CrossRef]
- Xu, L.; Chen, Z.; Bai, X.; Deng, J.; Zhao, X.; Jiang, H. Determination of aflatoxin B1 in peanuts based on millimetre wave. Food Chem. 2025, 464, 141867. [Google Scholar] [CrossRef] [PubMed]
- Weng, S.; Chu, Z.; Wang, M.; Han, K.; Zhu, G.; Liu, C.; Li, X.; Huang, L. Reflectance spectroscopy with operator difference for determination of behenic acid in edible vegetable oils by using convolutional neural network and polynomial correction. Food Chem. 2022, 367, 130668. [Google Scholar] [CrossRef] [PubMed]
- Silva, E.F.R.; da Silva Santos, B.R.; Minho, L.A.C.; Brandao, G.C.; de Jesus Silva, M.; Silva, M.V.L.; Dos Santos, W.N.L.; Dos Santos, A.M.P. Characterization of the chemical composition (mineral, lead and centesimal) in pine nut (Araucaria angustifolia (Bertol.) Kuntze) using exploratory data analysis. Food Chem. 2022, 369, 130672. [Google Scholar] [CrossRef] [PubMed]
- Alharbi, H.; Kahfi, J.; Dutta, A.; Jaremko, M.; Emwas, A.-H. The detection of adulteration of olive oil with various vegetable oils—A case study using high-resolution 700 MHz NMR spectroscopy coupled with multivariate data analysis. Food Control 2024, 166, 110679. [Google Scholar] [CrossRef]
- Wei, H.N.; Liu, X.Y.; Wang, C.C.; Feng, R.; Zhang, B. Characteristics of corn starch/polyvinyl alcohol composite film with improved flexibility and UV shielding ability by novel approach combining chemical cross-linking and physical blending. Food Chem. 2024, 456, 140051. [Google Scholar] [CrossRef] [PubMed]
- Taylor, J.N.; Bando, K.; Tsukagoshi, S.; Tanaka, L.; Fujita, K.; Fujita, S. Microscopic water dispersion and hydrogen-bonding structures in margarine spreads with Raman hyperspectral imaging and machine learning. Food Chem. 2025, 465, 142035. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Zhu, Z.; Sun, D.W. Visualization and quantification of content and hydrogen bonding state of water in apple and potato cells by confocal Raman microscopy: A comparison study. Food Chem. 2022, 385, 132679. [Google Scholar] [CrossRef]
| Indicators | Methods | Parameters | Train | Test | ||
|---|---|---|---|---|---|---|
| RMSEC (%) | RMSEP (%) | |||||
| Adulteration-A | Raw | Lvs = 12 | 0.9092 | 1.7557 | 0.8709 | 2.2281 |
| SNV | Lvs = 12 | 0.9029 | 1.8152 | 0.8790 | 2.1571 | |
| MSC | Lvs = 10 | 0.8940 | 1.8969 | 0.8761 | 2.1833 | |
| SG | Lvs = 11 | 0.8853 | 1.9727 | 0.8834 | 2.1175 | |
| Adulteration-B | Raw | Lvs = 8 | 0.9245 | 2.2643 | 0.9235 | 2.2738 |
| SNV | Lvs = 7 | 0.9353 | 2.0953 | 0.9311 | 2.1571 | |
| MSC | Lvs = 11 | 0.9313 | 2.1592 | 0.9275 | 2.2135 | |
| SG | Lvs = 10 | 0.9433 | 1.9614 | 0.9315 | 2.1510 | |
| Adulteration-C | Raw | Lvs = 7 | 0.9959 | 0.8360 | 0.9956 | 0.8866 |
| SNV | Lvs = 8 | 0.9960 | 0.8228 | 0.9957 | 0.8732 | |
| MSC | Lvs = 8 | 0.9972 | 0.6905 | 0.9957 | 0.8738 | |
| SG | Lvs = 9 | 0.9974 | 0.6648 | 0.9958 | 0.8695 | |
| Indicators | Models | Parameters | Train | Test | ||
|---|---|---|---|---|---|---|
| RMSEC (%) | RMSEP (%) | |||||
| Adulteration-A | PLSR | Lvs = 11 | 0.9365 | 1.6145 | 0.8911 | 2.0470 |
| SVM | c = 2.8284 g = 0.0221 | 0.9853 | 0.7051 | 0.9713 | 1.0518 | |
| BKA-SVM | c = 431.3487 g = 0.0405 | 0.9930 | 0.1520 | 0.9833 | 0.8026 | |
| Adulteration-B | PLSR | Lvs = 10 | 0.9815 | 1.3982 | 0.9375 | 2.0544 |
| SVM | c = 22.6274 g = 0.0028 | 0.9658 | 1.5203 | 0.9579 | 1.6909 | |
| BKA-SVM | c = 1020.2249 g = 0.0141 | 0.9990 | 0.2624 | 0.9893 | 0.8494 | |
| Adulteration-C | PLSR | Lvs = 9 | 0.9971 | 0.7033 | 0.9960 | 0.8014 |
| SVM | c = 64 g = 0.0009 | 0.9978 | 0.6180 | 0.9977 | 0.6225 | |
| BKA-SVM | c = 1020.7405 g = 0.0018 | 0.9991 | 0.4003 | 0.9987 | 0.4801 | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Luo, W.; Deng, J.; Li, C.; Jiang, H. Quantitative Analysis of Peanut Skin Adulterants by Fourier Transform Near-Infrared Spectroscopy Combined with Chemometrics. Foods 2025, 14, 466. https://doi.org/10.3390/foods14030466
Luo W, Deng J, Li C, Jiang H. Quantitative Analysis of Peanut Skin Adulterants by Fourier Transform Near-Infrared Spectroscopy Combined with Chemometrics. Foods. 2025; 14(3):466. https://doi.org/10.3390/foods14030466
Chicago/Turabian StyleLuo, Wangfei, Jihong Deng, Chenxi Li, and Hui Jiang. 2025. "Quantitative Analysis of Peanut Skin Adulterants by Fourier Transform Near-Infrared Spectroscopy Combined with Chemometrics" Foods 14, no. 3: 466. https://doi.org/10.3390/foods14030466
APA StyleLuo, W., Deng, J., Li, C., & Jiang, H. (2025). Quantitative Analysis of Peanut Skin Adulterants by Fourier Transform Near-Infrared Spectroscopy Combined with Chemometrics. Foods, 14(3), 466. https://doi.org/10.3390/foods14030466







