Enhanced Bioactive Peptide Release from Pre-Hydrolysed Pea Protein: Impact of Pepsin Digestion on Antidiabetic and Antihypertensive Functions
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials and Chemicals
2.2. Characterisation of Pea Protein Products
2.3. Preparation of Crude Pea Protein Hydrolysates and <10 kDa Fractions
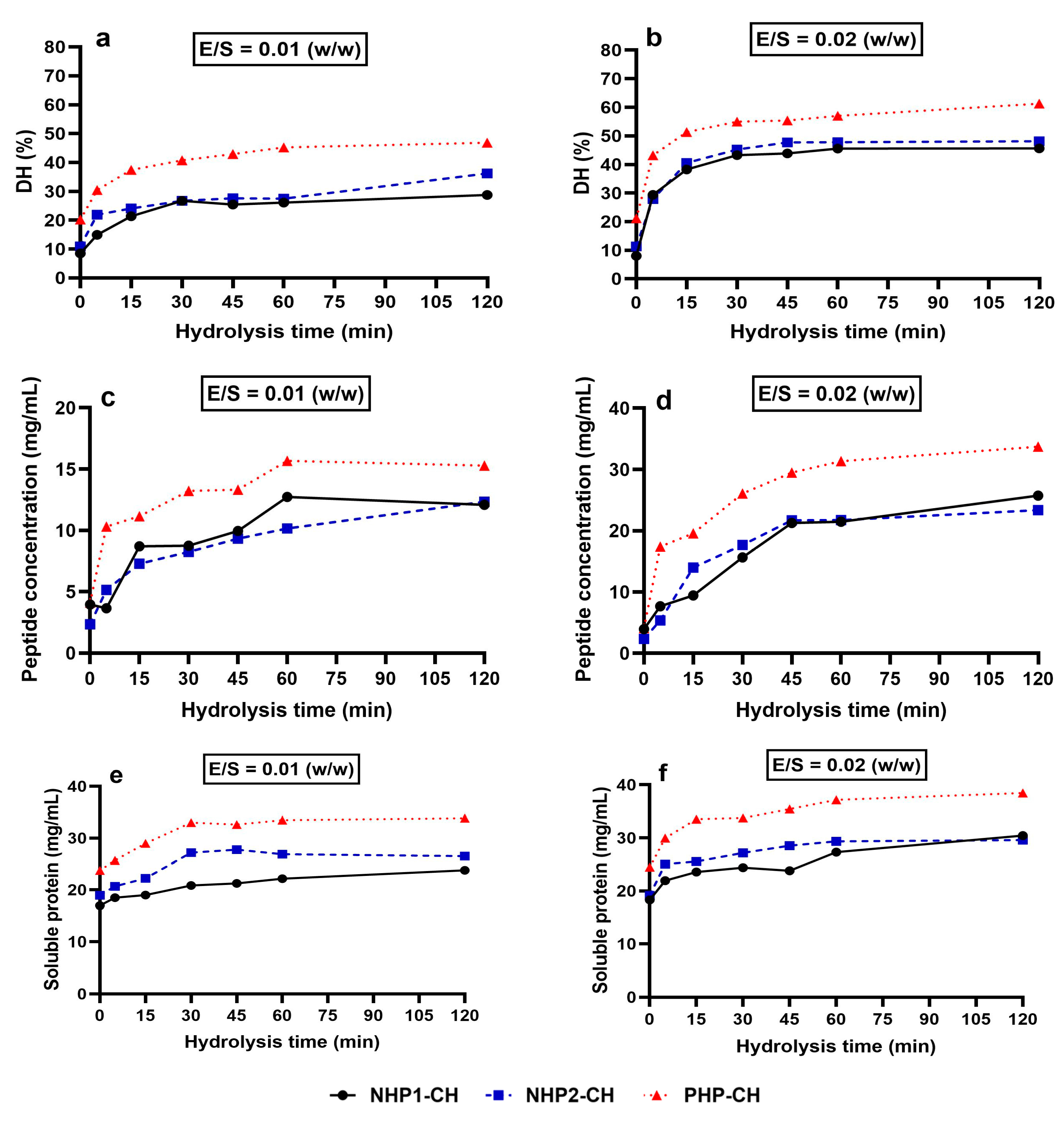
2.3.1. Assessment of Degree of Hydrolysis (DH)
2.3.2. Quantification of Released Peptides
2.3.3. Determination of Soluble Protein
2.4. Enzyme Inhibitory Properties
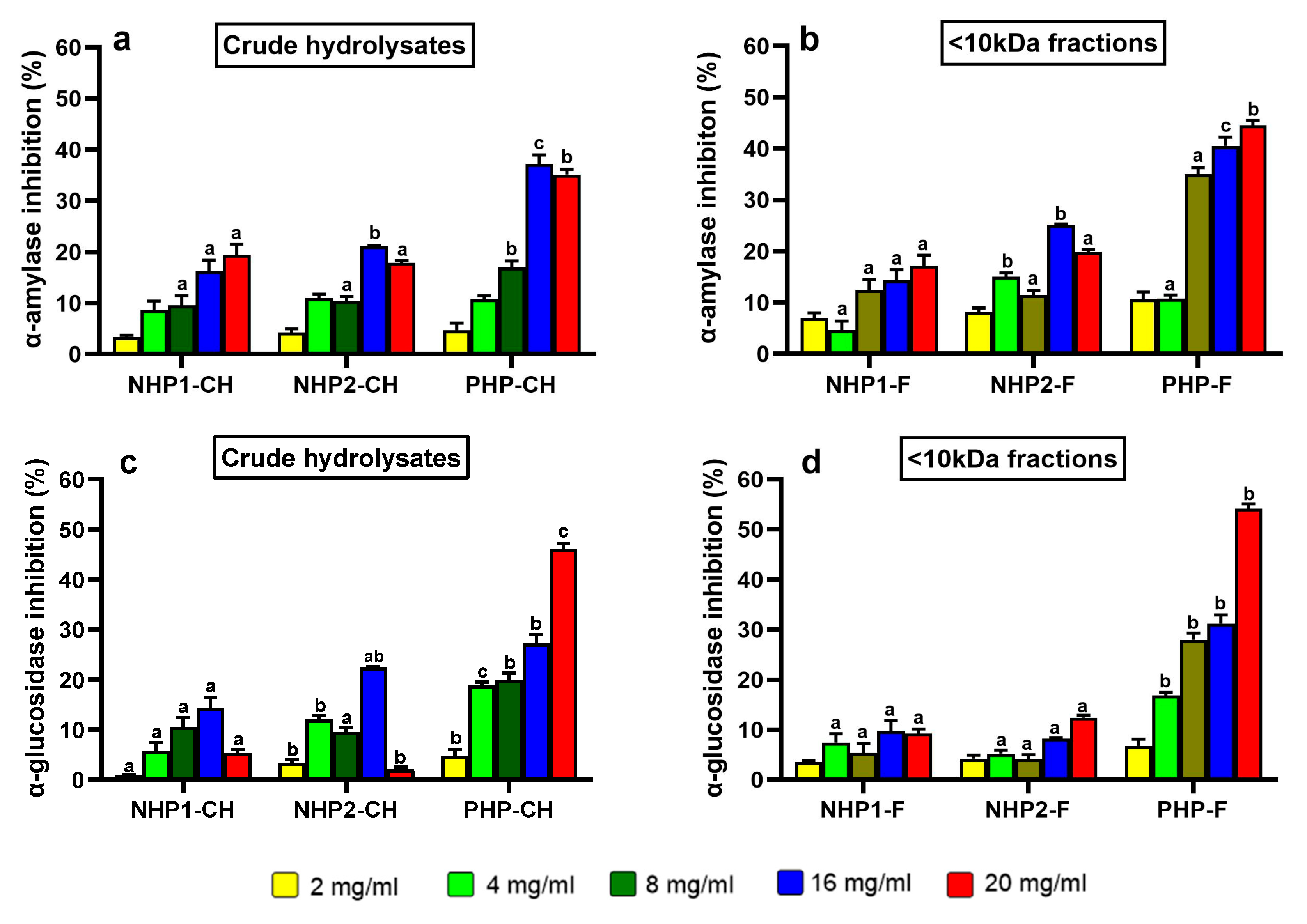
2.4.1. Measurement of α-Amylase Inhibitory Activity
2.4.2. Measurement of α-Glucosidase Inhibitory Activity
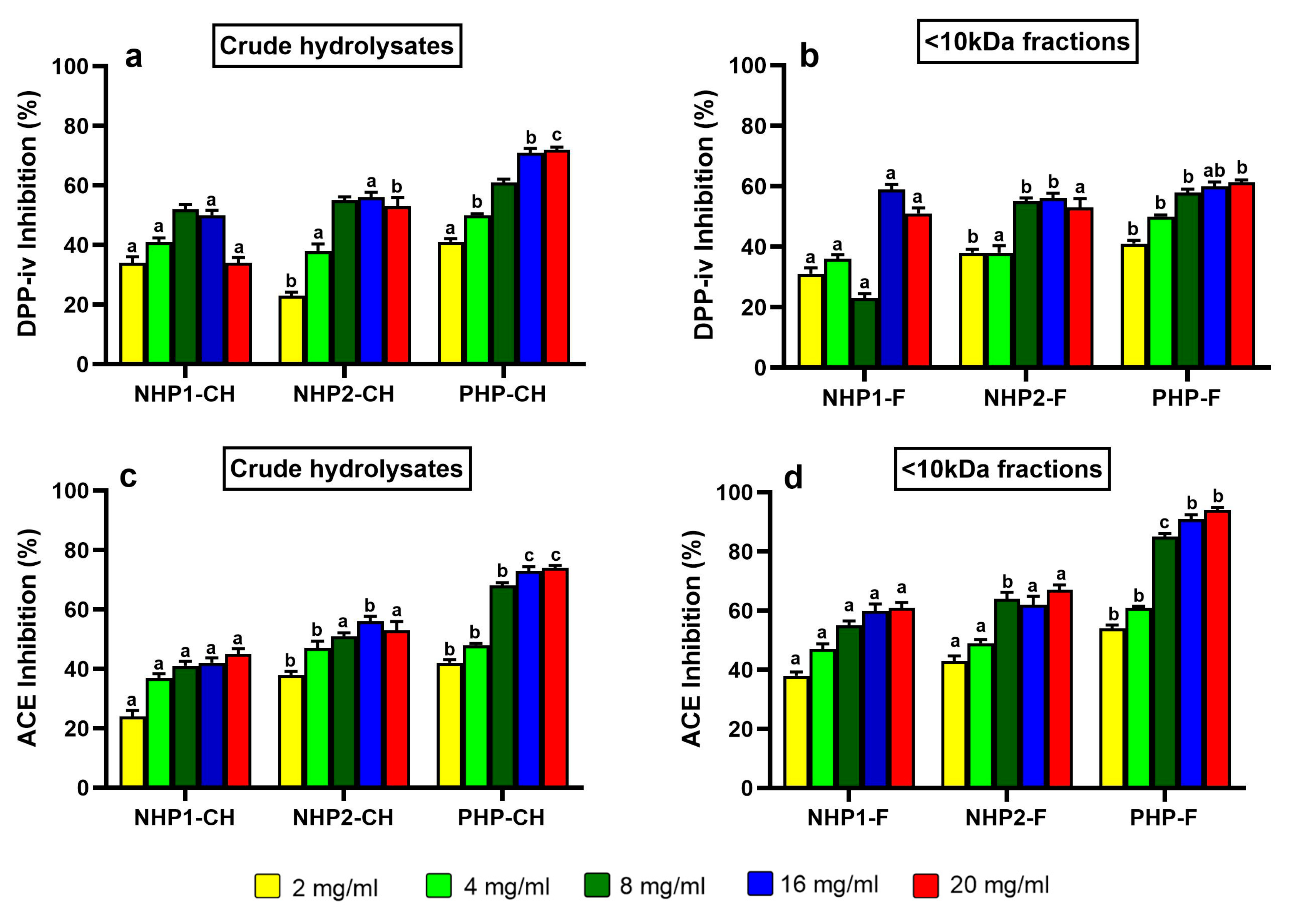
2.4.3. Measurement of DPP-IV Inhibitory Activity
2.4.4. Measurement of ACE Inhibitory Activity
2.5. Peptidomic Profiling of Pea Protein Hydrolysates/Fractions
2.6. In Silico Bioactivity Prediction of Peptides Identified from Pea Protein Hydrolysate/Fractions
2.7. Statistical Analysis
3. Results and Discussion
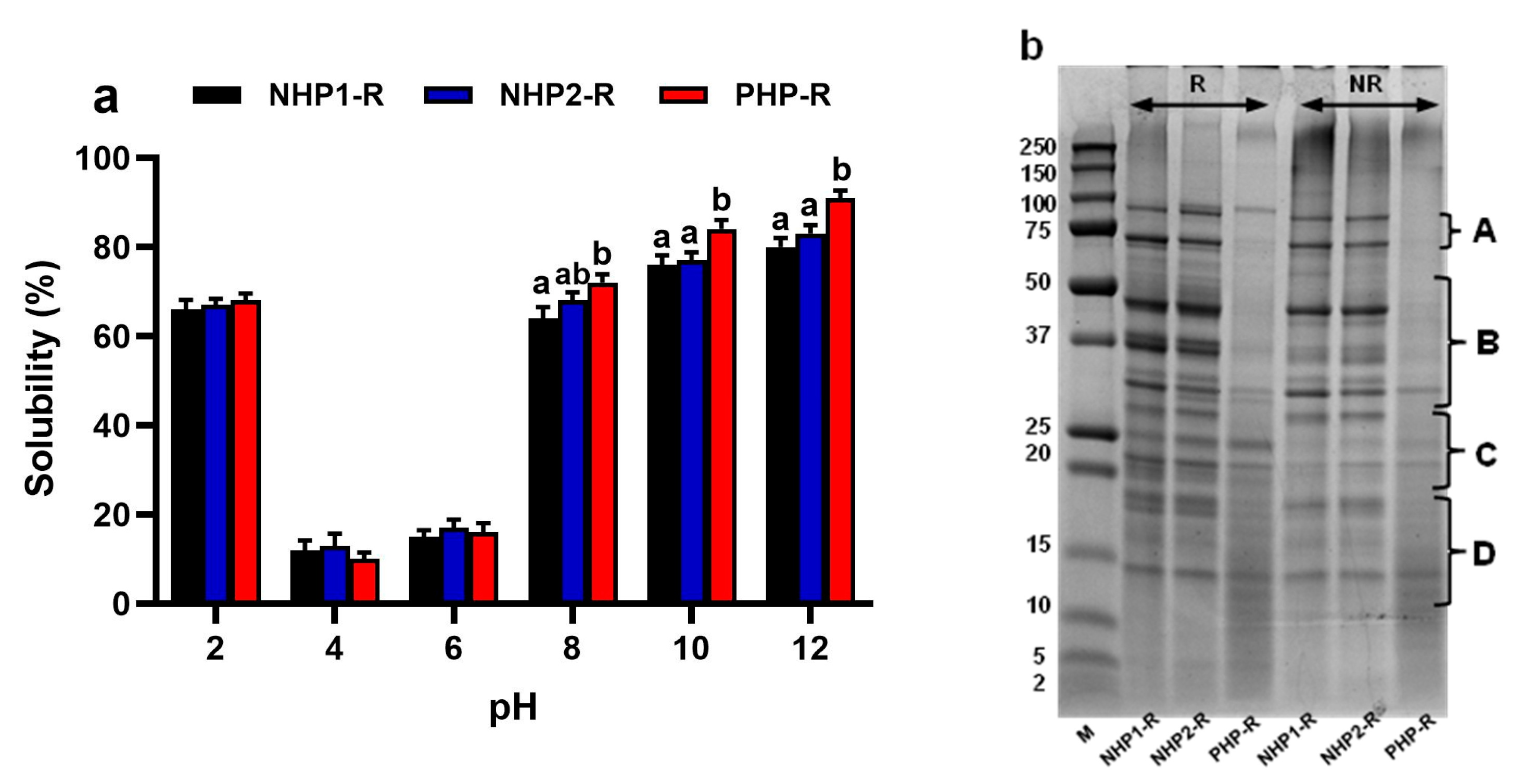
3.1. Characterisation of Raw Pea Protein Samples
3.2. PHP-CH Exhibits Enhanced Pepsin Digestibility, Peptide Release and Solubility
3.3. Inhibition of Carbohydrate-Digesting Enzymes by Pea Protein Hydrolysates and Peptide Fractions
3.4. DPP-IV Inhibitory Activity
3.5. ACE Inhibitory Activity
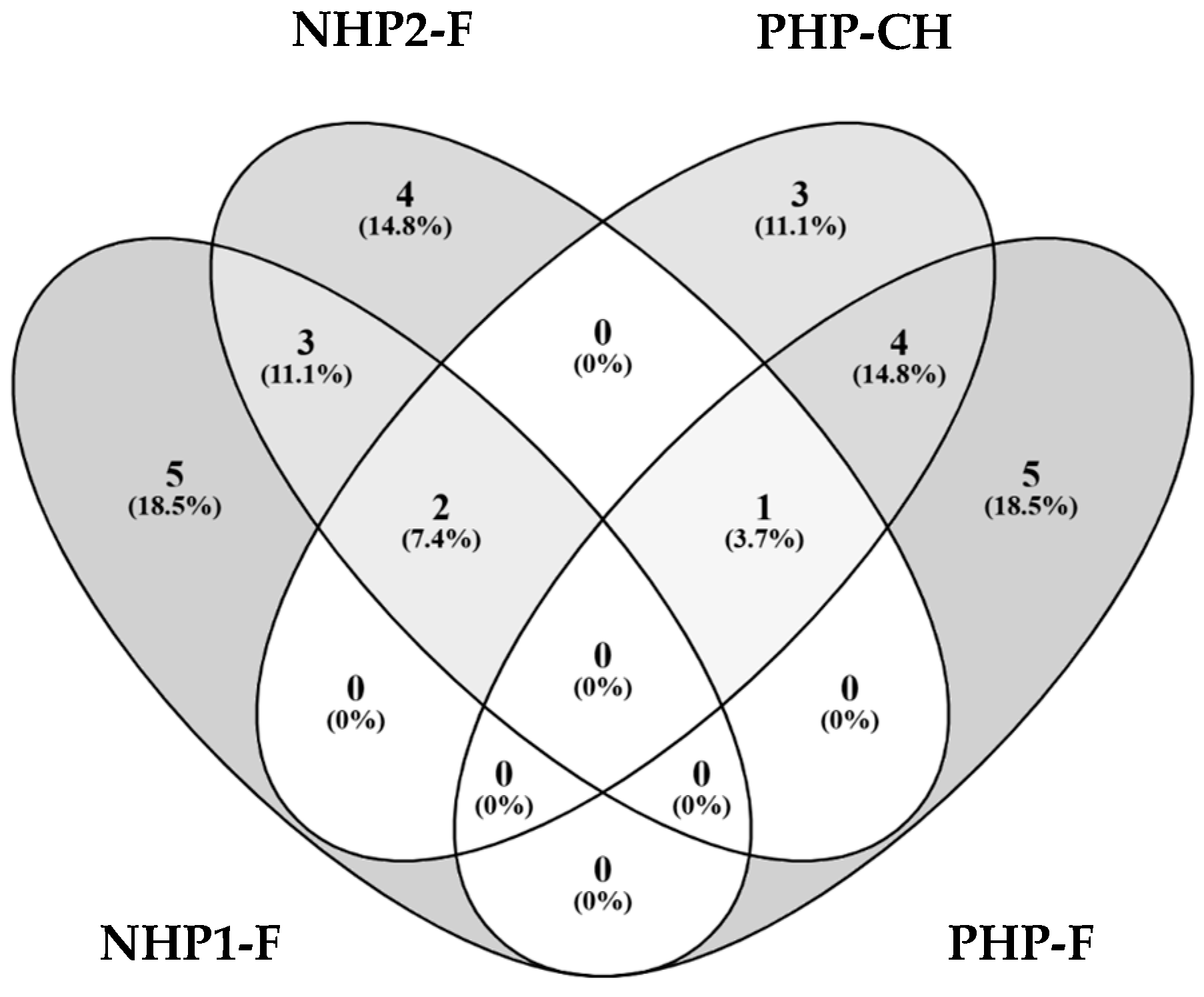
3.6. Enhanced Bioactive Peptide Profiles and Predicted Enzyme Inhibition in PHP-F
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| DPP-IV | Dipeptidyl peptidase |
| ACE | Angiotensin converting enzyme |
| DH | Degree of hydrolysis |
| PHP | Pre-hydrolysed pea protein |
| NHP | Non-hydrolysed pea protein |
References
- Nasri, R. Valorisation of plant-derived proteins for health benefits: A review. J. Funct. Foods 2017, 35, 123–134. [Google Scholar] [CrossRef]
- Patil, S.P.; Mandal, S.; Tomar, S.K.; Anand, S. Bioactive peptides as functional ingredients in management of chronic diseases. Nutr. Res. 2015, 35, 29–41. [Google Scholar] [CrossRef]
- Panyam, D.; Kilara, A. Enhancing the functionality of food proteins by enzymatic modification. J. Dairy Sci. 1996, 79, 1529–1536. [Google Scholar] [CrossRef]
- Tavano, O.L. Protein hydrolysis using proteases: An important tool for food biotechnology. J. Mol. Catal. B Enzym. 2013, 90, 1–11. [Google Scholar] [CrossRef]
- Sarmadi, B.H.; Ismail, A. Antioxidative peptides from food proteins: A review. Food Chem. 2010, 119, 315–326. [Google Scholar] [CrossRef]
- Araiza-Calahorra, A.; Mondor, M.; Boesch, C.; Orfila, C.; Goycoolea, F.M.; Hernández-Álvarez, A.J. Proteins, peptides, and protein hydrolysates as immunomodulatory and antioxidant agents for the formulation of functional foods. In Current Advances for Development of Functional Foods Modulating Inflammation and Oxidative Stress; Hernández-Álvarez, A.J., Ed.; Academic Press: Cambridge, MA, USA, 2022; pp. 137–164. [Google Scholar] [CrossRef]
- Alashi, A.M.; Blanchard, C.L.; Mailer, R.J.; Agboola, S.O.; Mawson, A.J.; He, R.; Girgih, A.T.; Aluko, R.E. Antioxidant and enzyme inhibitory properties of peptide fractions obtained from a pea protein isolate enzymatic hydrolysate. Food Chem. 2014, 160, 286–293. [Google Scholar] [CrossRef]
- Gong, M.; Wan, J.; Jiang, Y.; Zhang, Y.; Liu, Q.; Ma, M. Mechanism and kinetics of enzyme inhibitory peptides from pea protein hydrolysates. Food Bioprod. Process. 2020, 123, 151–162. [Google Scholar] [CrossRef]
- Tundis, R.; Loizzo, M.R.; Menichini, F. Natural products as α-amylase and α-glucosidase inhibitors and their hypoglycemic potential in the treatment of diabetes: An update. Mini-Rev. Med. Chem. 2010, 17, 1533–1545. [Google Scholar] [CrossRef]
- Awosika, T.O.; Aluko, R.E. Inhibition of the in vitro activities of α-amylase, α-glucosidase and pancreatic lipase by yellow field pea (Pisum sativum L.) protein hydrolysates. Int. J. Food Sci. Technol. 2019, 54, 2021–2034. [Google Scholar] [CrossRef]
- Aluko, R.E. Functional and bioactive properties of pea protein hydrolysates. J. Food Sci. 2013, 78, C307–C313. [Google Scholar] [CrossRef]
- Pownall, T.L.; Udenigwe, C.C.; Aluko, R.E. Amino acid composition and antioxidant properties of pea seed (Pisum sativum L.) enzymatic protein hydrolysate fractions. J. Agric. Food Chem. 2010, 58, 4712–4718. [Google Scholar] [CrossRef]
- Mojica, L.; de Mejía, E.G. Optimization of enzymatic production of anti-diabetic peptides from black bean (Phaseolus vulgaris L.) protein using response surface methodology. Food Funct. 2016, 7, 713–727. [Google Scholar] [CrossRef]
- Kristinsson, H.G.; Rasco, B.A. Fish protein hydrolysates: Production, biochemical, and functional properties. Crit. Rev. Food Sci. Nutr. 2000, 40, 43–81. [Google Scholar] [CrossRef]
- Korhonen, H.; Pihlanto, A. Bioactive peptides: Production and functionality. Int. Dairy J. 2006, 16, 945–960. [Google Scholar] [CrossRef]
- Nongonierma, A.B.; FitzGerald, R.J. The scientific evidence for the role of milk protein-derived bioactive peptides in humans: A review. J. Food Biochem. 2015, 39, 165–173. [Google Scholar] [CrossRef]
- Udenigwe, C.C.; Aluko, R.E. Influence of peptide size on the antioxidant properties of enzymatic pea protein hydrolysate fractions. J. Food Sci. 2012, 77, R11–R24. [Google Scholar] [CrossRef]
- Iwaniak, A.; Minkiewicz, P.; Darewicz, M. Food-originating peptides as DPP-IV inhibitors. Trends Food Sci. Technol. 2014, 35, 284–297. [Google Scholar] [CrossRef]
- Mojica, L.; Chen, K.; de Mejía, E.G. Impact of commercial enzyme selection on the production of anti-diabetic peptides from black bean protein. J. Funct. Foods 2017, 29, 112–123. [Google Scholar] [CrossRef]
- Ma, Z.; Mondor, M.; Valencia, F.G.; Hernández-Álvarez, A.J. The current state of insect proteins: Extraction technologies, bioactive peptides, and allergenicity of edible insect proteins. Food Funct. 2023, 14, 8129–8156. [Google Scholar] [CrossRef] [PubMed]
- Carrasco-Castilla, J.; Hernández-Álvarez, A.J.; Jiménez-Martínez, C.; Gutiérrez-López, G.F.; Dávila-Ortiz, G. Use of proteomics and peptidomics methods in food bioactive peptide science and engineering. Food Eng. Rev. 2012, 4, 224–243. [Google Scholar] [CrossRef]
- FitzGerald, R.J.; Mora, L.; Hayes, M. Application of peptidomics to the discovery of bioactive peptides in foods. Nutrients 2020, 12, 1503. [Google Scholar] [CrossRef]
- AOAC. Official Methods of Analysis, 16th ed.; AOAC International: Washington, DC, USA, 1995. [Google Scholar]
- Al-Tubuly, A.A. SDS-PAGE and Western Blotting. In Diagnostic and Therapeutic Antibodies; Wild, M.A., Ed.; Humana Press: Totowa, NJ, USA, 2000; pp. 391–405. [Google Scholar] [CrossRef]
- Bio-Rad Laboratories. A Guide to Polyacrylamide Gel Electrophoresis and Detection: Bulletin 6040 Rev B; Bio-Rad: Hercules, CA, USA, 2012. [Google Scholar]
- Betschart, A.A.; Irving, D.W.; Shepherd, A.D. Protein digestibility of legumes as affected by processing and storage. J. Food Sci. 1974, 39, 327–330. [Google Scholar] [CrossRef]
- Thermo Fisher Scientific. Pierce™ BCA Protein Assay Kit Instruction Manual; Thermo Fisher Scientific: Waltham, MA, USA, 2023. [Google Scholar]
- Ma, Z.; Mondor, M.; Boesch, C.; Sanchez-Velazquez, O.A.; Dowle, A.A.; Hernández-Álvarez, A.J. The transepithelial transport of peptides derived from insects (Galleria mellonella and Alphitobius diaperinus) through static in vitro digestion (INFOGEST) and their ability to mitigate oxidative stress. Food Chem. 2025, 481, 144036. [Google Scholar] [CrossRef]
- Thermo Fisher Scientific. Pierce™ Fluorometric Peptide Assay Kit Manual; Thermo Fisher Scientific: Waltham, MA, USA, 2020. [Google Scholar]
- Preston, G.W.; Brochocka, A. Quantification of a peptide standard using the intrinsic fluorescence of tyrosine. J. Peptide Sci. 2016, 24, e3113. [Google Scholar] [CrossRef]
- Zulfiqar, S.; Blando, F.; Orfila, C.; Boesch, C. Chromogenic assay is more efficient in identifying α-amylase inhibitory properties of anthocyanin-rich samples when compared to the 3,5-dinitrosalicylic acid (DNS) assay. Molecules 2023, 28, 6399. [Google Scholar] [CrossRef] [PubMed]
- Zulfiqar, S.; Marshall, L.J.; Boesch, C. Hibiscus sabdariffa inhibits α-glucosidase activity in vitro and lowers postprandial blood glucose response in humans. Hum. Nutr. Metab. 2022, 30, 200164. [Google Scholar] [CrossRef]
- Han, R.; Hernández-Álvarez, A.J.; Maycock, J.; Murray, B.S.; Boesch, C. Comparison of alcalase- and pepsin-treated oilseed protein hydrolysates: Experimental validation of predicted antioxidant, antihypertensive and antidiabetic properties. Curr. Res. Food Sci. 2021, 4, 141–149. [Google Scholar] [CrossRef] [PubMed]
- Ma, Z.; Mondor, M.; Dowle, A.A.; Goycoolea, F.M.; Hernández-Álvarez, A.J. Buffalo worm (Alphitobius diaperinus) proteins: Structural properties, proteomics and nutritional benefits. Food Chem. 2025, 464, 141757. [Google Scholar] [CrossRef] [PubMed]
- FitzGerald, R.J.; Cermeño, M.; Khalesi, M.; Kleekayai, T.; Amigo Benavent, M. Application of in silico approaches for the generation of milk protein-derived bioactive peptides. J. Funct. Foods 2020, 64, 103636. [Google Scholar] [CrossRef]
- Damodaran, S.; Parkin, K.L. Fennema’s Food Chemistry, 5th ed.; CRC Press: Boca Raton, FL, USA, 2017. [Google Scholar]
- Tang, C.H.; Sun, X.; Zhang, B. Physicochemical and functional properties of hemp (Cannabis sativa L.) protein isolate. J. Agric. Food Chem. 2006, 54, 8945–8950. [Google Scholar] [CrossRef] [PubMed]
- Stone, A.K.; Karalash, A.; Tyler, R.T.; Warkentin, T.D.; Nickerson, M.T. Functional attributes of pea protein isolates prepared using different extraction methods and cultivars. Food Res. Int. 2015, 76, 31–38. [Google Scholar] [CrossRef]
- Udenigwe, C.C.; Aluko, R.E. Food protein-derived bioactive peptides: Production, processing, and potential health benefits. J. Food Sci. 2012, 77, R11–R24. [Google Scholar] [CrossRef]
- Kim, S.-K.; Mendis, E. Bioactive compounds from marine processing byproducts: A review. Food Res. Int. 2006, 39, 383–393. [Google Scholar] [CrossRef]
- Karami, Z.; Akbari-Adergani, B. Bioactive food-derived peptides: A review on correlation between structure of bioactive peptides and their functional properties. J. Food Sci. Technol. 2019, 56, 535–547. [Google Scholar] [CrossRef]
- García Arteaga, V.; Demand, V.; Kern, K.; Strube, A.; Szardenings, M.; Muranyi, I.; Eisner, P.; Schweiggert-Weisz, U. Enzymatic hydrolysis and fermentation of pea protein isolate and its effects on antigenic proteins, functional properties, and sensory profile. Foods 2022, 11, 118. [Google Scholar] [CrossRef] [PubMed]
- Shevkani, K.; Kaur, A.; Kumar, S.; Singh, N. Physicochemical, functional and nutraceutical characteristics of pigeon pea and its flour fractions. LWT-Food Sci. Technol. 2015, 63, 1042–1049. [Google Scholar] [CrossRef]
- Shevkani, K.; Singh, N.; Kaur, A.; Rana, J.C. Structural and functional characterization of kidney bean and field pea protein isolates: A comparative study. Food Hydrocoll. 2015, 43, 679–689. [Google Scholar] [CrossRef]
- Carbonaro, M.; Maselli, P.; Nucara, A. Relationship between digestibility and secondary structure of raw and thermally treated legume proteins: A Fourier transform infrared (FT-IR) spectroscopic study. Amino Acids 2012, 43, 911–921. [Google Scholar] [CrossRef] [PubMed]
- Padial-Domínguez, M.; Espejo-Carpio, F.J.; Pérez-Gálvez, R.; Guadix, A.; Guadix, E.M. Optimization of enzymatic hydrolysis for production of chickpea protein hydrolysates with improved functional properties. LWT-Food Sci. Technol. 2020, 117, 108646. [Google Scholar] [CrossRef]
- He, R.; Girgih, A.T.; Malomo, S.; Ju, X.; Aluko, R.E. Antioxidant activities of enzymatic rapeseed protein hydrolysates and the membrane fractions from ultrafiltration. Food Chem. 2013, 139, 546–552. [Google Scholar] [CrossRef]
- Bamdad, F.; Shin, S.H.; Suh, J.W.; Jeon, Y.J. Evaluation of immunomodulatory activities of enzymatic protein hydrolysates from Spirulina maxima. Food Chem. 2017, 210, 519–526. [Google Scholar] [CrossRef]
- Hartmann, R.; Meisel, H. Food-derived peptides with biological activity: From research to food applications. Curr. Opin. Biotechnol. 2007, 18, 163–169. [Google Scholar] [CrossRef]
- Chabanon, G.; Chevalot, I.; Framboisier, X.; Chenu, S.; Marc, I. Hydrolysis of rapeseed protein isolates: Kinetics, characterization and functional properties of hydrolysates. Process Biochem. 2007, 42, 1419–1428. [Google Scholar] [CrossRef]
- Liu, R.; Cheng, J.; Wu, H.; Wang, Y.; Wang, Y. Structure–activity relationship of bioactive peptides derived from food proteins: A review. Trends Food Sci. Technol. 2020, 99, 21–35. [Google Scholar] [CrossRef]
- Gong, B.; Yuan, L.; Wang, H.; Qiu, Y.; Wang, Y.; Zhang, Y. Inhibitory activities of chickpea protein hydrolysates against α-glucosidase and α-amylase: Identification and molecular docking study. J. Food Biochem. 2020, 44, e13361. [Google Scholar] [CrossRef]
- Hong, L.; Fan, L.; Wu, J.; Yang, J.; Hou, D.; Yao, Y.; Zhou, S. Pulse proteins and their hydrolysates: A comprehensive review of their beneficial effects on metabolic syndrome and the gut microbiome. Nutrients 2024, 16, 1845. [Google Scholar] [CrossRef] [PubMed]
- Nongonierma, A.B.; FitzGerald, R.J. Dipeptidyl peptidase IV inhibitory and insulinotropic properties of milk protein hydrolysates. Peptides 2015, 62, 197–204. [Google Scholar] [CrossRef]
- Miguel, M.; Aleixandre, A.; Recio, I.; López-Fandiño, R. Bioactive peptides derived from egg proteins. World’s Poult. Sci. J. 2005, 61, 365–377. [Google Scholar] [CrossRef]
- Wu, J.; Ding, X. Hypotensive and physiological effect of angiotensin-converting enzyme inhibitory peptides derived from food proteins. J. Food Sci. 2001, 66, 614–621. [Google Scholar] [CrossRef]
- He, R.; Malomo, S.A.; Alashi, A.; Girgih, A.T.; Ju, X.; Aluko, R.E. Purification and hypotensive activity of rapeseed protein-derived peptides. Food Chem. 2013, 141, 153–159. [Google Scholar] [CrossRef]
- Rudolph, S.; Lunow, D.; Kaiser, S.; Henle, T. Identification and quantification of ACE-inhibiting peptides in enzymatic hydrolysates of plant proteins. Food Chem. 2017, 224, 19–25. [Google Scholar] [CrossRef] [PubMed]
- He, R.; Aluko, R.E.; Ju, X. ACE-inhibitory and antioxidant peptides derived from pea protein. J. Sci. Food Agric. 2013, 93, 2622–2631. [Google Scholar] [CrossRef]
- Wang, X.; Li, J.; Han, J.; Wang, H. Characterization of α-glucosidase inhibitory peptides from mung bean proteins. LWT-Food Sci. Technol. 2020, 126, 109332. [Google Scholar] [CrossRef]
- Silva, S.V.; Alashi, A.M.; Roy, F.; Aluko, R.E. Pea protein-derived peptides: Molecular weight profile and impact on absorption. J. Food Biochem. 2021, 45, e13828. [Google Scholar] [CrossRef]
- Nongonierma, A.B.; FitzGerald, R.J. Strategies for the discovery and identification of food protein-derived biologically active peptides. Trends Food Sci. Technol. 2015, 46, 274–283. [Google Scholar] [CrossRef]
- Udenigwe, C.C.; Mohan, A. Mechanisms of food protein-derived antihypertensive peptides other than ACE inhibition. J. Funct. Foods 2014, 8, 45–52. [Google Scholar] [CrossRef]
- Nongonierma, A.B.; FitzGerald, R.J. Dipeptidyl peptidase IV inhibitory and antioxidative properties of milk protein-derived peptides. J. Funct. Foods 2013, 5, 1940–1951. [Google Scholar] [CrossRef]
- Lacroix, I.M.E.; Li-Chan, E.C.Y. DPP-IV inhibitory activity of pea protein hydrolysate fractions. J. Funct. Foods 2013, 5, 235–243. [Google Scholar] [CrossRef]
- Han, R.; He, R.; Ju, X. In silico bioactivity prediction of legume protein-derived peptides and experimental validation. Food Res. Int. 2019, 116, 835–845. [Google Scholar] [CrossRef]
| Protein Sample | Peptide Sequence | ACE Inhibition | DPP-IV Inhibition | α-Amylase Inhibition | α-Glucosidase Inhibition |
|---|---|---|---|---|---|
| NHP1-F | FVPHYNL, RSRNPIYSNKFGKF, RGIIPLEN, RSDQENPFIF, LAIPVNKPGQL, YKSKPHTIF, YRNGIYAPHW, YENENGHIRL, IRRTIDPNGLHL, FENENGHIRL | All | All | None | None |
| NHP2-F | NENGHIRL, RSRNPIYSNKFGKF, RGIIPLEN, FVPHYNL, LAIPVNRPGQL, YKSKPHTIF, LVIPVNGPGKF, LAIPVNKPGQL, KVLYGPTPVRDGF, YRAKPHTIF | All | All | None | None |
| PHP-CH | FVPHYNL, RSRNPIYSNKFGKF, LVIPVNGPGKF, FHMPPSSGSAPVNL, DQMPRRFY, VQRYEARL, EITPEKNQQL, QRNALRRPYYSNAPQE, SHKPEYSNKFGKL, NIGPSSSPDIYNPEAGRIKTVTS | All | All | DQMPRRFY | FHMPPSSGSAPVNL, NIGPSSSPDIYNPEAGRIKTVTS, DQMPRRFY, VQRYEARL, EITPEKNQQL, QRNALRRPYYSNAPQE, SHKPEYSNKFGKL |
| PHP-F | LVIPVNGPGKF, AIPVNKPGQLQ, DQMPRRFY, FHMPPSSGSAPVNL, NIGPSSSPDIYNPEAGRIKTVTS, SHKPEYSNKFGKL, TETWNPNHPELK, WEAKGQTPLF, PRRFYRP, KPEYPYSN | All | All | DQMPRRFY PRRFYRP | AIPVNKPGQLQ, DQMPRRFY, FHMPPSSGSAPVNL, NIGPSSSPDIYNPEAGRIKTVTS, SHKPEYSNKFGKL, TETWNPNHPELK, WEAKGQTPLF, KPEYPYSN, PRRFYRP |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Elbira, A.; Hernández-Álvarez, A.J.; Boesch, C. Enhanced Bioactive Peptide Release from Pre-Hydrolysed Pea Protein: Impact of Pepsin Digestion on Antidiabetic and Antihypertensive Functions. Foods 2025, 14, 3306. https://doi.org/10.3390/foods14193306
Elbira A, Hernández-Álvarez AJ, Boesch C. Enhanced Bioactive Peptide Release from Pre-Hydrolysed Pea Protein: Impact of Pepsin Digestion on Antidiabetic and Antihypertensive Functions. Foods. 2025; 14(19):3306. https://doi.org/10.3390/foods14193306
Chicago/Turabian StyleElbira, Arig, Alan Javier Hernández-Álvarez, and Christine Boesch. 2025. "Enhanced Bioactive Peptide Release from Pre-Hydrolysed Pea Protein: Impact of Pepsin Digestion on Antidiabetic and Antihypertensive Functions" Foods 14, no. 19: 3306. https://doi.org/10.3390/foods14193306
APA StyleElbira, A., Hernández-Álvarez, A. J., & Boesch, C. (2025). Enhanced Bioactive Peptide Release from Pre-Hydrolysed Pea Protein: Impact of Pepsin Digestion on Antidiabetic and Antihypertensive Functions. Foods, 14(19), 3306. https://doi.org/10.3390/foods14193306







