Selenium Biotransformation and Fractionation of Selenopeptide from Germinated Perilla (Perilla frutescens) Seeds
Abstract
1. Introduction
2. Material and Methods
2.1. Materials
2.2. Selenium Enrichment in Perilla frutescens Seeds
2.3. Selenoprotein Extraction and Enzymatic Hydrolyzation
2.4. Partial Purification of Se-Peptides
2.4.1. Ultrafiltration
2.4.2. Preparative High-Performance Liquid Chromatography (Prep-HPLC)
2.4.3. Selenopeptide Sequences Identification
2.5. Determinations of Se-Enriched Perilla frutescens
2.5.1. Protein Analysis
2.5.2. Total Selenium Content
2.5.3. Degree of Hydrolysis (DH)
2.5.4. Antioxidant Activity
ABTS Radical Scavenging Activity Assay
Ferric Reducing Antioxidant Power (FRAP) Assay
2.5.5. ACE Inhibitory Activity
2.5.6. Cell Viability Assay
2.6. Statistical Analysis
3. Results and Discussion
3.1. Selenium Biotransformation in Se-Enriched Perilla Seeds
3.2. Selenopeptide Preparation and Antioxidant Activities
3.2.1. Effect of Protein Extraction Time on Protein Yield, Protein Content, and Se Content
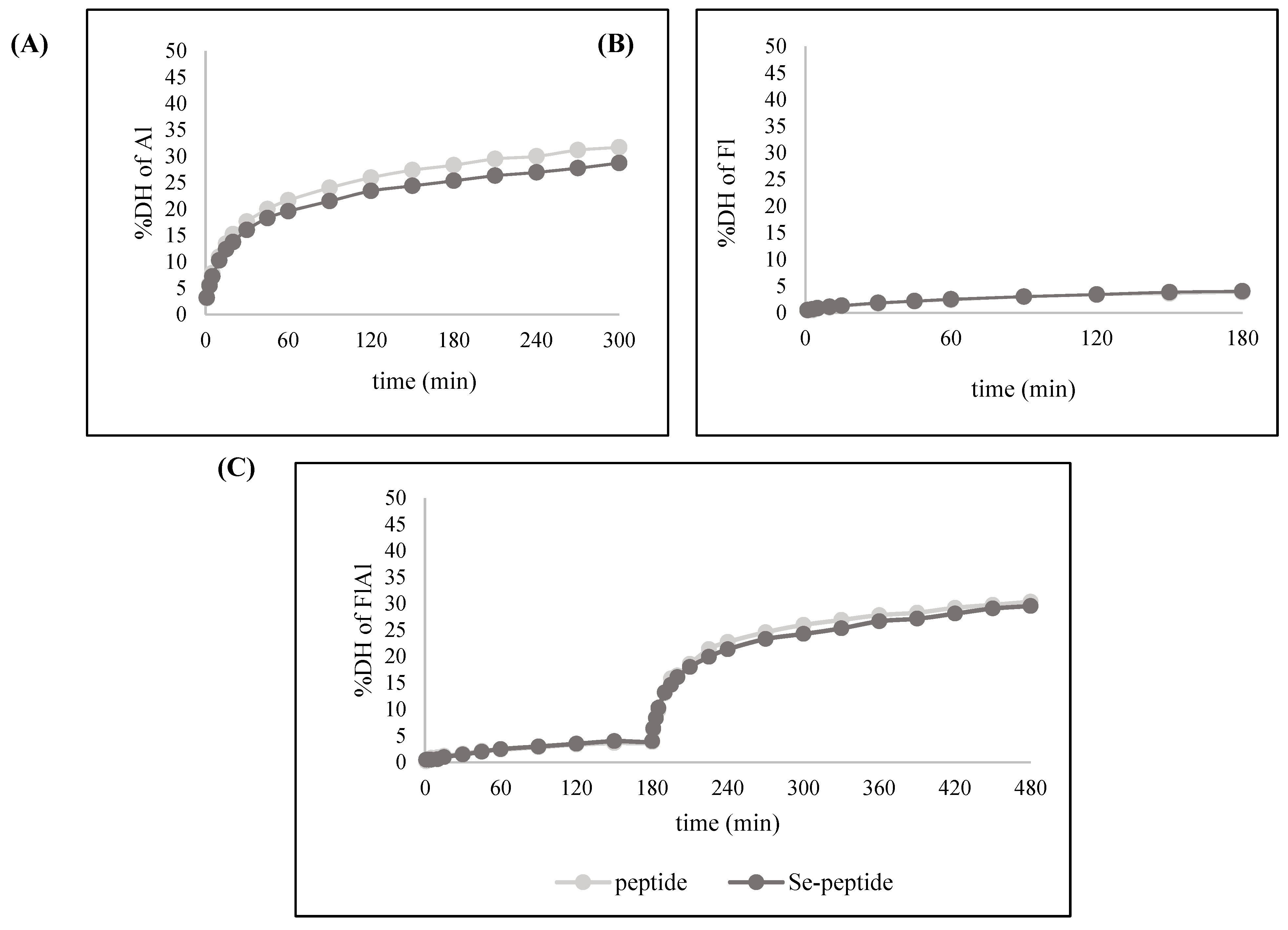
3.2.2. Effects of Enzymatic Hydrolysates on DH and Se Content of Se-Peptide
3.2.3. Antioxidant Activity of Se-Peptides
3.3. Membrane Ultrafiltration of Se-Peptides
3.3.1. Antioxidant Activities of Se-Peptides Fractions
3.3.2. ACE Inhibitory and Anticancer Activities of Se-Peptides Fractions
3.4. Purification, Antioxidants, ACE Inhibition, and Anticancer Activities of Se-Peptide Using Size-Exclusion Chromatography
3.5. Identification of Amino Acid Sequence in Se-Peptides from Se-Enriched Perilla frutescens
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| Al | Alcalase |
| Fl | Flavourzyme |
| AlFl | Combined enzymes |
| PPs | Perilla peptides |
| PSePs | Perilla Se-peptides |
| SPs | Perilla peptides after size exclusion |
| SSePs | Perilla Se-peptides after size exclusion |
References
- Pusadee, T.; Prom-u-thai, C.; Yimyam, N.; Jamjod, S.; Rerkasem, B. Phenotypic and Genetic Diversity of Local Perilla (Perilla frutescens (L.) Britt.) from Northern Thailand. Econ. Bot. 2017, 71, 175–187. [Google Scholar] [CrossRef]
- Paradee, N.; Howes, M.-J.R.; Utama-ang, N.; Chaikitwattna, A.; Hider, R.C.; Srichairatanakool, S. A chemically characterized ethanolic extract of Thai Perilla frutescens (L.) Britton fruits (nutlets) reduces oxidative stress and lipid peroxidation in human hepatoma (HuH7) cells. Phytother. Res. 2019, 33, 2064–2074. [Google Scholar] [CrossRef]
- Dhyani, A.; Chopra, R.; Garg, M. A Review On Nutritional Value, Functional Properties and Pharmacological Application of Perilla (Perilla frutescens L.). Biomed. Pharmacol. J. 2019, 12, 649–660. [Google Scholar] [CrossRef]
- Guan, L.; Zhu, L.; Zhang, X.; Han, Y.; Wang, K.; Ji, N.; Yao, X.; Zhou, Y.; Li, B.; Chen, Q.; et al. Perilla Seed Oil and Protein: Composition, Health Benefits, and Potential Applications in Functional Foods. Molecules 2024, 29, 5258. [Google Scholar] [CrossRef]
- Souphannavong, C.; Arjin, C.; Sartsook, A.; Yosen, T.; Thongkham, M.; Seel-audom, M.; Mekchay, S.; Sringarm, K. Nutritional values and nutrient digestibility of ground perilla cake (Perilla frutescens) in growing pig diets. Vet. Integr. Sci. 2021, 19, 423–438. [Google Scholar] [CrossRef]
- Hu, Y.; Luo, H.; Netala, V.R.; Li, H.; Zhang, Z.; Hou, T. Comprehensive Review of Biological Functions and Therapeutic Potential of Perilla Seed Meal Proteins and Peptides. Foods 2024, 14, 47. [Google Scholar] [CrossRef] [PubMed]
- Mukhamedov, N.; Wubulikasimu, A.; Rustamova, N.; Nuerxiati, R.; Mirzaakhmedov, S.; Ishimov, U.; Ziyavitdinov, J.; Yili, A.; Aisa, H.A. Synthesis and Characterization of Novel Chickpea Protein Hydrolysate-Vanadium Complexes Having Cell Inhibitory Effects on Lung Cancer A549 Cells Lines. Protein J. 2021, 40, 721–730. [Google Scholar] [CrossRef] [PubMed]
- Yin, J.; Liu, W.; Wu, M.; Chen, M.; Pei, X.; He, Y.; Shen, F.; Zhang, R.; He, J. Characterization of selenium-containing broccoli (Brassica oleracea L. var. italica planch) proteins and evaluation of antioxidant activity by electron spin resonance. Food Chem. 2024, 456, 140065. [Google Scholar] [CrossRef]
- Zhao, L.; Zhao, G.; Zhao, Z.; Chen, P.; Tong, J.; Hu, X. Selenium distribution in a Se-enriched mushroom species of the genus Ganoderma. J. Agric. Food Chem. 2004, 52, 3954–3959. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Fan, B.; Lei, N.; Xiong, Y.; Liu, Y.; Tong, L.; Wang, F.; Maesen, P.; Blecker, C. Selenium Biofortification of Soybean Sprouts: Effects of Selenium Enrichment on Proteins, Protein Structure, and Functional Properties. Front. Nutr. 2022, 9, 849928. [Google Scholar] [CrossRef] [PubMed]
- Serrano-Sandoval, S.N.; Guardado-Félix, D.; Gutiérrez-Uribe, J.A. Changes in digestibility of proteins from chickpeas (Cicer arietinum L.) germinated in presence of selenium and antioxidant capacity of hydrolysates. Food Chem. 2019, 285, 290–295. [Google Scholar] [CrossRef]
- Zhang, X.; He, H.; Xiang, J.; Yin, H.; Hou, T. Selenium-Containing Proteins/Peptides from Plants: A Review on the Structures and Functions. J. Agric. Food Chem. 2020, 68, 15061–15073. [Google Scholar] [CrossRef] [PubMed]
- Celus, I.; Brijs, K.; Delcour, J.A. Enzymatic Hydrolysis of Brewers’ Spent Grain Proteins and Technofunctional Properties of the Resulting Hydrolysates. J. Agric. Food Chem. 2007, 55, 8703–8710. [Google Scholar] [CrossRef] [PubMed]
- Hunsakul, K.; Laokuldilok, T.; Sakdatorn, V.; Klangpetch, W.; Brennan, C.S.; Utama-ang, N. Optimization of enzymatic hydrolysis by alcalase and flavourzyme to enhance the antioxidant properties of jasmine rice bran protein hydrolysate. Sci. Rep. 2022, 12, 12582. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.-Y.; Wang, C.-Y.; Wang, S.-T.; Li, Y.-Q.; Mo, H.-Z.; He, J.-X. Physicochemical properties and antioxidant activities of tree peony (Paeonia suffruticosa Andr.) seed protein hydrolysates obtained with different proteases. Food Chem. 2021, 345, 128765. [Google Scholar] [CrossRef]
- He, R.; Girgih, A.T.; Malomo, S.A.; Ju, X.; Aluko, R.E. Antioxidant activities of enzymatic rapeseed protein hydrolysates and the membrane ultrafiltration fractions. J. Funct. Foods 2013, 5, 219–227. [Google Scholar] [CrossRef]
- Lintschinger, J.; Fuchs, N.; Moser, J.; Kuehnelt, D.; Goessler, W. Selenium-Enriched Sprouts. A Raw Material for Fortified Cereal-Based Diets. J. Agric. Food Chem. 2000, 48, 5362–5368. [Google Scholar] [CrossRef]
- Xin, Y.; Xu, M.; Chen, L.; Wang, G.; Lu, W.; Liu, Z.; Shang, R.; Li, Y.; Wang, Z.; Sun, H.; et al. Effects of Different Defatting Methods of Black Soldier Fly (Hermetia illucens) Larvae Meal on the Metabolic Energy and Nutrient Digestibility in Young Laying Hens. Animals 2024, 14, 2521. [Google Scholar] [CrossRef]
- Hijona, E.; Hijona, L.; Larzabal, M.; Sarasqueta, C.; Aldazabal, P.; Arenas, J.; Bujanda, L. Biochemical determination of lipid content in hepatic steatosis by the Soxtec method. World J. Gastroenterol. 2010, 16, 1495–1499. [Google Scholar] [CrossRef]
- Kim, J.M.; Liceaga, A.M.; Yoon, K.Y. Purification and identification of an antioxidant peptide from perilla seed (Perilla frutescens) meal protein hydrolysate. Food Sci. Nutr. 2019, 7, 1645–1655. [Google Scholar] [CrossRef]
- Rezvankhah, A.; Yarmand, M.S.; Ghanbarzadeh, B. The effects of combined enzymatic and physical modifications of lentil protein applying Alcalase, Flavourzyme, microbial transglutaminase, and ultrasound: Antioxidant, antihypertension, and antidiabetic activities. J. Food Meas. Charact. 2022, 16, 3743–3759. [Google Scholar] [CrossRef]
- Clemente, A.; Vioque, J.; Sanchez-Vioque, R.; Pedroche, J.; Millán, F. Production of Extensive Chickpea (Cicer arietinum L.) Protein Hydrolysates with Reduced Antigenic Activity. J. Agric. Food Chem. 1999, 47, 3776–3781. [Google Scholar] [CrossRef] [PubMed]
- Park, Y.B.; Yoon, Y.K. Biological activity of enzymatic hydrolysates and the membrane ultrafiltration fractions from perilla seed meal protein. Czech J. Food Sci. 2019, 37, 180–185. [Google Scholar] [CrossRef]
- Fashakin, O.O.; Tangjaidee, P.; Unban, K.; Klangpetch, W.; Khumsap, T.; Sringarm, K.; Rawdkuen, S.; Phongthai, S. Isolation and Identification of Antioxidant Peptides Derived from Cricket (Gryllus bimaculatus) Protein Fractions. Insects 2023, 14, 674. [Google Scholar] [CrossRef]
- Krobthong, S.; and Yingchutrakul, Y. Identification and enhancement of antioxidant P1-peptide isolated from Ganoderma lucidum hydrolysate. Food Biotechnol. 2020, 34, 338–351. [Google Scholar] [CrossRef]
- Ewles, M.; and Goodwin, L. Bioanalytical Approaches to Analyzing Peptides and Proteins by LC–MS/MS. Bioanalysis 2011, 3, 1379–1397. [Google Scholar] [CrossRef]
- ACC, A.M. American Association of Cereal Chemists. Method 66–50 26–10A 26.41 66 2000. 41. Available online: https://www.apsnet.org/members/leadership/apsleadership/leadershipresources/Documents/ScisocOverview.pdf (accessed on day month year).
- Kamboj, A.; Sahil; Chopra, R.; Prabhakar, P.K. Perilla protein isolate exhibits synergistic techno-functionality through modification via sequential dynamic high-pressure microfluidization and enzymatic hydrolysis. Innov. Food Sci. Emerg. Technol. 2024, 94, 103683. [Google Scholar] [CrossRef]
- Mezeyová, I.; Hegedűsová, A.; Hegedűs, O.; Vargová, A.; Timoracká, M.; Šlosár, M.; Andrejiová, A.; Juríková, T.; Mezey, J. Basil seeds as a source of antioxidants affected by fortification with selenium. Folia Hortic. 2020, 32, 11–20. [Google Scholar] [CrossRef]
- Zhang, H.; Zhang, Z.; He, D.; Li, S.; Xu, Y. Optimization of Enzymatic Hydrolysis of Perilla Meal Protein for Hydrolysate with High Hydrolysis Degree and Antioxidant Activity. Molecules 2022, 27, 1079. [Google Scholar] [CrossRef]
- Hong, J.; Chen, T.-T.; Hu, P.; Yang, J.; Wang, S.-Y. Purification and characterization of an antioxidant peptide (GSQ) from Chinese leek (Allium tuberosum Rottler) seeds. J. Funct. Foods 2014, 10, 144–153. [Google Scholar] [CrossRef]
- León Madrazo, A.; Segura Campos, M.R. Antioxidant potential of peptides derived from chia seeds (Salvia hispanica L.) as natural preservatives. Food Chem. 2025, 465, 141968. [Google Scholar] [CrossRef]
- Candra, I.; Imalia Dwi, P.; Maman, S.; Andita, U.; Ismail; Ratna Komala, P.; Anisa, L.; Andrean Nur, P. Antioxidant Activity of DPPH, CUPRAC, and FRAP Methods, as well as Activity of Alpha-Glucosidase Inhibiting Enzymes from Tinospora crispa (L.) Stem Ultrasonic Extract. Pharmacogn. J. 2022, 14. [Google Scholar] [CrossRef]
- Fujimura, Y.; Shimura, M.; Nagai, H.; Hamada-Sato, N. Evaluation of angiotensin-converting enzyme-inhibitory activity in abalone viscera fermented by Lactobacillus casei 001. J. Funct. Foods 2021, 82, 104474. [Google Scholar] [CrossRef]
- Hankittichai, P.; Buacheen, P.; Pitchakarn, P.; Na Takuathung, M.; Wikan, N.; Smith, D.R.; Potikanond, S.; Nimlamool, W. Artocarpus lakoocha Extract Inhibits LPS-Induced Inflammatory Response in RAW 264.7 Macrophage Cells. Int. J. Mol. Sci. 2020, 21, 1355. [Google Scholar] [CrossRef]
- Liao, R. Selenium Accumulation Characteristics of Perilla frutescens. In IOP Conference Series: Earth and Environmental Science; IOP Publishing: Bristol, UK, 2018; Volume 199, p. 042023. [Google Scholar] [CrossRef]
- Liu, K.; Chen, F.; Zhao, Y.; Gu, Z.; Yang, H. Selenium accumulation in protein fractions during germination of Se-enriched brown rice and molecular weights distribution of Se-containing proteins. Food Chem. 2011, 127, 1526–1531. [Google Scholar] [CrossRef]
- Liu, K.; Gu, Z. Selenium Accumulation in Different Brown Rice Cultivars and Its Distribution in Fractions. J. Agric. Food Chem. 2009, 57, 695–700. [Google Scholar] [CrossRef]
- D’Amato, R.; Fontanella, M.C.; Falcinelli, B.; Beone, G.M.; Bravi, E.; Marconi, O.; Benincasa, P.; Businelli, D. Selenium Biofortification in Rice (Oryza sativa L.) Sprouting: Effects on Se Yield and Nutritional Traits with Focus on Phenolic Acid Profile. J. Agric. Food Chem. 2018, 66, 4082–4090. [Google Scholar] [CrossRef] [PubMed]
- Tangjaidee, P.; Swedlund, P.; Xiang, J.; Yin, H.; Quek, S.Y. Selenium-enriched plant foods: Selenium accumulation, speciation, and health functionality. Front. Nutr. 2023, 9, 962312. [Google Scholar] [CrossRef]
- Gupta, M.; Gupta, S. An Overview of Selenium Uptake, Metabolism, and Toxicity in Plants. Front. Plant Sci. 2016, 7, 2074. [Google Scholar] [CrossRef]
- Van Hoewyk, D. A tale of two toxicities: Malformed selenoproteins and oxidative stress both contribute to selenium stress in plants. Ann. Bot. 2013, 112, 965–972. [Google Scholar] [CrossRef]
- Trolove, S.N.; Tan, Y.; Morrison, S.C.; Feng, L.; Eason, J. Development of a method for producing selenium-enriched radish sprouts. LWT 2018, 95, 187–192. [Google Scholar] [CrossRef]
- Shen, L.; Wang, X.; Wang, Z.; Wu, Y.; Chen, J. Studies on tea protein extraction using alkaline and enzyme methods. Food Chem. 2008, 107, 929–938. [Google Scholar] [CrossRef]
- Hadidi, M.; Aghababaei, F.; McClements, D.J. Enhanced alkaline extraction techniques for isolating and modifying plant-based proteins. Food Hydrocoll. 2023, 145, 109132. [Google Scholar] [CrossRef]
- Yao, S.; Li, W.; Martin, G.J.O.; Ashokkumar, M. An Investigation into the Mechanism of Alkaline Extraction-Isoelectric Point Precipitation (AE-IEP) of High-Thiol Plant Proteins. Appl. Sci. 2023, 13, 6469. [Google Scholar] [CrossRef]
- Ha, H.Y.; Alfulaij, N.; Berry, M.J.; Seale, L.A. From Selenium Absorption to Selenoprotein Degradation. Biol. Trace Elem. Res. 2019, 192, 26–37. [Google Scholar] [CrossRef] [PubMed]
- Ma, S.; Caprioli, R.M.; Hill, K.E.; Burk, R.F. Loss of selenium from selenoproteins: Conversion of selenocysteine to dehydroalanine in vitro. J. Am. Soc. Mass Spectrom. 2003, 14, 593–600. [Google Scholar] [CrossRef]
- Huang, Y.; Lei, N.; Xiong, Y.; Liu, Y.; Tong, L.; Wang, F.; Fan, B.; Maesen, P.; Blecker, C. Influence of Selenium Biofortification of Soybeans on Speciation and Transformation during Seed Germination and Sprouts Quality. Foods 2022, 11, 1200. [Google Scholar] [CrossRef] [PubMed]
- Tacias-Pascacio, V.G.; Morellon-Sterling, R.; Siar, E.-H.; Tavano, O.; Berenguer-Murcia, Á.; Fernandez-Lafuente, R. Use of Alcalase in the production of bioactive peptides: A review. Int. J. Biol. Macromol. 2020, 165, 2143–2196. [Google Scholar] [CrossRef]
- Hu, F.; Ci, A.-T.; Wang, H.; Zhang, Y.-Y.; Zhang, J.-G.; Thakur, K.; Wei, Z.-J. Identification and hydrolysis kinetic of a novel antioxidant peptide from pecan meal using Alcalase. Food Chem. 2018, 261, 301–310. [Google Scholar] [CrossRef] [PubMed]
- Najafian, L.; Babji, A.S. Production of bioactive peptides using enzymatic hydrolysis and identification antioxidative peptides from patin (Pangasius sutchi) sarcoplasmic protein hydolysate. J. Funct. Foods 2014, 9, 280–289. [Google Scholar] [CrossRef]
- Zheng, L.; Zhao, M.; Xiao, C.; Zhao, Q.; Su, G. Practical problems when using ABTS assay to assess the radical-scavenging activity of peptides: Importance of controlling reaction pH and time. Food Chem. 2016, 192, 288–294. [Google Scholar] [CrossRef]
- Kimatu, B.M.; Zhao, L.; Biao, Y.; Ma, G.; Yang, W.; Pei, F.; Hu, Q. Antioxidant potential of edible mushroom (Agaricus bisporus) protein hydrolysates and their ultrafiltration fractions. Food Chem. 2017, 230, 58–67. [Google Scholar] [CrossRef]
- Liu, J.; Zhang, D.; Zhu, Y.; Wang, Y.; He, S.; Zhang, T. Enhancing the in vitro Antioxidant Capacities via the interaction of amino acids. Emir. J. Food Agric. (EJFA) 2018, 30, 224–231. [Google Scholar]
- Cheng, C.; Li, Q.; Yi, Y.; Yang, H.; Coldea, T.E.; Zhao, H. Selenium biofortification during barley (Hordeum vulgare L.) germination: Comparative analysis of selenate, selenite, and selenomethionine on se-protein accumulation and phenolic acid profile. Food Chem. 2025, 485, 144548. [Google Scholar] [CrossRef]
- Phongthai, S.; D’Amico, S.; Schoenlechner, R.; Homthawornchoo, W.; Rawdkuen, S. Fractionation and antioxidant properties of rice bran protein hydrolysates stimulated by in vitro gastrointestinal digestion. Food Chem. 2018, 240, 156–164. [Google Scholar] [CrossRef] [PubMed]
- Olagunju, A.I.; Omoba, O.S.; Enujiugha, V.N.; Alashi, A.M.; Aluko, R.E. Pigeon pea enzymatic protein hydrolysates and ultrafiltration peptide fractions as potential sources of antioxidant peptides: An in vitro study. LWT 2018, 97, 269–278. [Google Scholar] [CrossRef]
- Olalere, O.A.; Yap, P.-G.; Gan, C.-Y. Comprehensive review on some food-derived bioactive peptides with anti-hypertension therapeutic potential for angiotensin-converting enzyme (ACE) inhibition. J. Proteins Proteom. 2023, 14, 129–161. [Google Scholar] [CrossRef]
- Mansinhbhai, C.H.; Sakure, A.; Maurya, R.; Bishnoi, M.; Kondepudi, K.K.; Das, S.; Hati, S. Significance of whey protein hydrolysate on anti-oxidative, ACE-inhibitory and anti-inflammatory activities and release of peptides with biofunctionality: An in vitro and in silico approach. J. Food Sci. Technol. 2022, 59, 2629–2642. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Li, W.; Li, H.; Liu, W.; Li, L.; Liu, X. Selenium-Enriched Soybean Peptides as Novel Organic Selenium Compound Supplements: Inhibition of Occupational Air Pollution Exposure-Induced Apoptosis in Lung Epithelial Cells. Nutrients 2023, 16, 71. [Google Scholar] [CrossRef] [PubMed]
- Triantis, T.M.; Yannakopoulou, E.; Nikokavoura, A.; Dimotikali, D.; Papadopoulos, K. Chemiluminescent studies on the antioxidant activity of amino acids. Anal. Chim. Acta 2007, 591, 106–111. [Google Scholar] [CrossRef]
- Patriarca, E.J.; Cermola, F.; D’Aniello, C.; Fico, A.; Guardiola, O.; De Cesare, D.; Minchiotti, G. The Multifaceted Roles of Proline in Cell Behavior. Front. Cell Dev. Biol. 2021, 9, 728576. [Google Scholar] [CrossRef] [PubMed]
- Naqash, S.Y.; Nazeer, R.A. In Vitro Antioxidant and Antiproliferative Activities of Bioactive Peptide Isolated from Nemipterus Japonicus Backbone. Int. J. Food Prop. 2012, 15, 1200–1211. [Google Scholar] [CrossRef]

| Se Concentration (ppm) | Fat Content ns (%) | Protein Content (%) | Se Content (µg/g Sample) |
|---|---|---|---|
| 0 (control) | 44.18 ± 0.63 | 37.34 ± 0.29 b | 1.11 ± 0.02 e |
| 20 | 43.97 ± 0.25 | 37.52 ± 0.02 b | 25.22 ± 0.42 d |
| 40 | 44.14 ± 1.26 | 37.72 ± 0.34 ab | 68.83 ± 0.74 c |
| 60 | 42.08 ± 0.94 | 38.15 ± 0.23 ab | 86.64 ± 0.87 b |
| 80 | 42.87 ± 0.21 | 39.05 ± 1.14 a | 164.07 ± 2.51 a |
| 100 | 44.02 ± 0.63 | 38.93 ± 0.10 a | 164.62 ± 4.80 a |
| Extraction Time (min) | Yield (%) | Protein Content (%) | Se Content (µg/g Sample) |
|---|---|---|---|
| 30 | 11.48 ± 0.44 b | 78.71 ± 0.00 b | 77.24 ± 5.06 a |
| 60 | 11.70 ± 0.10 ab | 79.50 ± 0.15 a | 66.67 ± 1.88 b |
| 90 | 12.33 ± 0.04 a | 79.37 ± 0.24 a | 64.15 ± 4.94 b |
| Enzymatic Hydrolysates | PP | PSeP | ||||
|---|---|---|---|---|---|---|
| Al | Fl | FlAl | Al | Fl | FlAl | |
| Protein (%) | 61.20 ± 5.70 ns | 68.08 ± 0.76 ns | 65.04 ± 0.92 ns | 62.79 ± 0.74 B | 68.79 ± 0.31 A | 59.22 ± 1.25 C |
| Se contents (µg/g sample) | 1.50 ± 0.08 b | 1.69 ± 0.10 a | 1.53 ± 0.06 ab | 89.30 ± 0.74 B | 97.17 ± 0.69 A | 84.60 ± 0.68 C |
| DH (%) | 31.73 ± 0.39 a | 3.87 ± 0.00 c | 30.39 ± 0.00 b | 28.75 ± 0.26 B | 4.04 ± 0.00 C | 29.58 ± 0.01 A |
| % inhibition of ABTS | 80.28 ± 1.05 a | 68.77 ± 0.35 b | 80.50 ± 1.70 a | 79.14 ± 0.64 A | 68.06 ± 1.00 B | 79.42 ± 0.50 A |
| % inhibition of FRAP | 54.05 ± 0.80 c | 70.34 ± 1.41 a | 57.30 ± 0.57 b | 52.97 ± 1.57 B | 64.86 ± 2.08 A | 52.12 ± 0.72 B |
| Fractions | Se Contents (µg/g Sample) | %Inhibition of ABTS | %Inhibition of FRAP | |
|---|---|---|---|---|
| Peptides | F1 (>10 kDa) | 2.61 ± 0.52 ns | 89.88 ± 0.10 a | 60.65 ± 1.33 a |
| F2 (5–10 kDa) | 2.59 ± 0.09 ns | 78.68 ± 2.05 b | 51.83 ± 0.85 b | |
| F3 (3–5 kDa) | 2.39 ± 0.17 ns | 74.63 ± 1.77 c | 40.24 ± 0.62 c | |
| F4 (<3 kDa) | 2.72 ± 0.00 ns | 76.71 ± 1.28 bc | 36.57 ± 0.81 d | |
| Se-peptides | F1 (>10 kDa) | 124.58 ± 6.74 A | 83.63 ± 2.24 A | 59.59 ± 1.06 A |
| F2 (5–10 kDa) | 76.48 ± 2.04 B | 81.67 ± 1.30 A | 53.98 ± 1.60 B | |
| F3 (3–5 kDa) | 60.33 ± 2.16 C | 75.19 ± 0.17 B | 39.92 ± 1.04 C | |
| F4 (<3 kDa) | 75.86 ± 8.00 B | 68.25 ± 0.29 C | 38.39 ± 1.29 C |
| SP | SSeP | |||
|---|---|---|---|---|
| % Inhibition | ABTS | 63.62 ± 0.85 b | 66.30 ± 0.91 a | |
| FRAP | 56.30 ± 0.33 ns | 54.93 ± 0.96 ns | ||
| ACE | 74.30 ± 1.19 b | 83.87 ± 0.75 a | ||
| %viability | A549 cells | UT | 100.06 ± 0.93 ns | 101.72 ± 2.67 ns |
| 0.25 mg/mL | 95.71 ± 0.12 b | 89.33 ± 0.23 a | ||
| 0.5 mg/mL | 90.82 ± 0.12 b | 87.77 ± 1.04 a | ||
| 1 mg/mL | 88.95 ± 1.36 b | 85.88 ± 3.72 a | ||
| Sample | Peptide | Amino Acid Sequences | Mass (Da) | m/z | De Novo Score (%) |
|---|---|---|---|---|---|
| peptides | FPPEEMEACL | Phe-Pro-Pro-Glu-Glu-Met-Glu-Ala-Cys-Leu | 1164.48 | 583.26 | 93 |
| RMVLPEETEEEEEERS | Arg-Met-Val-Leu-Pro-Glu-Glu-Thr-Glu-Glu-Glu-Glu-Glu-Glu-Arg-Ser | 1990.88 | 996.45 | 91 | |
| RMVLPEETEEEEERESR | Arg-Met-Val-Leu-Pro-Glu-Glu-Thr-Glu-Glu-Glu-Glu-Glu-Arg-Glu-Ser-Arg | 2146.98 | 716.67 | 91 | |
| LAGGREDMPPQ | Leu-Ala-Gly-Gly-Arg-Glu-Asp-Met-Pro-Pro-Gln | 1169.55 | 585.78 | 86 | |
| QGKEDDRGLMVR | Gln-Gly-Lys-Glu-Asp-Asp-Arg-Gly-Leu-Met-Val-Arg | 1402.70 | 468.57 | 86 | |
| GKEDDRGMLVR | Gly-Lys-Glu-Asp-Asp-Arg-Gly-Met-Leu-Val-Arg | 1274.64 | 425.89 | 83 | |
| MDLPEERTEEEEESER | Met-Asp-Leu-Pro-Glu-Glu-Arg-Thr-Glu-Glu-Glu-Glu-Glu-Ser-Glu-Arg | 2006.84 | 1004.44 | 82 | |
| CHEEEERRE | Cys-His-Glu-Glu-Glu-Glu-Arg-Arg-Glu | 1215.49 | 608.76 | 80 | |
| VMPEETESFEPEPP | Val-Met-Pro-Glu-Glu-Thr-Glu-Ser-Phe-Glu-Pro-Glu-Pro-Pro | 1616.69 | 809.36 | 80 | |
| Se-peptides | RMVLPEETEEEEEERS | Arg-Met-Val-Leu-Pro-Glu-Glu-Thr-Glu-Glu-Glu-Glu-Glu-Glu-Arg-Ser | 1990.88 | 996.45 | 93 |
| FPPEEMEACL | Phe-Pro-Pro-Glu-Glu-Met-Glu-Ala-Cys-Leu | 1164.48 | 583.26 | 91 | |
| LAGGREDMPPQ | Leu-Ala-Gly-Gly-Arg-Glu-Asp-Met-Pro-Pro-Gln | 1169.55 | 585.78 | 89 | |
| MDLPEERTEEEEESER | Met-Asp-Leu-Pro-Glu-Glu-Arg-Thr-Glu-Glu-Glu-Glu-Glu-Ser-Glu-Arg | 2006.84 | 1004.44 | 87 | |
| RMVLPEETEEEEERESR | Arg-Met-Val-Leu-Pro-Glu-Glu-Thr-Glu-Glu-Glu-Glu-Glu-Arg-Glu-Ser-Arg | 2146.98 | 716.67 | 87 | |
| VMPEETESFEPEPP | Val-Met-Pro-Glu-Glu-Thr-Glu-Ser-Phe-Glu-Pro-Glu-Pro-Pro | 1616.69 | 809.36 | 85 | |
| GKEDDRGMLVR | Gly-Lys-Glu-Asp-Asp-Arg-Gly-Met-Leu-Val-Arg | 1274.64 | 425.89 | 84 | |
| QGKEDDRGLMVR | Gln-Gly-Lys-Glu-Asp-Asp-Arg-Gly-Leu-Met-Val-Arg | 1402.70 | 468.57 | 84 | |
| CHEEEERRE | Cys-His-Glu-Glu-Glu-Glu-Arg-Arg-Glu | 1215.49 | 608.76 | 83 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Monkhai, T.; Rawdkuen, S.; Phongthai, S.; Pakdeebamrung, P.; Singhadechachai, N.; Chaikaew, A.; Rachtanapun, P.; Tangjaidee, P. Selenium Biotransformation and Fractionation of Selenopeptide from Germinated Perilla (Perilla frutescens) Seeds. Foods 2025, 14, 2988. https://doi.org/10.3390/foods14172988
Monkhai T, Rawdkuen S, Phongthai S, Pakdeebamrung P, Singhadechachai N, Chaikaew A, Rachtanapun P, Tangjaidee P. Selenium Biotransformation and Fractionation of Selenopeptide from Germinated Perilla (Perilla frutescens) Seeds. Foods. 2025; 14(17):2988. https://doi.org/10.3390/foods14172988
Chicago/Turabian StyleMonkhai, Tanaporn, Saroat Rawdkuen, Suphat Phongthai, Pornrawin Pakdeebamrung, Naphatsawan Singhadechachai, Apinya Chaikaew, Pornchai Rachtanapun, and Pipat Tangjaidee. 2025. "Selenium Biotransformation and Fractionation of Selenopeptide from Germinated Perilla (Perilla frutescens) Seeds" Foods 14, no. 17: 2988. https://doi.org/10.3390/foods14172988
APA StyleMonkhai, T., Rawdkuen, S., Phongthai, S., Pakdeebamrung, P., Singhadechachai, N., Chaikaew, A., Rachtanapun, P., & Tangjaidee, P. (2025). Selenium Biotransformation and Fractionation of Selenopeptide from Germinated Perilla (Perilla frutescens) Seeds. Foods, 14(17), 2988. https://doi.org/10.3390/foods14172988








