Aptamer-CRISPR/Cas12a-Based Lateral Flow Technique for Visualized Rapid Detection of Endogenous Damage Factor Neu5Gc in Red Meat
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Aptamer Truncation Optimization
2.3. Aptamer Optimization Selection and crRNA Design
2.4. Construction of Aptamer-Cas12a Test Strip System and Validation of Design Principles
2.5. Aptamer-Cas12a Test Strip System Optimization
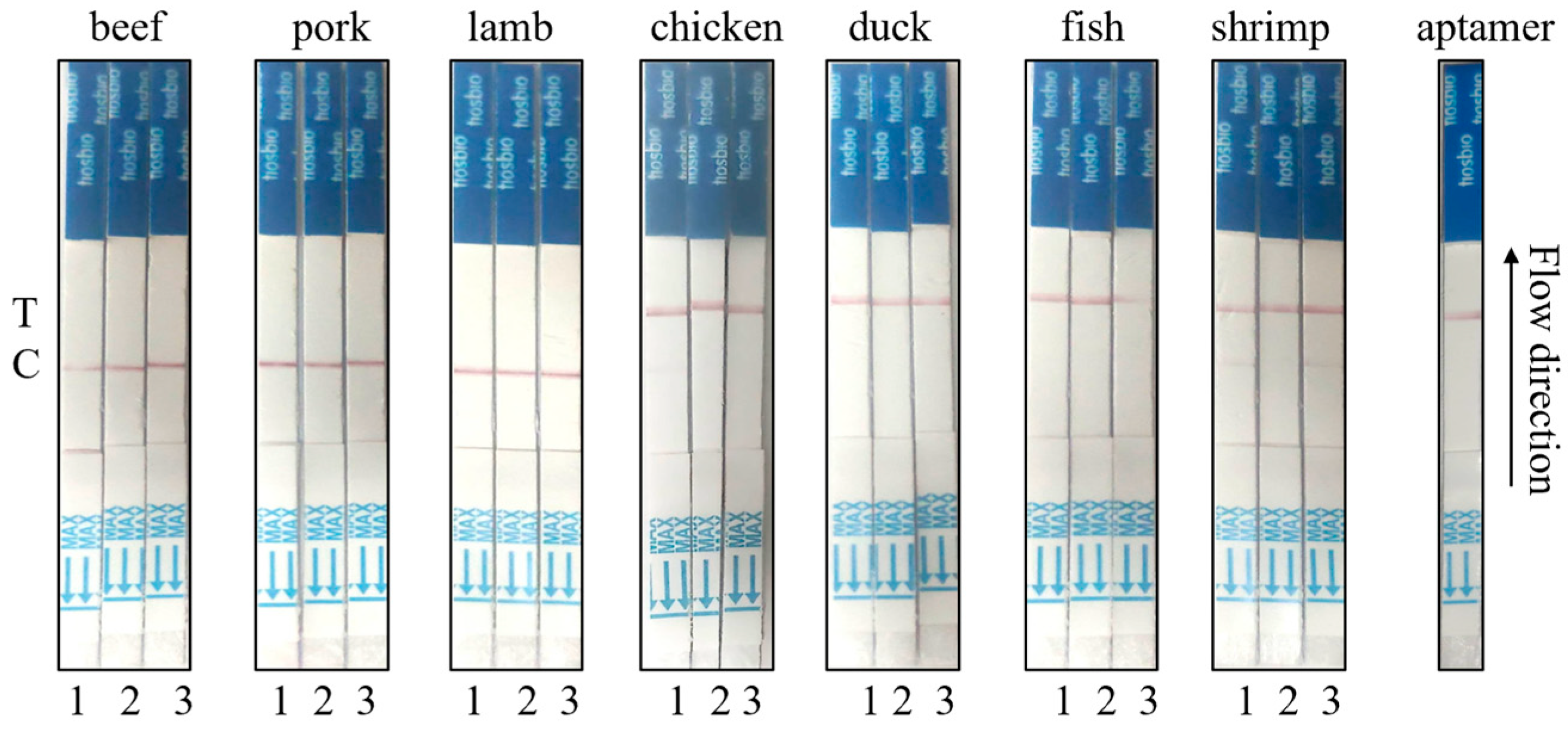
2.6. Specificity Determination of Neu5Gc Lateral Flow Detection Test Strips Based on Aptamer-Cas12a
2.7. Aptamer-Cas12a Test Strip Sensitivity Assay
2.8. Aptamer-Cas12a-Based Neu5Gc Lateral Flow Test Strip Spiking and Actual Sample Detection
3. Results
3.1. Analysis of Truncation Optimization of Nucleic Acid Aptamers
3.2. Proof of Principle Validation of Neu5Gc Lateral Flow Detection Test Strips Based on Aptamer-Cas12a
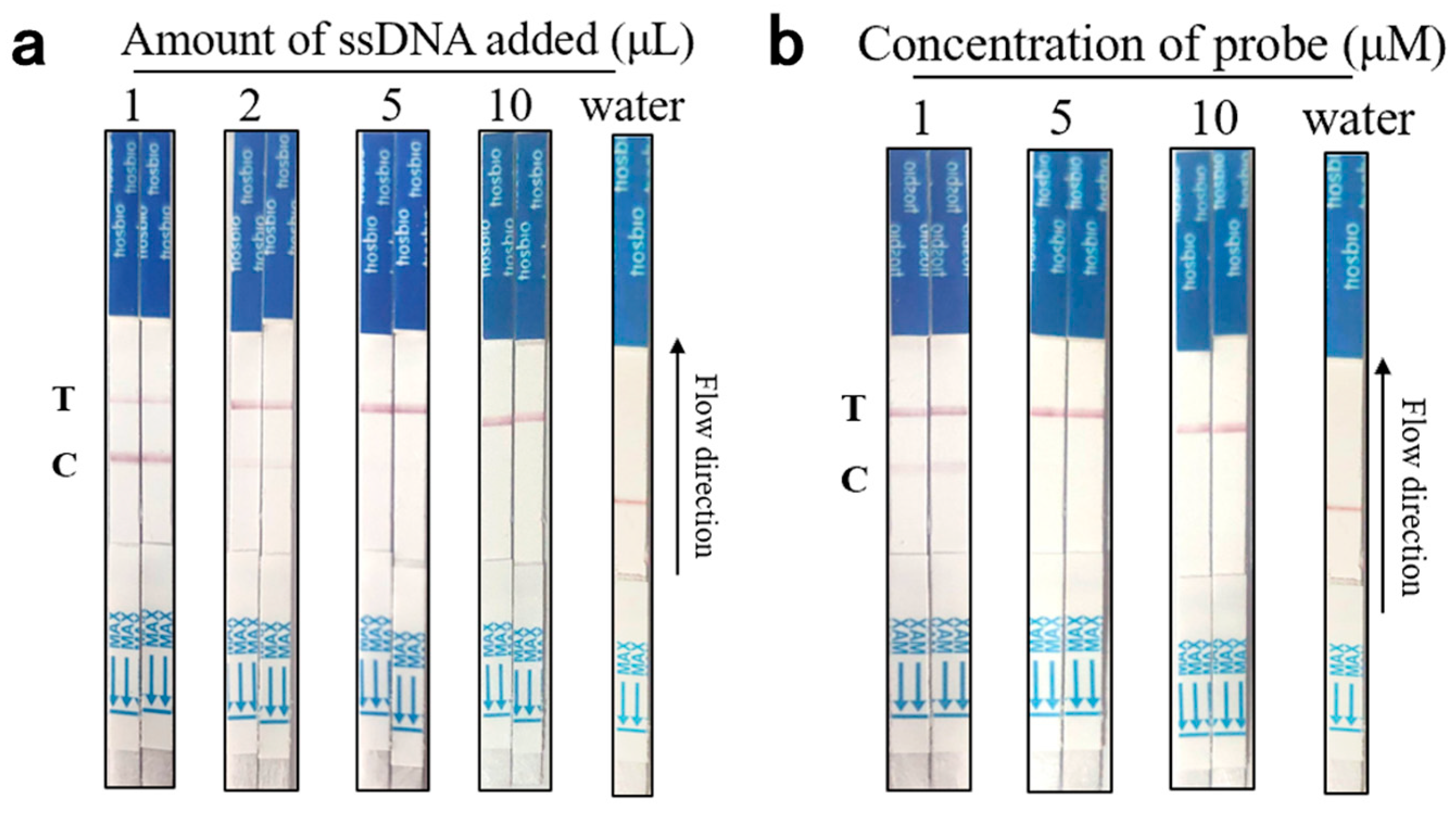
3.3. Optimization Results of Aptamer-Cas12a Test Strip Probe Concentration and Aptamer Content
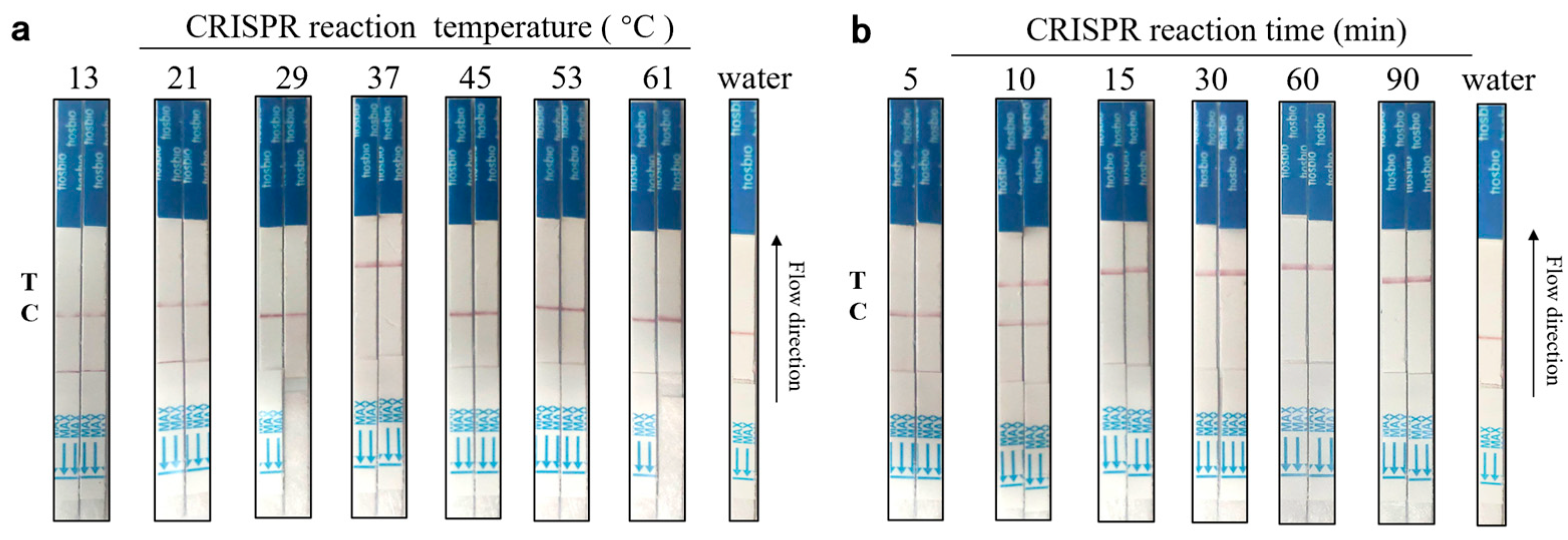
3.4. Aptamer-Cas12a Test Strip Reaction Time and Temperature Optimization Results
3.5. Based on the Specific Determination Results of the Lateral Flow Test Strip for Neu5Gc Using the Aptamer-Cas12a System
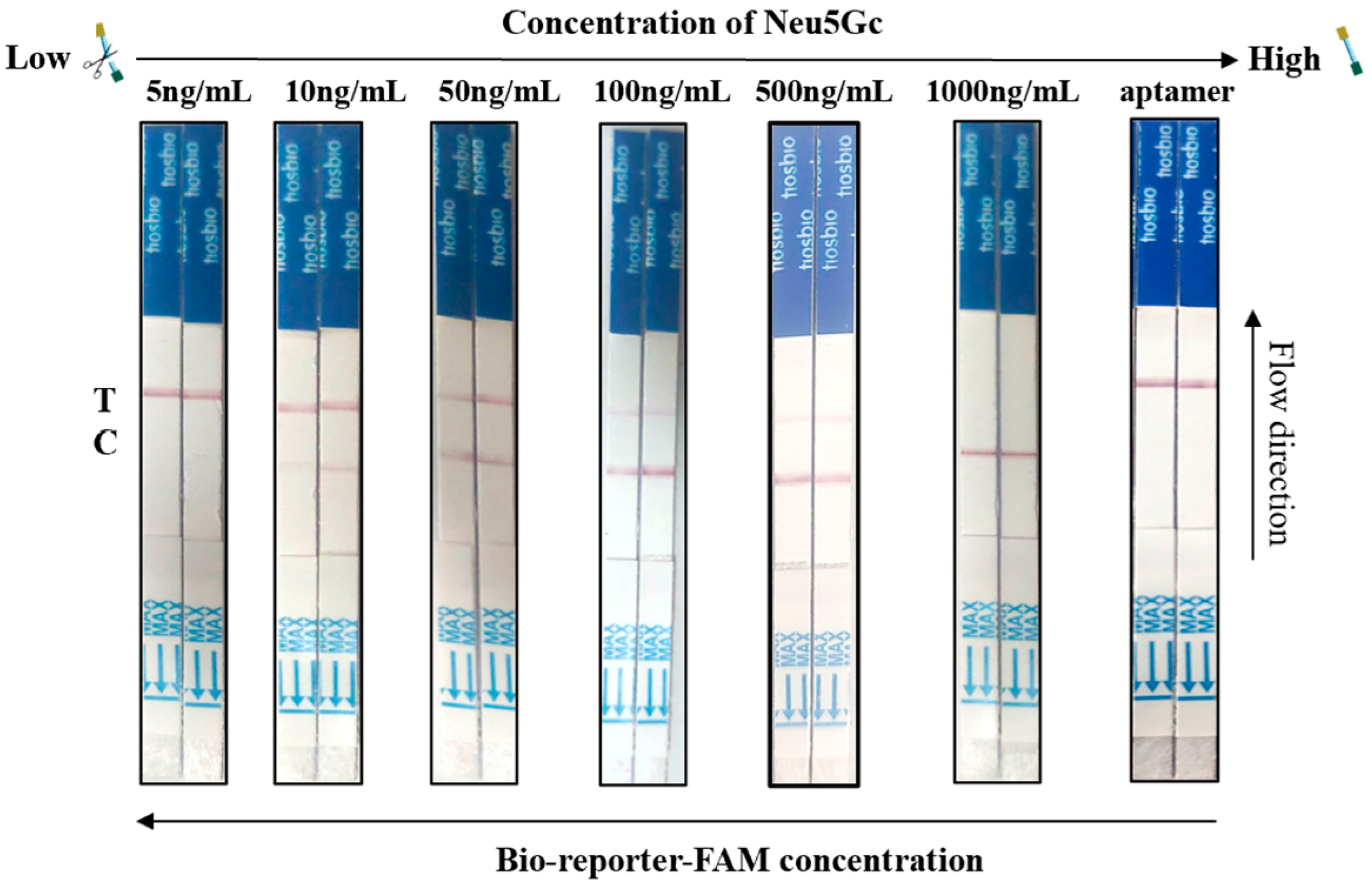
3.6. Aptamer-Cas12a Test Strip Sensitivity Assay Results
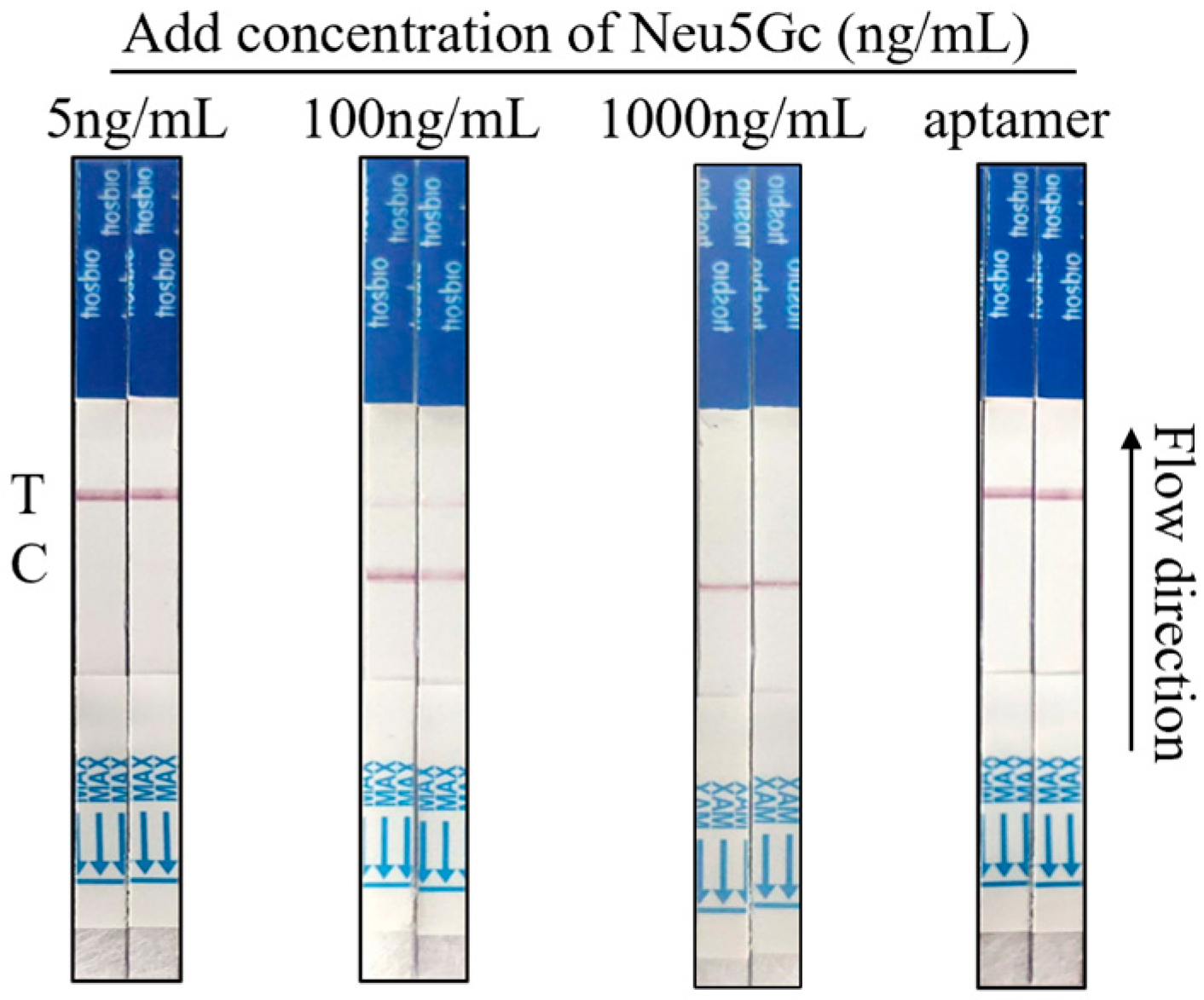
3.7. Results of Neu5Gc Lateral Flow Test Strip Spiking Assay Based on Aptamer-Cas12a
3.8. Actual Sample Detection of Neu5Gc Lateral Flow Test Strips Based on Aptamer-Cas12a
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Bashir, S.; Fezeu, L.K.; Ben-Arye, S.L.; Yehuda, S.; Reuven, E.M.; de Edelenyi, F.S.; Fellah-Hebia, I.; Le Tourneau, T.; Imbert-Marcille, B.M.; Drouet, E.B.; et al. Association between Neu5Gc carbohydrate and serum antibodies against it provides the molecular link to cancer: French NutriNet-Santé study. BMC Med. 2020, 18, 262. [Google Scholar] [CrossRef]
- Angeletti, A.; Bruschi, M.; Kajana, X.; Lugani, F.; Candiano, G.; Ghiggeri, G.M. Ghiggeri Anti-Neu5Gc Antibodies do not Affect Response to Human or Chimeric Monoclonal Anti-CD20 Antibodies in Children with Nephrotic Syndrome. J. Am. Soc. Nephrol. 2022, 33, 1985–1987. [Google Scholar] [CrossRef]
- Kawanishi, K.; Coker, J.K.; Grunddal, K.V.; Dhar, C.; Hsiao, J.; Zengler, K.; Varki, N.; Varki, A.; Gordts, P.L. Dietary Neu5Ac Intervention Protects Against Atherosclerosis Associated With Human-Like Neu5Gc Loss-Brief Report. Arter. Thromb. Vasc. Biol. 2021, 41, 2730–2739. [Google Scholar] [CrossRef]
- Liang, M.; Wu, J.; Li, H.; Zhu, Q. N-glycolylneuraminic acid in red meat and processed meat is a health concern: A review on the formation, health risk, and reduction. Compr. Rev. Food Sci. Food Saf. 2024, 23, e13314. [Google Scholar] [CrossRef]
- Liu, F.; van der Molen, J.; Kuipers, F.; van Leeuwen, S.S. Quantitation of bioactive components in infant formulas: Milk oligosaccharides, sialic acids and corticosteroids. Food Res. Int. 2023, 174, 113589. [Google Scholar] [CrossRef]
- Wang, X.; Lin, X.; Ouyang, Y.Y.; Liu, J.; Zhao, G.; Pan, A.; Hu, F.B. Red and processed meat consumption and mortality: Dose–response meta-analysis of prospective cohort studies. Public Health Nutr. 2016, 19, 893–905. [Google Scholar] [CrossRef]
- Feskens, E.J.M.; Sluik, D.; van Woudenbergh, G.J. Meat Consumption, Diabetes, and Its Complications. Curr. Diabetes Rep. 2013, 13, 298–306. [Google Scholar] [CrossRef] [PubMed]
- Zelber-Sagi, S.; Ivancovsky-Wajcman, D.; Isakov, N.F.; Webb, M.; Orenstein, D.; Shibolet, O.; Kariv, R. High red and processed meat consumption is associated with non-alcoholic fatty liver disease and insulin resistance. J. Hepatol. 2018, 68, 1239–1246. [Google Scholar] [CrossRef]
- Hashemian, M.; Merat, S.; Poustchi, H.; Jafari, E.; Radmard, A.-R.; Kamangar, F.; Freedman, N.; Hekmatdoost, A.; Sheikh, M.; Boffetta, P.; et al. Red Meat Consumption and Risk of Nonalcoholic Fatty Liver Disease in a Population With Low Meat Consumption: The Golestan Cohort Study. Off. J. Am. Coll. Gastroenterol. /ACG 2021, 116, 1667. [Google Scholar] [CrossRef] [PubMed]
- English, D.R.; MacInnis, R.J.; Hodge, A.M.; Hopper, J.L.; Haydon, A.M.; Giles, G.G. Red Meat, Chicken, and Fish Consumption and Risk of Colorectal Cancer. Cancer Epidemiol. Biomark. Prev. 2004, 13, 1509–1514. [Google Scholar] [CrossRef]
- Larsson, S.C.; Rafter, J.; Holmberg, L.; Bergkvist, L.; Wolk, A. Red meat consumption and risk of cancers of the proximal colon, distal colon and rectum: The Swedish Mammography Cohort. Int. J. Cancer 2005, 113, 829–834. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Lv, B.; Zhou, Y.; Ye, F. Meta-analysis of the Relationship between Meat Consumption and Breast Cancer. Public Health Prev. Med. 2016, 27, 72–75+80. [Google Scholar]
- Bonequi, P.; Meneses-González, F.; Correa, P.; Rabkin, C.S.; Camargo, M.C. Risk factors for gastric cancer in Latin America: A meta-analysis. Cancer Causes Control. 2013, 24, 217–231. [Google Scholar] [CrossRef]
- GRAS Substances (SCOGS) Database. Pharma Excipients. Available online: https://www.pharmaexcipients.com/excipient-sources/gras-substances-scogs-database/ (accessed on 25 July 2025).
- Ana, A.; Raquel, G.M.; Georgia, G.; Angelo, M.; Caroline, M.; Agnes, R.; Tobin, R. EFSA’s activities on emerging risks in 2019. EFSA Support. Publ. 2020, 17, 1924E. [Google Scholar] [CrossRef]
- Samraj, A.N.; Pearce, O.M.T.; Läubli, H.; Crittenden, A.N.; Bergfeld, A.K.; Banda, K.; Gregg, C.J.; Bingman, A.E.; Secrest, P.; Diaz, S.L.; et al. A red meat-derived glycan promotes inflammation and cancer progression. Proc. Natl. Acad. Sci. USA 2015, 112, 542–547. [Google Scholar] [CrossRef] [PubMed]
- Labrada, M.; Dorvignit, D.; Hevia, G.; Rodríguez-Zhurbenko, N.; Hernández, A.M.; Vázquez, A.M.; Fernández, L.E. GM3(Neu5Gc) ganglioside: An evolution fixed neoantigen for cancer immunotherapy. Semin. Oncol. 2018, 45, 41–51. [Google Scholar] [CrossRef]
- Li, Z.; Wang, X.; Deng, X.; Song, J.; Yang, T.; Liao, Y.; Gong, G.; Huang, L.; Lu, Y.; Wang, Z. High-sensitivity qualitative and quantitative analysis of human, bovine and goat milk glycosphingolipids using HILIC-MS/MS with internal standards. Carbohydr. Polym. 2023, 312, 120795. [Google Scholar] [CrossRef]
- Zhu, C.; Feng, Z.; Qin, H.; Chen, L.; Yan, M.; Li, L.; Qu, F. Recent progress of SELEX methods for screening nucleic acid aptamers. Talanta 2024, 266, 124998. [Google Scholar] [CrossRef]
- Cheng, L.; Yang, F.; Tang, L.; Qian, L.; Chen, X.; Guan, F.; Zhang, J.; Li, G. Electrochemical Evaluation of Tumor Development via Cellular Interface Supported CRISPR/Cas Trans-Cleavage. Research 2022, 2022, 9826484. [Google Scholar] [CrossRef]
- Ma, Y.; Wang, X.; Wang, Z.; Cong, P.; Xu, J.; Xue, C. Characterization of Gangliosides in Three Sea Urchin Species by HILIC-ESI-MS/MS. J. Agric. Food Chem. 2021, 69, 7641–7651. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.; Ren, H.; Wang, H.; Xiao, Y.; Wang, C.; Liu, M.; Duan, F.; Li, H.; Hu, P.; Li, Y.; et al. Characterization of an Aptamer Targeting Neu5Gc, as an Endogenous Pathogenic Factor Derived from Red Meat. Molecules 2024, 29, 1273. [Google Scholar] [CrossRef]
- Rockey, W.M.; Hernandez, F.J.; Huang, S.-Y.; Cao, S.; Howell, C.A.; Thomas, G.S.; Liu, X.Y.; Lapteva, N.; Spencer, D.M.; McNamara, J.O.; et al. Rational truncation of an RNA aptamer to prostate-specific membrane antigen using computational structural modeling. Nucleic Acid Ther. 2011, 21, 299–314. [Google Scholar] [CrossRef]
- Varki, A. Sialic acids in human health and disease. Trends Mol. Med. 2008, 14, 351–360. [Google Scholar] [CrossRef]
- Wang, J.Y.; Doudna, J.A. Doudna CRISPR technology: A decade of genome editing is only the beginning. Science 2023, 379, eadd8643. [Google Scholar] [CrossRef] [PubMed]
- Liu, G.; Lin, Q.; Jin, S.; Gao, C. The CRISPR-Cas toolbox and gene editing technologies. Mol. Cell 2022, 82, 333–347. [Google Scholar] [CrossRef]
- Sale, J.E.; Stoddard, B.L. Stoddard CRISPR in Nucleic Acids Research: The sequel. Nucleic Acids Res. 2024, 52, 3489–3492. [Google Scholar] [CrossRef]
- Reysenbach, A.-L.; Terns, M.P. CRISPR-influenced symbiosis. Nat. Microbiol. 2023, 8, 1611–1612. [Google Scholar] [CrossRef] [PubMed]
- Wimmer, F.; Mougiakos, I.; Englert, F.; Beisel, C.L. Beisel Rapid cell-free characterization of multi-subunit CRISPR effectors and transposons. Mol. Cell 2022, 82, 1210–1224.e6. [Google Scholar] [CrossRef]
- Broughton, J.P.; Deng, X.; Yu, G.; Fasching, C.L.; Servellita, V.; Singh, J.; Miao, X.; Streithorst, J.A.; Granados, A.; Sotomayor-Gonzalez, A.; et al. CRISPR-Cas12-based detection of SARS-CoV-2. Nat. Biotechnol. 2020, 38, 870–874. [Google Scholar] [CrossRef]
- Xu, X.; Chemparathy, A.; Zeng, L.; Kempton, H.R.; Shang, S.; Nakamura, M.; Qi, L.S. Engineered miniature CRISPR-Cas system for mammalian genome regulation and editing. Mol. Cell 2021, 81, 4333–4345.e4. [Google Scholar] [CrossRef] [PubMed]
- Cox, D.B.T.; Gootenberg, J.S.; Abudayyeh, O.O.; Franklin, B.; Kellner, M.J.; Joung, J.; Zhang, F. RNA editing with CRISPR-Cas13. Science 2017, 358, 1019–1027. [Google Scholar] [CrossRef]
- East-Seletsky, A.; O’Connell, M.R.; Knight, S.C.; Burstein, D.; Cate, J.H.D.; Tjian, R.; Doudna, J.A. Two distinct RNase activities of CRISPR-C2c2 enable guide-RNA processing and RNA detection. Nature 2016, 538, 270–273. [Google Scholar] [CrossRef] [PubMed]
- Van Dongen, J.E.; Berendsen, J.T.; Steenbergen, R.D.; Wolthuis, R.; Eijkel, J.C.; Segerink, L.I. Point-of-care CRISPR/Cas nucleic acid detection: Recent advances, challenges and opportunities. Biosens Bioelectron 2020, 166, 112445. [Google Scholar] [CrossRef] [PubMed]
- Chen, A.; Yang, S. Replacing antibodies with aptamers in lateral flow immunoassay. Biosens Bioelectron 2015, 71, 230–242. [Google Scholar] [CrossRef]
- Ivanov, A.V.; Safenkova, I.V.; Zherdev, A.V.; Dzantiev, B.B. Dzantiev DIRECT2: A novel platform for a CRISPR-Cas12-based assay comprising universal DNA-IgG probe and a direct lateral flow test. Biosens Bioelectron 2022, 208, 114227. [Google Scholar] [CrossRef] [PubMed]
- Lobato, I.M.; O’SUllivan, C.K. O’Sullivan Recombinase polymerase amplification: Basics, applications and recent advances. Trends Anal. Chem. 2018, 98, 19–35. [Google Scholar] [CrossRef]
- Storms, S.M.; Shisler, J.; Nguyen, T.H.; Zuckermann, F.A.; Lowe, J.F. Lateral flow paired with RT-LAMP: A speedy solution for Influenza A virus detection in swine. Vet. Microbiol. 2024, 296, 110174. [Google Scholar] [CrossRef]
- Cheng, M.; Tan, C.; Xiang, B.; Lin, W.; Cheng, B.; Peng, X.; Yang, Y.; Lin, Y. Chain hybridization-based CRISPR-lateral flow assay enables accurate gene visual detection. Anal. Chim. Acta 2023, 1270, 341437. [Google Scholar] [CrossRef]
- Li, Q.; Li, X.; Zhou, P.; Chen, R.; Xiao, R.; Pang, Y. Split aptamer regulated CRISPR/Cas12a biosensor for 17β-estradiol through a gap-enhanced Raman tags based lateral flow strategy. Biosens Bioelectron 2022, 215, 114548. [Google Scholar] [CrossRef]
- Shen, J.; Zhu, B.; Chen, Z.; Jia, L.; Sun, S. Precision Characterization of Site-Specific O-Acetylated Sialic Acids on N-Glycoproteins. Anal. Chem. 2023, 95, 1995–2003. [Google Scholar] [CrossRef]
- Safferthal, M.; Bechtella, L.; Zappe, A.; Vos, G.M.; Pagel, K. Labeling of Mucin-Type O-Glycans for Quantification Using Liquid Chromatography and Fluorescence Detection. ACS Meas. Sci. Au 2024, 4, 223–230. [Google Scholar] [CrossRef] [PubMed]
- Bashir, S.; Ben Arye, S.L.; Reuven, E.M.; Yu, H.; Costa, C.; Galiñanes, M.; Bottio, T.; Chen, X.; Padler-Karavani, V. Presentation Mode of Glycans Affect Recognition of Human Serum anti-Neu5Gc IgG Antibodies. Bioconjugate Chem. 2019, 30, 161–168. [Google Scholar] [CrossRef] [PubMed]
- Gong, S.; Ren, H.; Lin, C.; Hu, P.; Tian, R.; Liu, Z.; Li, Y.; Zhou, Y.; Yang, Y.; Lu, S. Immunochromatographic strip biosensor for the rapid detection of N-glycolylneuraminic acid based on aptamer-conjugated nanoparticle. Anal. Biochem. 2018, 561–562, 52–58. [Google Scholar] [CrossRef] [PubMed]
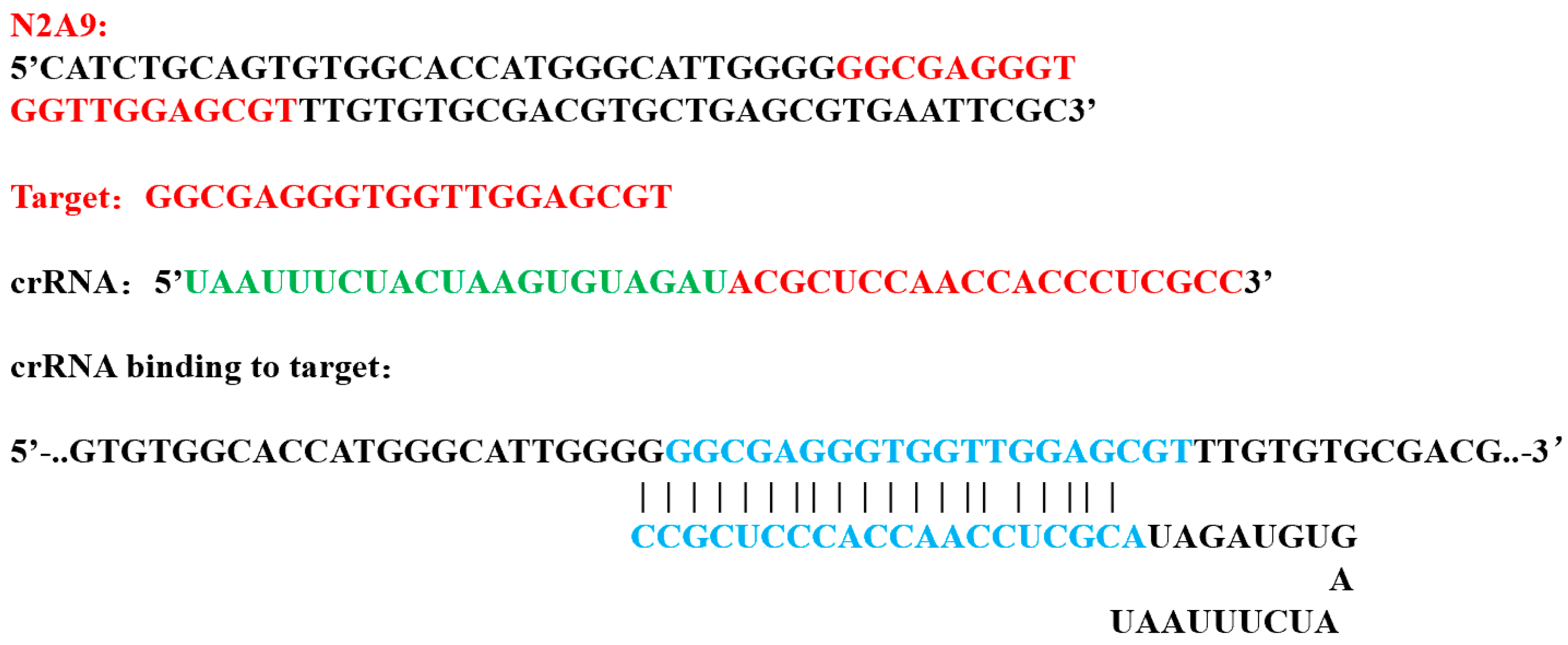
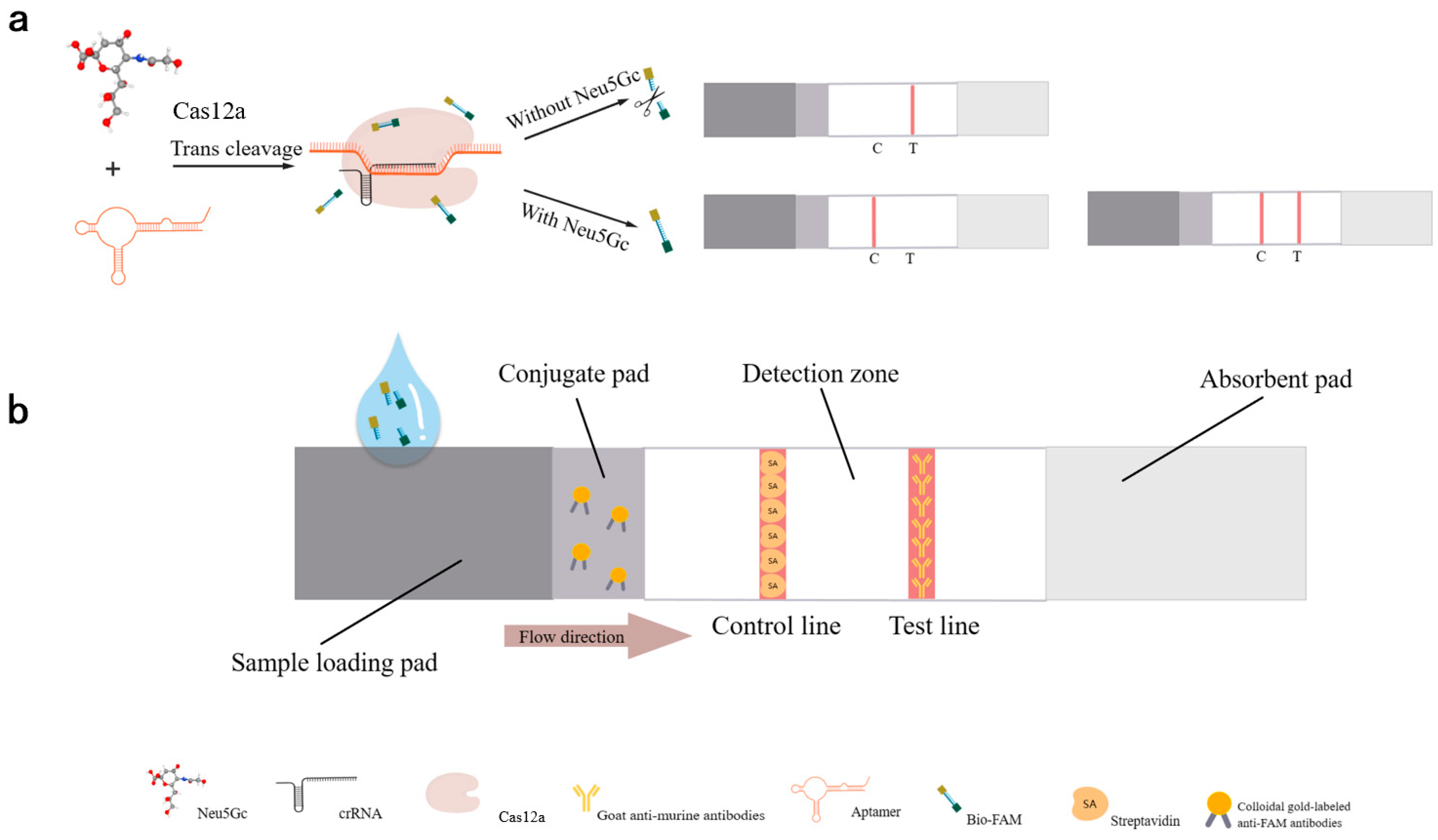
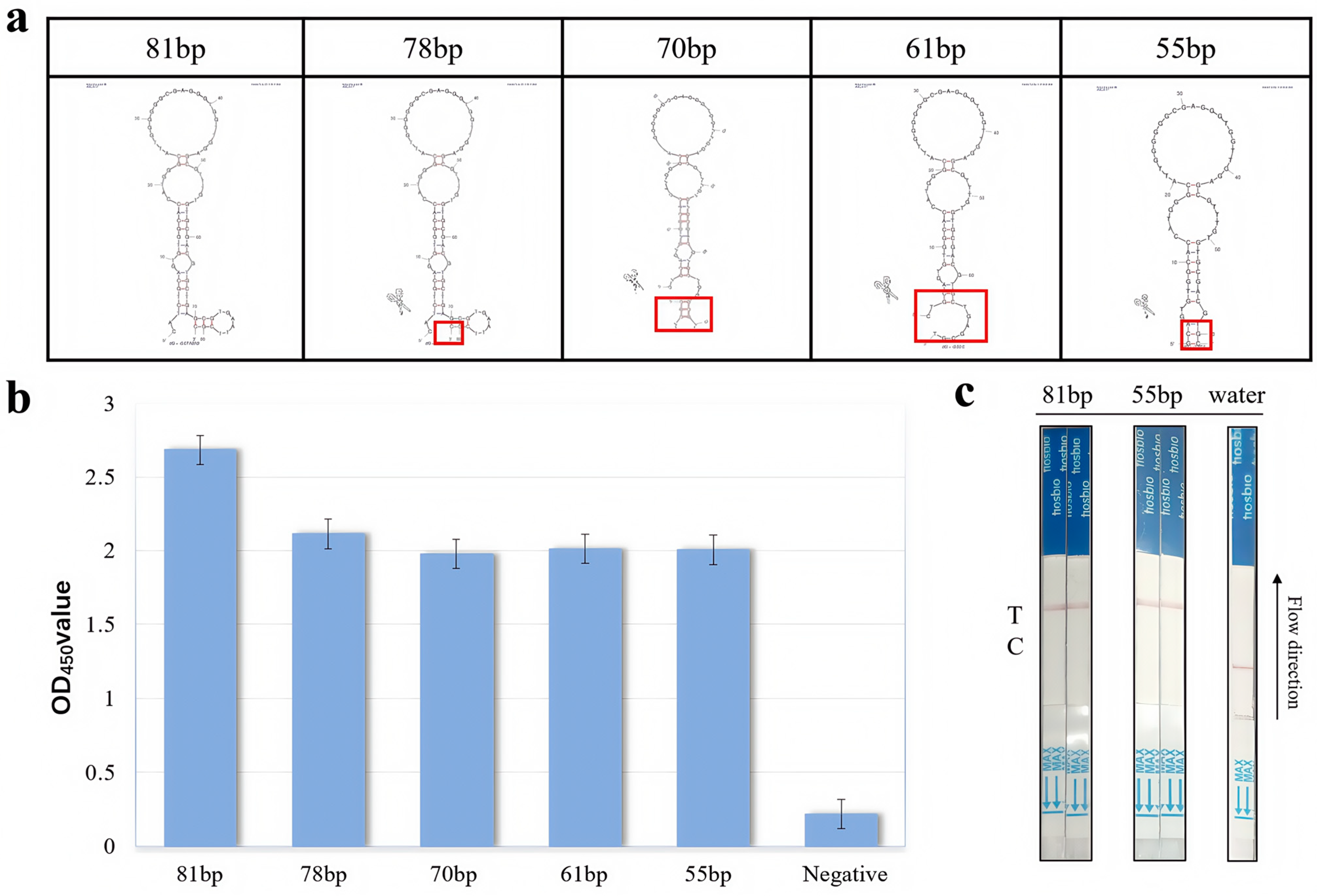
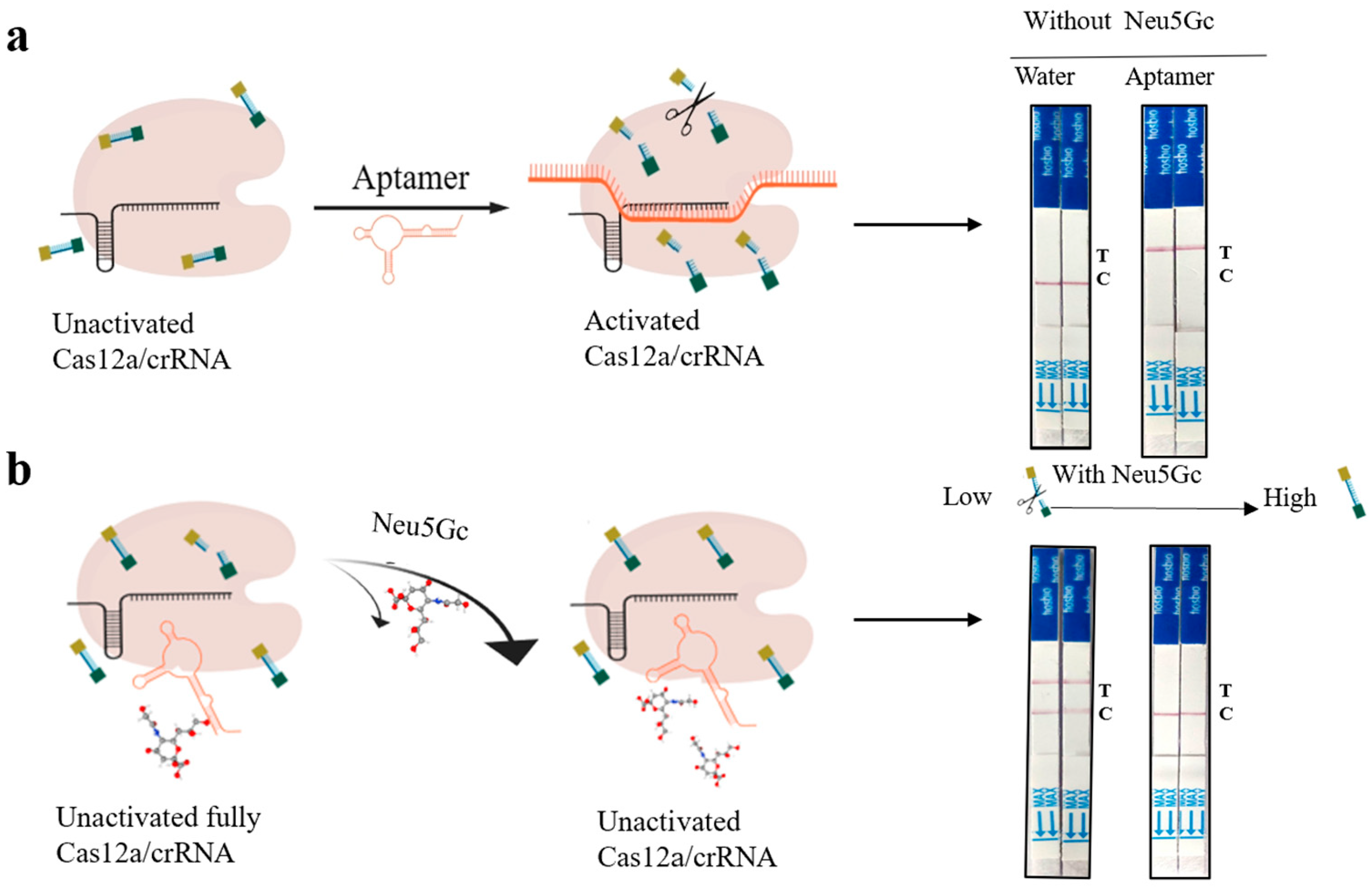

| Sequence Name | Sequences (5’–3’) | Sequence Lengths |
|---|---|---|
| N2A9.81 | CATCTGCAGTGTGGCACCATGGGCATTGGGGGGCGAGGGTGGTTGGAGCGTTTGTGTGCGACGTGCTGAGCGTGAATTCGC | 81 bp |
| N2A9.78 | CATCTGCAGTGTGGCACCATGGGCATTGGGGGGCGAGGGTGGTTGGAGCGTTTGTGTGCGACGTGCTGAGCGTGAATTCGC | 78 bp |
| N2A9.70 | CATCTGCAGTGTGGCACCATGGGCATTGGGGGGCGAGGGTGGTTGGAGCGTTTGTGTGCGACGTGCTGAGCGTGAATTCGC | 70 bp |
| N2A9.61 | CATCTGCAGTGTGGCACCATGGGCATTGGGGGGCGAGGGTGGTTGGAGCGTTTGTGTGCGACGTGCTGAGCGTGAATTCGC | 61 bp |
| N2A9.55 | CATCTGCAGTGTGGCACCATGGGCATTGGGGGGCGAGGGTGGTTGGAGCGTTTGTGTGCGACGTGCTGAGCGTGAATTCGC | 55 bp |
| Remark | Underscores are truncated removal sequences | |
| Reaction | System Name | Volumetric (µL) |
|---|---|---|
| 10 min at room temperature | Liquid to be examined | 5(0) |
| Aptamer100 nM | 5 | |
| CRISPR/Cas12a reaction 37 °C for 15 min | Buffer | 5 |
| LbCas12a, 1 µM | 5 | |
| crRNA, 1 µM | 5 | |
| Dual-labeled probes, 5 µM | 5 | |
| DEPC water | 20(25) | |
| Total | 50 |
| Detection Method | Detection Time | Detection Temperature | Minimum Detection Limit | Required Instrument |
|---|---|---|---|---|
| HPLC [41] | 10 h | 18–25 °C | 0.02 ng/μL | High Performance Liquid Chromatography |
| MS [42] | 12 h | 18–25 °C | 0.02pg/μL | Mass spectrometer |
| Aptamer-ELISA [22] | 17 h | 37 °C | 0.71 ng/mL | microplate reader |
| Aptamer-Cas12a test strip | 25 min | 37 °C | 10 ng/mL | Metal bath or Warm water bath |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Guo, Y.; Ren, H.; Wang, H.; Duan, X.; Qi, S.; Yang, X.; Shangguan, C.; Li, H.; Li, Y.; Hu, P.; et al. Aptamer-CRISPR/Cas12a-Based Lateral Flow Technique for Visualized Rapid Detection of Endogenous Damage Factor Neu5Gc in Red Meat. Foods 2025, 14, 2879. https://doi.org/10.3390/foods14162879
Guo Y, Ren H, Wang H, Duan X, Qi S, Yang X, Shangguan C, Li H, Li Y, Hu P, et al. Aptamer-CRISPR/Cas12a-Based Lateral Flow Technique for Visualized Rapid Detection of Endogenous Damage Factor Neu5Gc in Red Meat. Foods. 2025; 14(16):2879. https://doi.org/10.3390/foods14162879
Chicago/Turabian StyleGuo, Yuxi, Honglin Ren, Han Wang, Xuepeng Duan, Shuaihao Qi, Xi Yang, Chunyi Shangguan, Haosong Li, Yansong Li, Pan Hu, and et al. 2025. "Aptamer-CRISPR/Cas12a-Based Lateral Flow Technique for Visualized Rapid Detection of Endogenous Damage Factor Neu5Gc in Red Meat" Foods 14, no. 16: 2879. https://doi.org/10.3390/foods14162879
APA StyleGuo, Y., Ren, H., Wang, H., Duan, X., Qi, S., Yang, X., Shangguan, C., Li, H., Li, Y., Hu, P., Lu, Q., & Lu, S. (2025). Aptamer-CRISPR/Cas12a-Based Lateral Flow Technique for Visualized Rapid Detection of Endogenous Damage Factor Neu5Gc in Red Meat. Foods, 14(16), 2879. https://doi.org/10.3390/foods14162879





