Assessing Milk Authenticity Using Protein and Peptide Biomarkers: A Decade of Progress in Species Differentiation and Fraud Detection
Abstract
1. Introduction: Milk as a Nutritional Matrix and Target for Adulteration
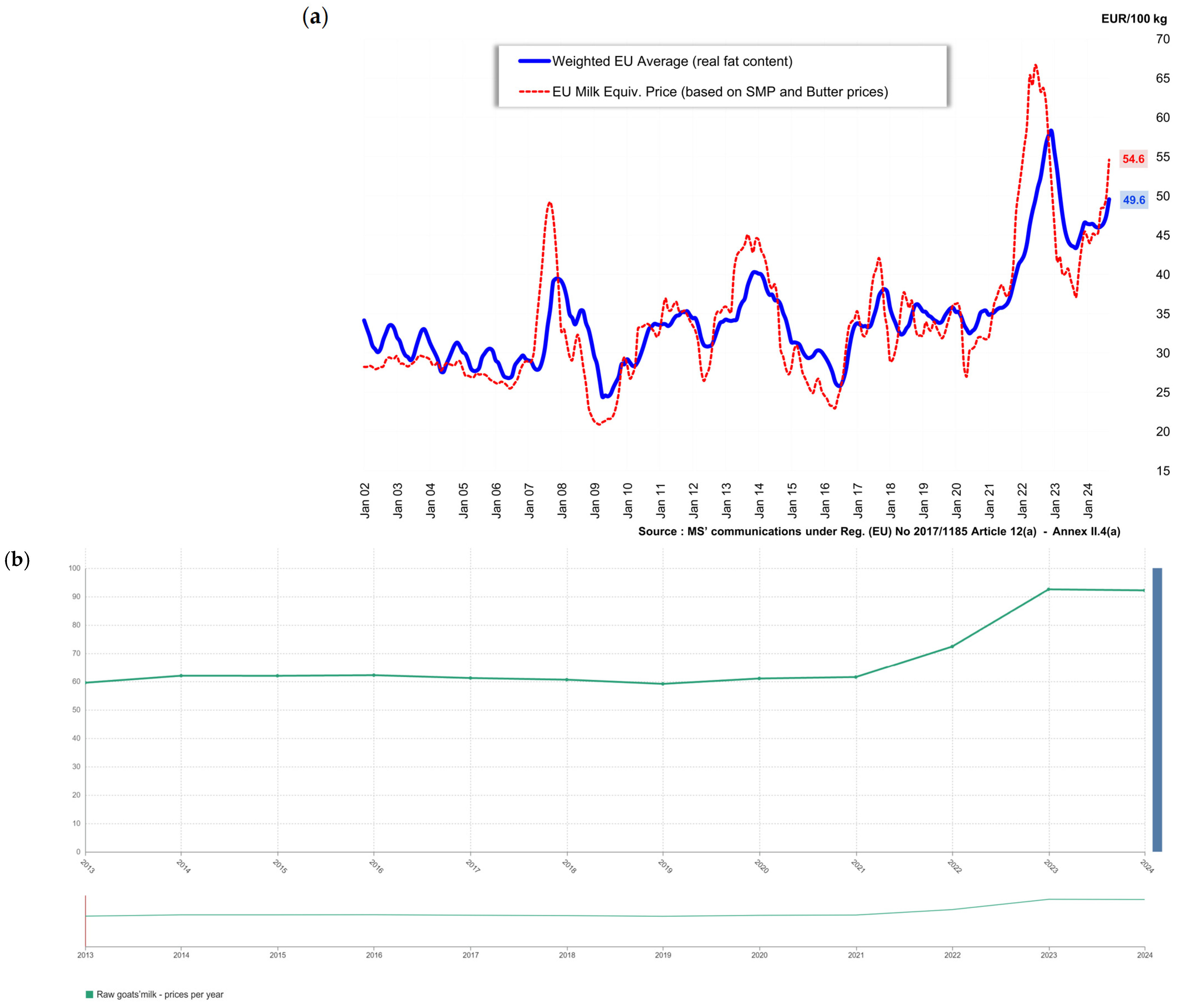
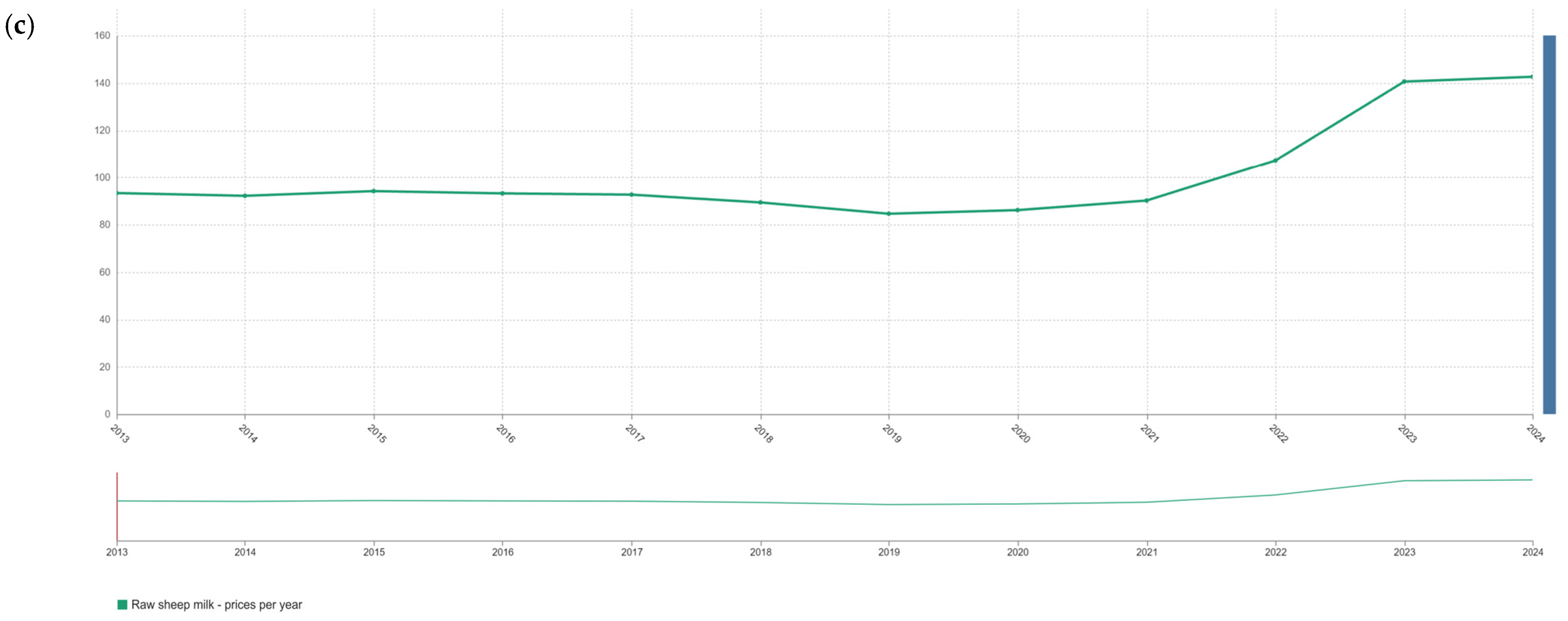
2. Economically Motivated Adulteration: Patterns and Implications in Milk from Different Species
3. Analytical Strategies for Milk Authentication: From Electrophoresis to Proteomics
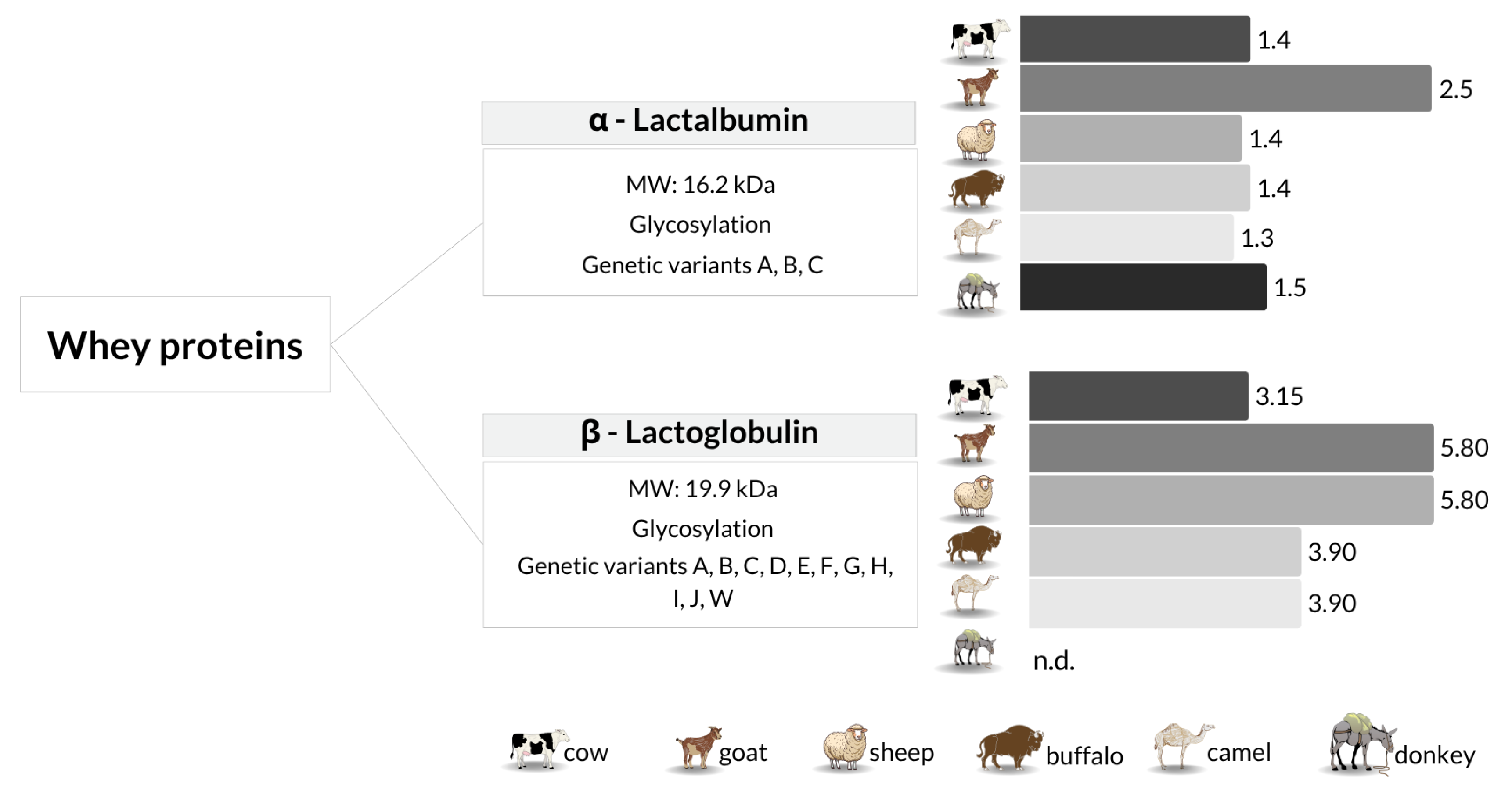
4. Protein Biomarkers for Detecting Milk Adulteration
Survey of Proteomic Studies on Milk Authentication Using Protein Biomarkers
5. Peptide Biomarkers in Milk Authentication
Application of Peptide Biomarkers in Milk Adulteration Studies
6. Chemometric Workflows for Interpreting Proteomic Data in Milk Authentication
6.1. Data Preprocessing Strategies in Proteomics-Based Milk Authentication
6.2. Feature Selection and Exploratory Analysis: Identifying Discriminative Biomarkers
6.3. Supervised Classification and Quantification Modeling
6.4. Model Validation and Performance Evaluation
6.5. Data Fusion Strategies
7. Conclusions and Future Directions
- Geographical Origin Profiling: Differentiating protein and peptide profiles based on geographical origin remains underexplored. While environmental factors including feed composition, climate, and farming practices are believed to influence protein and peptide profiles, systematic studies are lacking. Investigating variations of the same protein or peptide within the same animal species across different regions could identify subtle, consistent geographical markers. Such studies are essential for combating fraud and protecting products with geographical designations such as PDO and protected geographical indication (PGI) while promoting fair trade practices;
- Effect of Breed on Protein Profiles: The influence of animal breeds on milk protein and peptide profiles remains understudied. Detailed comparisons between breeds can provide valuable insights into intra-species variations, improving species differentiation and quality assessments. Such studies would establish comprehensive protein and peptide reference databases [135,136,137];
- Data Reliability and Sampling Protocols: Reliable biomarker development requires standardized methodologies for sample collection, storage, and analysis. Future studies should adopt longitudinal sampling approaches that account for lactation stages, seasonal variations, and the physiological states of animals [138,139,140]. Ensuring comprehensive and high-quality data will strengthen the reproducibility and validity of biomarker-based authentication methods;
- Integrative Multi-Omics and Data Fusion Strategies: The future of milk authentication lies in the integration of complementary omics technologies—such as proteomics, metabolomics, genomics, and isotopic profiling—within unified analytical frameworks [141]. While each omics platform provides distinct insights, their combination through structured data fusion approaches can substantially improve the sensitivity, specificity, and robustness of authenticity assessments. The integration of proteomics and metabolomics through structured data fusion has proven essential for advancing foodomics, particularly in complex matrices like milk. Complementing this, Blanchet and Smolinska (2016) proposed a two-step fusion framework where each omics dataset is first compressed to extract the most relevant variables and subsequently merged to allow joint analysis [131]. Together, these strategies enable a more holistic understanding of molecular mechanisms;
- Machine learning (ML): Recent advances in machine learning (ML)-assisted proteomics offer promising avenues to strengthen milk authentication workflows. Li et al. (2025) emphasized that deep learning models such as convolutional neural network (CNN)s and residual networks can enhance peptide identification and retention time prediction. However, challenges remain, including limited dataset diversity, insufficient multimodal integration, and the absence of standardized validation protocols. Future research should focus on developing robust, large-scale datasets and combining data-driven and mechanistic modeling approaches to improve accuracy, interpretability, and reproducibility [142].
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| AP-MALDI | Atmospheric Pressure–Matrix-Assisted Laser Desorption/Ionization |
| BSA | Bovine Serum Albumin |
| CE | Capillary Electrophoresis |
| cGMP | Casein Glycomacropeptide |
| CMP | Caseinomacropeptide |
| CN | Casein |
| DDA | Data-Dependent Acquisition |
| DIA | Data-Independent Acquisition |
| ELISA | Enzyme-Linked Immunosorbent Assay |
| EMA | Economically Motivated Adulteration |
| ESI-MS | Electrospray Ionization–Mass Spectrometry |
| EU | European Union |
| FAO | Food and Agriculture Organization of the United Nations |
| FDA | Food and Drug Administration |
| FTIR | Fourier-Transform Infrared Spectroscopy |
| GLM-Lasso | Generalized Linear Model with Lasso regularization |
| GMP | Glycomacropeptide |
| IEF | Isoelectric Focusing |
| IgG | Immunoglobulin G |
| LC-MS/MS | Liquid Chromatography–Mass Spectrometry |
| LDA | Linear Discriminant Analysis |
| LFIA | Lateral Flow Immunoassay |
| MALDI-TOF-MS | Matrix-Assisted Laser Desorption/Ionization–Time-of-Flight–Mass Spectrometry |
| MFGM | Milk Fat Globule Membrane |
| MID | Mid-Infrared Spectroscopy |
| MS | Mass Spectrometry |
| MMP | Mare Milk Powder |
| NIR | Near-Infrared Spectroscopy |
| nLC-MS/MS | Nano Liquid Chromatography–Mass Spectrometry |
| NLISA | Nanozyme-Linked Immunosorbent Assay |
| NMR | Nuclear Magnetic Resonance |
| OECD | Organization for Economic Co-operation and Development |
| P | Phosphorylation |
| PAGIF | Polyacrylamide Gel |
| PCA | Principal Component Analysis |
| PCR | Polymerase Chain Reaction |
| PDO | Protected Designation of Origin |
| PGI | Protected Geographical Indication |
| PMM | Pasteurized Mare Milk |
| PLS-DA | Partial Least Squares–Discriminant Analysis |
| PRISMA | Preferred Reporting Items for Systematic Reviews and Meta-Analyses |
| PTM | Post-Translational Modification |
| QCM | Quartz Crystal Microbalance |
| RSD | Relative Standard Deviation |
| SDS-PAGE | Sodium Dodecyl Sulfate Polyacrylamide Gel Electrophoresis |
| TSG | Traditional Specialty Guaranteed |
| UHPLC-ESI-QTOF-MS | Ultra-High Performance Liquid Chromatography–Electrospray Ionization–Quadrupole Time-of-Flight–Mass Spectrometry |
| UPLC-DAD | Ultra-Performance Liquid Chromatography–Diode Array Detector |
| α-La | Alpha-Lactalbumin |
| αs1-CN | Alpha-s1 Casein |
| αs2-CN | Alpha-s2 Casein |
| β-CN | Beta Casein |
| β-lg | Beta-Lactoglobulin |
| γ-CN | Gamma Casein |
| κ-CN | Kappa Casein |
References
- O’Mahony, J.A.; Fox, P.F. Milk: An Overview. In Milk Proteins; Elsevier: Amsterdam, The Netherlands, 2014; pp. 19–73. ISBN 978-0-12-405171-3. [Google Scholar]
- Smith, R. Regulation (EC) No 883/2004 of the European Parliament and of the Council of 29 April 2004. In Core EU Legislation; Macmillan Education UK: London, UK, 2015; pp. 288–318. ISBN 978-1-137-54501-5. [Google Scholar]
- Fox, P.F.; Guinee, T.P.; Cogan, T.M.; McSweeney, P.L.H. Fundamentals of Cheese Science; Springer US: Boston, MA, USA, 2017; ISBN 978-1-4899-7679-6. [Google Scholar]
- Goulding, D.A.; Fox, P.F.; O’Mahony, J.A. Milk Proteins: An Overview. In Milk Proteins; Elsevier: Amsterdam, The Netherlands, 2020; pp. 21–98. ISBN 978-0-12-815251-5. [Google Scholar]
- Stobiecka, M.; Król, J.; Brodziak, A.; Wajs, J. Characteristics of Milk from Different Species of Farm Animals with Special Emphasis on Health-Promoting Ingredients. Acta Sci. Pol. Zootech. 2022, 20, 85–96. [Google Scholar] [CrossRef]
- Warakaulle, S.; Mohamed, H.; Ranasinghe, M.; Shah, I.; Yanyang, X.; Chen, G.; Ayyash, M.M.; Vincent, D.; Kamal-Eldin, A. Advancement of Milk Protein Analysis: From Determination of Total Proteins to Their Identification and Quantification by Proteomic Approaches. J. Food Compos. Anal. 2024, 126, 105854. [Google Scholar] [CrossRef]
- Anagnostopoulos, A.K.; Katsafadou, A.I.; Pierros, V.; Kontopodis, E.; Fthenakis, G.C.; Arsenos, G.; Karkabounas, S.C.; Tzora, A.; Skoufos, I.; Tsangaris, G.T. Milk of Greek Sheep and Goat Breeds; Characterization by Means of Proteomics. J. Proteom. 2016, 147, 76–84. [Google Scholar] [CrossRef] [PubMed]
- Michailidou, S.; Tsangaris, G.; Fthenakis, G.C.; Tzora, A.; Skoufos, I.; Karkabounas, S.C.; Banos, G.; Argiriou, A.; Arsenos, G. Genomic Diversity and Population Structure of Three Autochthonous Greek Sheep Breeds Assessed with Genome-Wide DNA Arrays. Mol. Genet. Genom. 2018, 293, 753–768. [Google Scholar] [CrossRef]
- Dupont, D.; Croguennec, T.; Pochet, S. Milk Proteins—Analytical Methods. In Reference Module in Food Science; Elsevier: Amsterdam, The Netherlands, 2018; p. B9780081005965226164. ISBN 978-0-08-100596-5. [Google Scholar]
- Stastna, M. Advances in Separation and Identification of Biologically Important Milk Proteins and Peptides. Electrophoresis 2024, 45, 101–119. [Google Scholar] [CrossRef] [PubMed]
- Theodoridis, G.; Pechlivanis, A.; Thomaidis, N.; Spyros, A.; Georgiou, C.; Albanis, T.; Skoufos, I.; Kalogiannis, S.; Tsangaris, G.; Stasinakis, A.; et al. FoodOmicsGR_RI: A Consortium for Comprehensive Molecular Characterisation of Food Products. Metabolites 2021, 11, 74. [Google Scholar] [CrossRef]
- OECD. Food and Agriculture Organization of the United Nations. In OECD-FAO Agricultural Outlook 2024–2033; OECD-FAO Agricultural Outlook; OECD: Paris, France, 2024; ISBN 978-92-64-72259-0. [Google Scholar]
- Poonia, A.; Jha, A.; Sharma, R.; Singh, H.B.; Rai, A.K.; Sharma, N. Detection of Adulteration in Milk: A Review. Int. J. Dairy Tech. 2017, 70, 23–42. [Google Scholar] [CrossRef]
- Bernard, H.; Hazebrouck, S.; Gaiani, N.; Adel-Patient, K. Allergen Risk Assessment for Specific Allergy to Small Ruminant’s Milk: Development of Sensitive Immunoassays to Detect Goat’s and Sheep’s Milk Contaminations in Dairy Food Matrices. Front. Allergy 2021, 2, 733875. [Google Scholar] [CrossRef]
- Ferreira, M.M.; Marins-Gonçalves, L.; De Souza, D. An Integrative Review of Analytical Techniques Used in Food Authentication: A Detailed Description for Milk and Dairy Products. Food Chem. 2024, 457, 140206. [Google Scholar] [CrossRef]
- Tehrani, T.; Pont, L.; Benavente, F. Rapid Detection and Quantification of Milk Adulteration Using MALDI-MS Protein Profiling and Multivariate Calibration. J. Food Compos. Anal. 2024, 130, 106147. [Google Scholar] [CrossRef]
- Zhou, C.; Liu, L.; Chen, J.; Fu, Q.; Chen, Z.; Wang, J.; Sun, X.; Ai, L.; Xu, X.; Wang, J. Rapid Authentication of Characteristic Milk Powders by Recombinase Polymerase Amplification Assays. Food Chem. 2024, 443, 138540. [Google Scholar] [CrossRef] [PubMed]
- Ji, Z.; Zhang, J.; Deng, C.; Hu, Z.; Du, Q.; Guo, T.; Wang, J.; Fan, R.; Han, R.; Yang, Y. Identification of Mare Milk Adulteration with Cow Milk by Liquid Chromatography-High Resolution Mass Spectrometry Based on Proteomics and Metabolomics Approaches. Food Chem. 2023, 405, 134901. [Google Scholar] [CrossRef] [PubMed]
- Scopus Preview—Scopus—Welcome to Scopus. Available online: https://www.scopus.com/home.uri (accessed on 3 July 2025).
- Google Scholar. Available online: https://scholar.google.com/ (accessed on 3 July 2025).
- Gimonkar, S.; Van Fleet, E.; Boys, K.A. Dairy Product Fraud. In Food Fraud; Elsevier: Amsterdam, The Netherlands, 2021; pp. 249–279. ISBN 978-0-12-817242-1. [Google Scholar]
- Azad, T.; Ahmed, S. Common Milk Adulteration and Their Detection Techniques. Food Contam. 2016, 3, 22. [Google Scholar] [CrossRef]
- Patil, G.B.; Wani, S.P.; Bafna, P.S.; Bagul, V.S.; Kalaskar, M.G.; Mutha, R.E. Milk Adulteration: From Detection to Health Impact. Food Humanit. 2024, 3, 100339. [Google Scholar] [CrossRef]
- Nascimento, C.F.; Santos, P.M.; Pereira-Filho, E.R.; Rocha, F.R.P. Recent Advances on Determination of Milk Adulterants. Food Chem. 2017, 221, 1232–1244. [Google Scholar] [CrossRef]
- Mafra, I.; Honrado, M.; Amaral, J.S. Animal Species Authentication in Dairy Products. Foods 2022, 11, 1124. [Google Scholar] [CrossRef]
- European Commission. Joint Research Centre. Institute for Prospective Technological Studies. In Regional Economic Analysis of Milk Quota Reform in the EU; Publications Office: Luxembourg, 2009. [Google Scholar]
- European Commission. Joint Research Centre. Institute for Prospective Technological Studies. In Modelling and Analysis of the European Milk and Dairy Market; Publications Office: Luxembourg, 2009. [Google Scholar]
- Hemme, T.; Ndambi, A.; Schröer-Merker, E. Overview on Milk Prices and Production Costs World Wide; International Farm Comparison Network (IFCN): Kiel, Germany, 2012. [Google Scholar]
- Dubeuf, J.-P. Structural, Market and Organisational Conditions for Developing Goat Dairy Production Systems. Small Rumin. Res. 2005, 60, 67–74. [Google Scholar] [CrossRef]
- Sila, W.; Gachuiri, C.K.; Recha, J.W.; Audho, J.; Ojango, J.M.K. Adaptation and Returns from Improved Indigenous Small Ruminants in Climatically Challenged Smallholder Systems of Kenya. Sustainability 2021, 13, 9629. [Google Scholar] [CrossRef]
- Camillo, F.; Rota, A.; Biagini, L.; Tesi, M.; Fanelli, D.; Panzani, D. The Current Situation and Trend of Donkey Industry in Europe. J. Equine Vet. Sci. 2018, 65, 44–49. [Google Scholar] [CrossRef]
- Pizzano, R.; Salimei, E. Isoelectric Focusing and ELISA for Detecting Adulteration of Donkey Milk with Cow Milk. J. Agric. Food Chem. 2014, 62, 5853–5858. [Google Scholar] [CrossRef]
- European Commission. EU Evolutive Raw Milk Prices Evolution (up to September 2024); Directorate-General for Agriculture and Rural Development: Brussels, Belgium, 2024. [Google Scholar]
- Eurostat Selling Prices of Animal Products (Absolute Prices)—Annual Price (from 2000 Onwards). Available online: https://ec.europa.eu/eurostat/databrowser/product/page/APRI_AP_ANOUTA (accessed on 16 June 2025).
- Regulation—1151/2012—EN—EUR-Lex. Available online: https://eur-lex.europa.eu/eli/reg/2012/1151/oj/eng (accessed on 14 July 2025).
- European Commission 2023 Annual Report—Alert and Cooperation Network. Available online: https://ec.europa.eu/newsroom/sante/items/847722/en (accessed on 16 June 2025).
- European Union. Regulation (EU) No 1151/2012 of the European Parliament and of the Council of 21 November 2012 on Quality Schemes for Agricultural Products and Foodstuffs; European Union: Brussels, Belgium, 2012. [Google Scholar]
- Laaninen, T. Mandatory Origin-Labelling Schemes in Member States; European Parliamentary Research Service: Brussels, Belgium, 2018. [Google Scholar]
- FDA. Food Safety Modernization Act (FSMA); FDA: Silver Spring, MD, USA, 2011. [Google Scholar]
- European Union Regulation—1169/2011—EN—Food Information to Consumers Regulation—EUR-Lex. Available online: https://eur-lex.europa.eu/eli/reg/2011/1169/oj/eng (accessed on 16 June 2025).
- Moore, J.C.; Spink, J.; Lipp, M. Development and Application of a Database of Food Ingredient Fraud and Economically Motivated Adulteration from 1980 to 2010. J. Food Sci. 2012, 77, R118–R126. [Google Scholar] [CrossRef]
- Everstine, K.; Spink, J.; Kennedy, S. Economically Motivated Adulteration (EMA) of Food: Common Characteristics of EMA Incidents. J. Food Prot. 2013, 76, 723–735. [Google Scholar] [CrossRef]
- Robson, K.; Dean, M.; Haughey, S.; Elliott, C. A Comprehensive Review of Food Fraud Terminologies and Food Fraud Mitigation Guides. Food Control 2021, 120, 107516. [Google Scholar] [CrossRef]
- Masci, M.; Zoani, C.; Nevigato, T.; Turrini, A.; Jasionowska, R.; Caproni, R.; Ratini, P. Authenticity Assessment of Dairy Products by Capillary Electrophoresis. Electrophoresis 2022, 43, 340–354. [Google Scholar] [CrossRef]
- Sharma, N.; Sharma, R.; Rajput, Y.S.; Mann, B.; Singh, R.; Gandhi, K. Separation Methods for Milk Proteins on Polyacrylamide Gel Electrophoresis: Critical Analysis and Options for Better Resolution. Int. Dairy J. 2021, 114, 104920. [Google Scholar] [CrossRef]
- Büyükköroğlu, G.; Dora, D.D.; Özdemir, F.; Hızel, C. Techniques for Protein Analysis. In Omics Technologies and Bio-Engineering; Elsevier: Amsterdam, The Netherlands, 2018; pp. 317–351. ISBN 978-0-12-804659-3. [Google Scholar]
- Richter, W.; Krause, I.; Graf, C.; Sperrer, I.; Schwarzer, C.; Klostermeyer, H. An Indirect Competitive ELISA forthe Detection of Cows’ Milkand Caseinate in Goats’ andEwes’ Milk and Cheese Using Polyclonal Antibodies against Bovine γ-Caseins. Z. Leb. Forsch. A 1997, 204, 21–26. [Google Scholar] [CrossRef]
- Anguita, G.; Martín, R.; García, T.; Morales, P.; Haza, A.I.; González, I.; Sanz, B.; Hernández, P.E. A Competitive Enzyme-Linked Immunosorbent Assay for Detection of Bovine Milk in Ovine and Caprine Milk and Cheese Using a Monoclonal Antibody against Bovine β-Casein. J. Food Prot. 1997, 60, 64–66. [Google Scholar] [CrossRef]
- Mayer, H.K.; Heidler, D.; Rockenbauer, C. Determination of the Percentages of Cows’, Ewes’ and Goats’ Milk in Cheese by Isoelectric Focusing and Cation-Exchange HPLC of γ- and Para-κ-Caseins. Int. Dairy J. 1997, 7, 619–628. [Google Scholar] [CrossRef]
- Nagraik, R.; Sharma, A.; Kumar, D.; Chawla, P.; Kumar, A.P. Milk Adulterant Detection: Conventional and Biosensor Based Approaches: A Review. Sens. Bio-Sens. Res. 2021, 33, 100433. [Google Scholar] [CrossRef]
- Perez-Riverol, Y.; Alpi, E.; Wang, R.; Hermjakob, H.; Vizcaíno, J.A. Making Proteomics Data Accessible and Reusable: Current State of Proteomics Databases and Repositories. Proteomics 2015, 15, 930–950. [Google Scholar] [CrossRef]
- Mascot Search Engine|Protein Identification Software for Mass Spec Data. Available online: https://www.matrixscience.com/ (accessed on 3 July 2025).
- PEAKS Studio. Available online: https://www.bioinfor.com/peaks-studio/ (accessed on 3 July 2025).
- UniProt. Available online: https://www.uniprot.org (accessed on 3 July 2025).
- Vande Moortele, T.; Devlaminck, B.; Van De Vyver, S.; Van Den Bossche, T.; Martens, L.; Dawyndt, P.; Mesuere, B.; Verschaffelt, P. Unipept in 2024: Expanding Metaproteomics Analysis with Support for Missed Cleavages, Semi-Tryptic and Non-Tryptic Peptides. J. Proteome Res. 2025, 24, 949–954. [Google Scholar] [CrossRef]
- BLAST: Basic Local Alignment Search Tool. Available online: https://blast.ncbi.nlm.nih.gov/Blast.cgi (accessed on 3 July 2025).
- HMMER. Available online: http://hmmer.org/ (accessed on 3 July 2025).
- Perez-Riverol, Y.; Bandla, C.; Kundu, D.J.; Kamatchinathan, S.; Bai, J.; Hewapathirana, S.; John, N.S.; Prakash, A.; Walzer, M.; Wang, S.; et al. The PRIDE Database at 20 Years: 2025 Update. Nucleic Acids Res. 2025, 53, D543–D553. [Google Scholar] [CrossRef]
- Deutsch, E.W.; Bandeira, N.; Perez-Riverol, Y.; Sharma, V.; Carver, J.J.; Mendoza, L.; Kundu, D.J.; Wang, S.; Bandla, C.; Kamatchinathan, S.; et al. The ProteomeXchange Consortium at 10 Years: 2023 Update. Nucleic Acids Res. 2023, 51, D1539–D1548. [Google Scholar] [CrossRef]
- PeptideAtlas. Available online: https://peptideatlas.org/ (accessed on 3 July 2025).
- Kamal, M.; Karoui, R. Analytical Methods Coupled with Chemometric Tools for Determining the Authenticity and Detecting the Adulteration of Dairy Products: A Review. Trends Food Sci. Technol. 2015, 46, 27–48. [Google Scholar] [CrossRef]
- Hebling E Tavares, J.P.; Da Silva Medeiros, M.L.; Barbin, D.F. Near-infrared Techniques for Fraud Detection in Dairy Products: A Review. J. Food Sci. 2022, 87, 1943–1960. [Google Scholar] [CrossRef]
- Chaudhary, V.; Kajla, P.; Dewan, A.; Pandiselvam, R.; Socol, C.T.; Maerescu, C.M. Spectroscopic Techniques for Authentication of Animal Origin Foods. Front. Nutr. 2022, 9, 979205. [Google Scholar] [CrossRef]
- Angeletti, R.; Gioacchini, A.M.; Seraglia, R.; Piro, R.; Traldi, P. The Potential of Matrix-Assisted Laser Desorption/Ionization Mass Spectrometry in the Quality Control of Water Buffalo Mozzarella Cheese. J. Mass. Spectrom. 1998, 33, 525–531. [Google Scholar] [CrossRef]
- Karamoutsios, A.; Oikonomou, E.D.; Voidarou, C. (Chrysa); Hatzizisis, L.; Fotou, K.; Nikolaou, K.; Gouva, E.; Gkiza, E.; Giannakeas, N.; Skoufos, I.; et al. Innovations in Proteomic Technologies and Artificial Neural Networks: Unlocking Milk Origin Identification. BioTech 2025, 14, 33. [Google Scholar] [CrossRef]
- Zhang, H.; Abdallah, M.F.; Zhang, J.; Yu, Y.; Zhao, Q.; Tang, C.; Qin, Y.; Zhang, J. Comprehensive Quantitation of Multi-Signature Peptides Originating from Casein for the Discrimination of Milk from Eight Different Animal Species Using LC-HRMS with Stable Isotope Labeled Peptides. Food Chem. 2022, 390, 133126. [Google Scholar] [CrossRef]
- Kumar, P.; Rani, A.; Singh, S.; Kumar, A. Recent Advances on DNA and Omics-based Technology in Food Testing and Authentication: A Review. J. Food Saf. 2022, 42, e12986. [Google Scholar] [CrossRef]
- Plath, A.; Krause, I.; Einspanier, R. Species Identification in Dairy Products by Three Different DNA-Based Techniques. Z. Für Leb. Und-Forsch. A 1997, 205, 437–441. [Google Scholar] [CrossRef]
- Nehra, M.; Lettieri, M.; Dilbaghi, N.; Kumar, S.; Marrazza, G. Nano-Biosensing Platforms for Detection of Cow’s Milk Allergens: An Overview. Sensors 2019, 20, 32. [Google Scholar] [CrossRef]
- Xu, S.; Zhao, C.; Deng, X.; Zhang, R.; Qu, L.; Wang, M.; Ren, S.; Wu, H.; Yue, Z.; Niu, B. Determining the Geographical Origin of Milk by Multivariate Analysis Based on Stable Isotope Ratios, Elements and Fatty Acids. Anal. Methods 2021, 13, 2537–2548. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Wang, Y.; He, C.; Wu, B.; Zhang, T.; Sun, J. Several Feature Extraction Methods Combined with Near-Infrared Spectroscopy for Identifying the Geographical Origins of Milk. Foods 2024, 13, 1783. [Google Scholar] [CrossRef]
- Cardin, M.; Cardazzo, B.; Mounier, J.; Novelli, E.; Coton, M.; Coton, E. Authenticity and Typicity of Traditional Cheeses: A Review on Geographical Origin Authentication Methods. Foods 2022, 11, 3379. [Google Scholar] [CrossRef]
- McSweeney, P.L.H. Advanced Dairy Chemistry: Volume 1A: Proteins: Basic Aspects, 4th ed.; McSweeney, P.L.H., Fox, P.F., Eds.; Springer US: Boston, MA, USA, 2013; ISBN 978-1-4614-4713-9. [Google Scholar]
- Das, A.; Giri, K.; Behera, R.N.; Maity, S.; Ambatipudi, K. BoMiProt 2.0: An Update of the Bovine Milk Protein Database. J. Proteom. 2022, 267, 104696. [Google Scholar] [CrossRef]
- Delosière, M.; Pires, J.A.A.; Bernard, L.; Cassar-Malek, I.; Bonnet, M. Dataset Reporting 4654 Cow Milk Proteins Listed According to Lactation Stages and Milk Fractions. Data Brief. 2020, 29, 105105. [Google Scholar] [CrossRef]
- Nielsen, S.D.; Beverly, R.L.; Qu, Y.; Dallas, D.C. Milk Bioactive Peptide Database: A Comprehensive Database of Milk Protein-Derived Bioactive Peptides and Novel Visualization. Food Chem. 2017, 232, 673–682. [Google Scholar] [CrossRef]
- Selvaggi, M.; Laudadio, V.; Dario, C.; Tufarelli, V. Major Proteins in Goat Milk: An Updated Overview on Genetic Variability. Mol. Biol. Rep. 2014, 41, 1035–1048. [Google Scholar] [CrossRef]
- Martin, P.; Szymanowska, M.; Zwierzchowski, L.; Leroux, C. The Impact of Genetic Polymorphisms on the Protein Composition of Ruminant Milks. Reprod. Nutr. Dev. 2002, 42, 433–459. [Google Scholar] [CrossRef]
- Selvaggi, M.; Laudadio, V.; Dario, C.; Tufarelli, V. Investigating the Genetic Polymorphism of Sheep Milk Proteins: A Useful Tool for Dairy Production. J. Sci. Food Agric. 2014, 94, 3090–3099. [Google Scholar] [CrossRef]
- Caroli, A.M.; Chessa, S.; Erhardt, G.J. Invited Review: Milk Protein Polymorphisms in Cattle: Effect on Animal Breeding and Human Nutrition. J. Dairy Sci. 2009, 92, 5335–5352. [Google Scholar] [CrossRef] [PubMed]
- Fox, P.F.; Uniacke-Lowe, T.; McSweeney, P.L.H.; O’Mahony, J.A. Dairy Chemistry and Biochemistry; Springer International Publishing: Cham, Switzerland, 2015; ISBN 978-3-319-14891-5. [Google Scholar]
- Runthala, A.; Mbye, M.; Ayyash, M.; Xu, Y.; Kamal-Eldin, A. Caseins: Versatility of Their Micellar Organization in Relation to the Functional and Nutritional Properties of Milk. Molecules 2023, 28, 2023. [Google Scholar] [CrossRef] [PubMed]
- Bhat, M.Y.; Dar, T.A.; Singh, L.R. Casein Proteins: Structural and Functional Aspects. In Milk Proteins—From Structure to Biological Properties and Health Aspects; Gigli, I., Ed.; InTech: Houston, TX, USA, 2016; ISBN 978-953-51-2536-5. [Google Scholar]
- Jabeen, M.; Anwar, M.; Fatima, W.; Saleem, A.; Rehman, K.; Masood, M.; Iqbal, N.; Anjum, S.; Madeeha Zafar, A.; Aslam, N. Quantitative Estimation of Casein in Different Milk Samples. SJAC 2020, 8, 107. [Google Scholar] [CrossRef]
- Rafiq, S.; Huma, N.; Pasha, I.; Sameen, A.; Mukhtar, O.; Khan, M.I. Chemical Composition, Nitrogen Fractions and Amino Acids Profile of Milk from Different Animal Species. Asian Australas. J. Anim. Sci. 2015, 29, 1022–1028. [Google Scholar] [CrossRef]
- Pan, F.; Li, J.; Zhao, L.; Tuersuntuoheti, T.; Mehmood, A.; Zhou, N.; Hao, S.; Wang, C.; Guo, Y.; Lin, W. A Molecular Docking and Molecular Dynamics Simulation Study on the Interaction between Cyanidin—3-O-glucoside and Major Proteins in Cow’s Milk. J. Food Biochem. 2021, 45. [Google Scholar] [CrossRef]
- Swaisgood, H.E. Review and Update of Casein Chemistry. J. Dairy Sci. 1993, 76, 3054–3061. [Google Scholar] [CrossRef]
- Simpson, K.J.; Nicholas, K.R. The Comparative Biology of Whey Proteins. J. Mammary Gland. Biol. Neoplasia 2003, 7, 313–326. [Google Scholar] [CrossRef]
- Hernández-Ledesma, B.; Recio, I.; Amigo, L. β-Lactoglobulin as Source of Bioactive Peptides. Amino Acids 2008, 35, 257–265. [Google Scholar] [CrossRef]
- Zhang, G.; Annan, R.S.; Carr, S.A.; Neubert, T.A. Overview of Peptide and Protein Analysis by Mass Spectrometry. CP Protein Sci. 2010, 62. [Google Scholar] [CrossRef]
- Sassi, M.; Arena, S.; Scaloni, A. MALDI-TOF-MS Platform for Integrated Proteomic and Peptidomic Profiling of Milk Samples Allows Rapid Detection of Food Adulterations. J. Agric. Food Chem. 2015, 63, 6157–6171. [Google Scholar] [CrossRef]
- Ji, Z.; Zhang, J.; Deng, C.; Guo, T.; Han, R.; Yang, Y.; Zang, C.; Chen, Y. Identification of Pasteurized Mare Milk and Powder Adulteration with Bovine Milk Using Quantitative Proteomics and Metabolomics Approaches. Food Chem. X 2024, 22, 101265. [Google Scholar] [CrossRef] [PubMed]
- Fan, R.; Xie, S.; Wang, S.; Yu, Z.; Sun, X.; Du, Q.; Yang, Y.; Han, R. Identification Markers of Goat Milk Adulterated with Bovine Milk Based on Proteomics and Metabolomics. Food Chem. X 2023, 17, 100601. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Wang, J.; Li, M.; Yang, Y.; Wang, Z.; Miao, J.; Zhao, Z.; Yang, J. Detection of the Adulteration of Camel Milk Powder with Cow Milk by Ultra-High Performance Liquid Chromatography (UPLC). Int. Dairy J. 2021, 121, 105117. [Google Scholar] [CrossRef]
- Santos, A.S.D.O.D.; Pereira, H.P.; Fogaça, G.N.; Meurer, V.M.; Furtado, M.A.M.; Borges, C.A.V.; Weller, M.M.D.C.A.; Martins, M.F. Separation and Quantification of Milk Proteins with the Addition of Cheese Whey by Lab-on-a-Chip. Pesq. Agropec. Bras. 2023, 58, e03099. [Google Scholar] [CrossRef]
- Trimboli, F.; Morittu, V.M.; Cicino, C.; Palmieri, C.; Britti, D. Rapid Capillary Electrophoresis Approach for the Quantification of Ewe Milk Adulteration with Cow Milk. J. Chromatogr. A 2017, 1519, 131–136. [Google Scholar] [CrossRef]
- Zhang, L.; Hou, J.; Zhou, H.; Nawaz, M.A.H.; Li, Y.; Huang, H.; Yu, C. Identification of Milk Adulteration by a Sensor Array Based on Cationic Polymer Induced Aggregation of a Perylene Probe. Food Chem. 2021, 343, 128492. [Google Scholar] [CrossRef]
- Yasmin, I.; Iqbal, R.; Liaqat, A.; Khan, W.A.; Nadeem, M.; Iqbal, A.; Chughtai, M.F.J.; Rehman, S.J.U.; Tehseen, S.; Mehmood, T.; et al. Characterization and Comparative Evaluation of Milk Protein Variants from Pakistani Dairy Breeds. Food Sci. Anim. Resour. 2020, 40, 689–698. [Google Scholar] [CrossRef]
- Ren, Q.R.; Zhang, H.; Guo, H.Y.; Jiang, L.; Tian, M.; Ren, F.Z. Detection of Cow Milk Adulteration in Yak Milk by ELISA. J. Dairy Sci. 2014, 97, 6000–6006. [Google Scholar] [CrossRef]
- Groves, M.L. Some Minor Components of Casein and Other Phosphoproteins in Milk. A Review. J. Dairy Sci. 1969, 52, 1155–1165. [Google Scholar] [CrossRef]
- Regulation—273/2008—EN—EUR-Lex. Available online: https://eur-lex.europa.eu/eli/reg/2008/273/oj/eng (accessed on 16 June 2025).
- Angelopoulou, Μ.; Botsialas, A.; Salapatas, A.; Petrou, P.S.; Haasnoot, W.; Makarona, E.; Jobst, G.; Goustouridis, D.; Siafaka-Kapadai, A.; Raptis, I.; et al. Assessment of Goat Milk Adulteration with a Label-Free Monolithically Integrated Optoelectronic Biosensor. Anal. Bioanal. Chem. 2015, 407, 3995–4004. [Google Scholar] [CrossRef] [PubMed]
- Sakti, S.P.; Chabibah, N.; Ayu, S.P.; Padaga, M.C.; Aulanni’am, A. Development of QCM Biosensor with Specific Cow Milk Protein Antibody for Candidate Milk Adulteration Detection. J. Sens. 2016, 2016, 1807647. [Google Scholar] [CrossRef]
- Trimboli, F.; Costanzo, N.; Lopreiato, V.; Ceniti, C.; Morittu, V.M.; Spina, A.; Britti, D. Detection of Buffalo Milk Adulteration with Cow Milk by Capillary Electrophoresis Analysis. J. Dairy Sci. 2019, 102, 5962–5970. [Google Scholar] [CrossRef] [PubMed]
- Seddaoui, N.; Attaallah, R.; Amine, A. Development of an Optical Immunoassay Based on Peroxidase-Mimicking Prussian Blue Nanoparticles and a Label-Free Electrochemical Immunosensor for Accurate and Sensitive Quantification of Milk Species Adulteration. Microchim. Acta 2022, 189, 209. [Google Scholar] [CrossRef]
- Galan-Malo, P.; Mendiara, I.; Razquin, P.; Mata, L. Validation of a Rapid Lateral Flow Method for the Detection of Cows’ Milk in Water Buffalo, Sheep or Goat Milk. Food Addit. Contam. Part. A 2018, 35, 609–614. [Google Scholar] [CrossRef]
- Ruiz-Valdepeñas Montiel, V.; Povedano, E.; Benedé, S.; Mata, L.; Galán-Malo, P.; Gamella, M.; Reviejo, A.J.; Campuzano, S.; Pingarrón, J.M. Disposable Amperometric Immunosensor for the Detection of Adulteration in Milk through Single or Multiplexed Determination of Bovine, Ovine, or Caprine Immunoglobulins G. Anal. Chem. 2019, 91, 11266–11274. [Google Scholar] [CrossRef]
- Tsakali, E.; Chatzilazarou, A.; Houhoula, D.; Koulouris, S.; Tsaknis, J.; Van Impe, J. A Rapid HPLC Method for the Determination of Lactoferrin in Milk of Various Species. J. Dairy Res. 2019, 86, 238–241. [Google Scholar] [CrossRef]
- Zhang, L.-G.; Zhang, X.; Ni, L.-J.; Xue, Z.-B.; Gu, X.; Huang, S.-X. Rapid Identification of Adulterated Cow Milk by Non-Linear Pattern Recognition Methods Based on near Infrared Spectroscopy. Food Chem. 2014, 145, 342–348. [Google Scholar] [CrossRef]
- Vera-Bravo, R.; Hernández, A.V.; Peña, S.; Alarcón, C.; Loaiza, A.E.; Celis, C.A. Cheese Whey Milk Adulteration Determination Using Casein Glycomacropeptide as an Indicator by HPLC. Foods 2022, 11, 3201. [Google Scholar] [CrossRef]
- Motta, T.M.C.; Hoff, R.B.; Barreto, F.; Andrade, R.B.S.; Lorenzini, D.M.; Meneghini, L.Z.; Pizzolato, T.M. Detection and Confirmation of Milk Adulteration with Cheese Whey Using Proteomic-like Sample Preparation and Liquid Chromatography–Electrospray–Tandem Mass Spectrometry Analysis. Talanta 2014, 120, 498–505. [Google Scholar] [CrossRef]
- Zhang, J.; Wei, L.; Miao, J.; Yu, Y.; Yu, N.; Hu, Q.; Chen, H.; Chen, Y. Authenticity Identification of Animal Species in Characteristic Milk by Integration of Shotgun Proteomics and Scheduled Multiple Reaction Monitoring (MRM) Based on Tandem Mass Spectrometry. Food Chem. 2024, 436, 137736. [Google Scholar] [CrossRef]
- Russo, R.; Rega, C.; Chambery, A. Rapid Detection of Water Buffalo Ricotta Adulteration or Contamination by Matrix-assisted Laser Desorption/Ionisation Time-of-flight Mass Spectrometry. Rapid Comm. Mass. Spectrom. 2016, 30, 497–503. [Google Scholar] [CrossRef]
- Hao, X.; Fu, L.; Shao, L.; Chen, Q.; Dorus, B.; Cao, X.; Fang, F. Quantification of Major Milk Proteins Using Ultra-Performance Liquid Chromatography Tandem Triple Quadrupole Mass Spectrometry and Its Application in Milk Authenticity Analysis. Food Control 2022, 131, 108455. [Google Scholar] [CrossRef]
- Ke, X.; Zhang, J.; Lai, S.; Chen, Q.; Zhang, Y.; Jiang, Y.; Mo, W.; Ren, Y. Quantitative Analysis of Cow Whole Milk and Whey Powder Adulteration Percentage in Goat and Sheep Milk Products by Isotopic Dilution-Ultra-High Performance Liquid Chromatography-Tandem Mass Spectrometry. Anal. Bioanal. Chem. 2017, 409, 213–224. [Google Scholar] [CrossRef]
- Nardiello, D.; Natale, A.; Palermo, C.; Quinto, M.; Centonze, D. Milk Authenticity by Ion-Trap Proteomics Following Multi-Enzyme Digestion. Food Chem. 2018, 244, 317–323. [Google Scholar] [CrossRef]
- Lu, Y.; Dai, J.; Zhang, S.; Qiao, J.; Lian, H.; Mao, L. Identification of Characteristic Peptides of Casein in Cow Milk Based on MALDI-TOF MS for Direct Adulteration Detection of Goat Milk. Foods 2023, 12, 1519. [Google Scholar] [CrossRef]
- Hassanin, A.A.; Osman, A.; Atallah, O.O.; El-Saadony, M.T.; Abdelnour, S.A.; Taha, H.S.A.; Awad, M.F.; Elkashef, H.; Ahmed, A.E.; Abd El-Rahim, I.; et al. Phylogenetic Comparative Analysis: Chemical and Biological Features of Caseins (Alpha-S-1, Alpha-S-2, Beta- and Kappa-) in Domestic Dairy Animals. Front. Vet. Sci. 2022, 9, 952319. [Google Scholar] [CrossRef]
- Piras, C.; Hale, O.J.; Reynolds, C.K.; Jones, A.K.; Taylor, N.; Morris, M.; Cramer, R. Speciation and Milk Adulteration Analysis by Rapid Ambient Liquid MALDI Mass Spectrometry Profiling Using Machine Learning. Sci. Rep. 2021, 11, 3305. [Google Scholar] [CrossRef]
- Smit, S.; Hoefsloot, H.C.J.; Smilde, A.K. Statistical Data Processing in Clinical Proteomics. J. Chromatogr. B 2008, 866, 77–88. [Google Scholar] [CrossRef]
- Musfiroh, I.; Maritha, V.; Harlina, P.W.; Muchtaridi, M.; Bakar, N.K.A.; Rohman, A.; Dachriyanus, D.; Ikram, N.K.K.; Windarsih, A. Chemometrics Applications in Omics Studies (Metabolomics, Lipidomics, and Proteomics) for Halal Authentication in Food and Pharmaceutical Products. Appl. Food Res. 2025, 5, 100770. [Google Scholar] [CrossRef]
- Suppers, A.; Gool, A.J.V.; Wessels, H.J.C.T. Integrated Chemometrics and Statistics to Drive Successful Proteomics Biomarker Discovery. Proteomes 2018, 6, 20. [Google Scholar] [CrossRef]
- Rysova, L.; Cejnar, P.; Hanus, O.; Legarova, V.; Havlik, J.; Nejeschlebova, H.; Nemeckova, I.; Jedelska, R.; Bozik, M. Use of MALDI-TOF MS Technology to Evaluate Adulteration of Small Ruminant Milk with Raw Bovine Milk. J. Dairy Sci. 2022, 105, 4882–4894. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.; He, Z.; Yu, W. Comparison of Public Peak Detection Algorithms for MALDI Mass Spectrometry Data Analysis. BMC Bioinform. 2009, 10, 4. [Google Scholar] [CrossRef] [PubMed]
- Di Girolamo, F.; Masotti, A.; Salvatori, G.; Scapaticci, M.; Muraca, M.; Putignani, L. A Sensitive and Effective Proteomic Approach to Identify She-Donkey’s and Goat’s Milk Adulterations by MALDI-TOF MS Fingerprinting. IJMS 2014, 15, 13697–13719. [Google Scholar] [CrossRef]
- Yang, J.; Zheng, N.; Soyeurt, H.; Yang, Y.; Wang, J. Detection of Plant Protein in Adulterated Milk Using Nontargeted Nano-high-performance Liquid Chromatography–Tandem Mass Spectroscopy Combined with Principal Component Analysis. Food Sci. Nutr. 2019, 7, 56–64. [Google Scholar] [CrossRef] [PubMed]
- Chi, S.-X.; Liu, B.-H.; Zhang, B.; Wang, B.-R.; Zhou, J.; Li, L.; Zhang, Y.-H.; Mu, Z. Development of an ELISA Method to Determine Adulterated Cow Milk in Camel Milk. Int. Dairy J. 2024, 155, 105953. [Google Scholar] [CrossRef]
- Du, L.; Lu, W.; Zhang, Y.; Gao, B.; Yu, L. Detection of Milk Powder in Liquid Whole Milk Using Hydrolyzed Peptide and Intact Protein Mass Spectral Fingerprints Coupled with Data Fusion Technologies. Food Sci. Nutr. 2020, 8, 1471–1479. [Google Scholar] [CrossRef]
- Liu, Y.; Hu, X.; Voglmeir, J.; Liu, L. N-Glycan Profiles as a Tool in Qualitative and Quantitative Analysis of Goat Milk Adulteration. Food Chem. 2023, 423, 136116. [Google Scholar] [CrossRef]
- Piras, C.; Ceniti, C.; Hartmane, E.; Costanzo, N.; Morittu, V.M.; Roncada, P.; Britti, D.; Cramer, R. Rapid Liquid AP-MALDI MS Profiling of Lipids and Proteins from Goat and Sheep Milk for Speciation and Colostrum Analysis. Proteomes 2020, 8, 20. [Google Scholar] [CrossRef]
- Blanchet, L.; Smolinska, A. Data Fusion in Metabolomics and Proteomics for Biomarker Discovery. In Statistical Analysis in Proteomics; Jung, K., Ed.; Methods in Molecular Biology; Springer: New York, NY, USA, 2016; Volume 1362, pp. 209–223. ISBN 978-1-4939-3105-7. [Google Scholar]
- Borad, S.G.; Kumar, A.; Singh, A.K. Effect of Processing on Nutritive Values of Milk Protein. Crit. Rev. Food Sci. Nutr. 2017, 57, 3690–3702. [Google Scholar] [CrossRef]
- Jensen, H.B.; Holland, J.W.; Poulsen, N.A.; Larsen, L.B. Milk Protein Genetic Variants and Isoforms Identified in Bovine Milk Representing Extremes in Coagulation Properties. J. Dairy Sci. 2012, 95, 2891–2903. [Google Scholar] [CrossRef] [PubMed]
- Kilara, A.; Panyam, D. Peptides From Milk Proteins and Their Properties. Crit. Rev. Food Sci. Nutr. 2003, 43, 607–633. [Google Scholar] [CrossRef]
- Amalfitano, N.; Stocco, G.; Maurmayr, A.; Pegolo, S.; Cecchinato, A.; Bittante, G. Quantitative and Qualitative Detailed Milk Protein Profiles of 6 Cattle Breeds: Sources of Variation and Contribution of Protein Genetic Variants. J. Dairy Sci. 2020, 103, 11190–11208. [Google Scholar] [CrossRef]
- Gustavsson, F.; Buitenhuis, A.J.; Johansson, M.; Bertelsen, H.P.; Glantz, M.; Poulsen, N.A.; Lindmark Månsson, H.; Stålhammar, H.; Larsen, L.B.; Bendixen, C.; et al. Effects of Breed and Casein Genetic Variants on Protein Profile in Milk from Swedish Red, Danish Holstein, and Danish Jersey Cows. J. Dairy Sci. 2014, 97, 3866–3877. [Google Scholar] [CrossRef]
- Tacoma, R.; Fields, J.; Ebenstein, D.B.; Lam, Y.-W.; Greenwood, S.L. Characterization of the Bovine Milk Proteome in Early-Lactation Holstein and Jersey Breeds of Dairy Cows. J. Proteom. 2016, 130, 200–210. [Google Scholar] [CrossRef]
- Li, S.; Ye, A.; Singh, H. Seasonal Variations in Composition, Properties, and Heat-Induced Changes in Bovine Milk in a Seasonal Calving System. J. Dairy Sci. 2019, 102, 7747–7759. [Google Scholar] [CrossRef]
- Bernabucci, U.; Basiricò, L.; Morera, P.; Dipasquale, D.; Vitali, A.; Piccioli Cappelli, F.; Calamari, L. Effect of Summer Season on Milk Protein Fractions in Holstein Cows. J. Dairy Sci. 2015, 98, 1815–1827. [Google Scholar] [CrossRef]
- Morton, J.M.; Auldist, M.J.; Douglas, M.L.; Macmillan, K.L. Associations between Milk Protein Concentration at Various Stages of Lactation and Reproductive Performance in Dairy Cows. J. Dairy Sci. 2016, 99, 10044–10056. [Google Scholar] [CrossRef]
- Shi, J.; Liu, Y.; Xu, Y.-J. MS Based Foodomics: An Edge Tool Integrated Metabolomics and Proteomics for Food Science. Food Chem. 2024, 446, 138852. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Mo, H.; Song, Y.-C.; Chen, G.; Wu, C.-E.; Zhu, F.-Y. The Integration of Machine Learning into Proteomics Advances Food Authentication and Adulteration Control. Trends Food Sci. Technol. 2025, 161, 105029. [Google Scholar] [CrossRef]


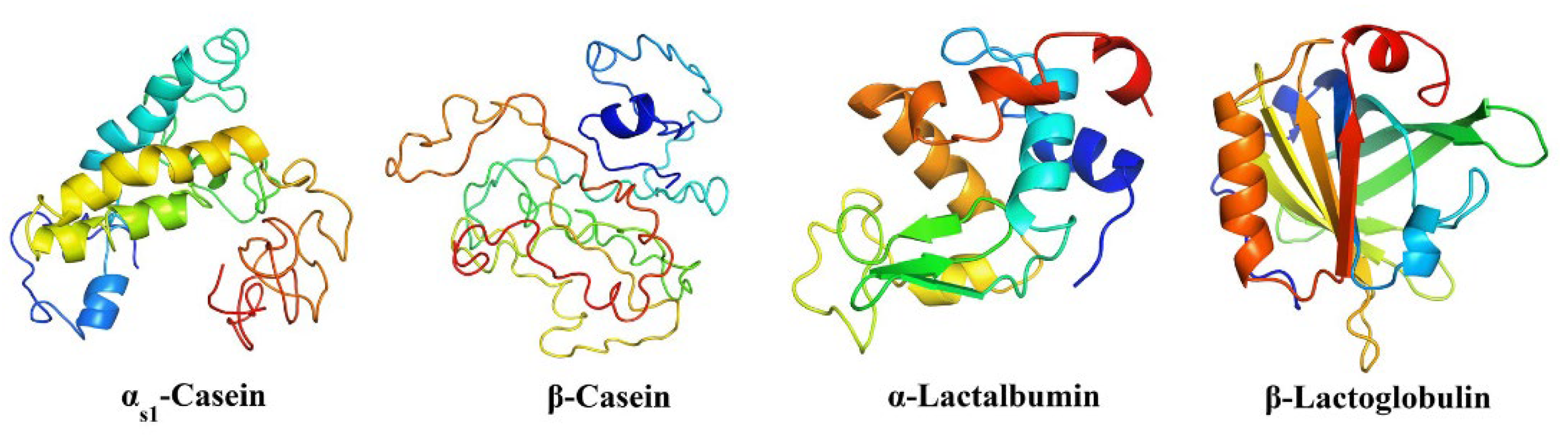


| Technique | Advantages | Limitations |
|---|---|---|
| Electrophoretic techniques (SDS-PAGE/IEF/2D-PAGE) [44,45] | Cost effective; useful for initial protein separation; can resolve isoforms | Low specificity; overlapping bands; limited in processed samples; requires standards for IEF |
| Immunoassays (e.g., ELISA) [25,50] | High sensitivity and specificity; fast; suitable for routine screening | Cross-reactivity; ineffective if target proteins are degraded |
| Mass Spectrometry-Based Proteomics (LC-MS/MS, HPLC-QTOF, and MALDI-TOF) [51,52,53,54,55,56,57,58,59,60,61]. | High multiplexing and quantification capability; species-level resolution | High cost; requires advanced instruments, skilled personnel, and complex data processing |
| Spectroscopic Methods (e.g., FTIR, NIR, and NMR) | Rapid; non-destructive; high throughput; some methods are portable | Dependent on large spectral databases; expensive for high-resolution platforms |
| DNA-Based Methods (PCR, qPCR, and RFLP) [66,67] | High specificity; good for detecting species substitution | Not suited for quantification; affected by DNA degradation in processed milk |
| Isotopic and Elemental Fingerprinting [70,71] | Accurate for geographical origin authentication | High equipment cost; may require large datasets; indirect biological relevance |
| Biosensors [69] | Low cost; fast; portable; user-friendly | Often qualitative; sensitivity limitations; limited multiplexing |
| Species | Casein | Whey Proteins | Total |
|---|---|---|---|
| Buffalo | 3.5–4.2 | 0.92 | 4.42–5.12 |
| Camel (bactrian) | 2.9 | 1.0 | 3.9 |
| Cow | 2.8 | 0.6 | 3.4 |
| Donkey | 1.0 | 1.0 | 2.0 |
| Goat | 2.5 | 0.4 | 2.9 |
| Horse | 1.3 | 1.2 | 2.54 |
| Casein (Gene) | Animal S.N. (En.) Names | Total Number of Amino Acids | M.W. kDa |
|---|---|---|---|
| α-S1-casein (CSN1S1) | Camelus dromedaries (Arabian camels) | 222 | 25.843 |
| Ovis aries (sheep) | 214 | 24.315 | |
| Capra hircus (goats) | 213 | 24.13 | |
| Bos taurus (cattle) | 214 | 24.435 | |
| Bubalus bubalis (water buffalos) | 214 | 24.312 | |
| α-S2-casein (CSN1S2) | Camelus dromedaries (Arabian camels) | 193 | 22.964 |
| Ovis aries (sheep) | 223 | 26.331 | |
| Capra hircus (goats) | 223 | 26.341 | |
| Bos taurus (cattle) | 222 | 26.12 | |
| Bubalus bubalis (water buffalos) | 222 | 26.223 | |
| β-casein (CSN2) | Camelus dromedaries (Arabian camels) | 232 | 26.217 |
| Ovis aries (sheep) | 222 | 24.946 | |
| Capra hircus (goats) | 223 | 24.992 | |
| Bos taurus (cattle) | 223 | 25.098 | |
| Bubalus bubalis (water buffalos) | 224 | 25.101 | |
| κ-casein (CSN3) | Camelus dromedaries (Arabian camels) | 219 | 24.717 |
| Ovis aries (sheep) | 162 | 17.899 | |
| Capra hircus (goats) | 162 | 17.896 | |
| Bos taurus (cattle) | 190 | 21.269 | |
| Bubalus bubalis (water buffalos) | 190 | 21.397 |
| Marker Proteins | Marker Peptide Sequence | Milk Origin | Aim of the Study | Analytical Technique | Detection Limit | Ref. |
|---|---|---|---|---|---|---|
| αs1-casein | FFVAPFPEVFGK (cow) | Camel | Analyze major camel and cow milk proteins through selected stable marker peptides and detect adulteration with cow milk | Identification of digested peptides: UPLC-ESI-TOF-MS (+); Quantitative analysis of peptides: UPLC-ESI-QQQMS (MRM) | 0.101 ng/mL | [114] |
| FFVAPFPEVFGK (38–49) (cow) | Cow, Goat, Sheep | Quantify cow’s whey and whole-milk powder percentage in goat or sheep milk products, including infant formula | UHPLC-ESI-TOF-MS (+) | 0.01–0.05 g/100 g cow’s whey and whole-milk powder in goat’s or sheep’s milk products, including infant formula | [115] | |
| FVVAPFPEVFR (38–48) (Goat and Sheep) | Cow, Goat, Sheep | Quantify cow’s whey and whole-milk powder percentage in goat or sheep milk products, including infant formula | UHPLC-ESI-TOF-MS (+) | 0.01–0.05 g/100 g cow’s whey and whole-milk powder in goat’s or sheep’s milk products, including infant formula | [115] | |
| VNELSK (52–57) (cow) | Cow, Goat, Sheep | Quantify cow’s whey and whole-milk powder percentage in goat or sheep milk products, including infant formula | UHPLC-ESI-TOF-MS (+) | 0.01–0.05 g/100 g cow’s whey and whole-milk powder in goat’s or sheep’s milk products, including infant formula | [115] | |
| ENINELSK (50–57) (Goat and Sheep) | Cow, Goat, Sheep | Quantify cow’s whey and whole-milk powder percentage in goat or sheep milk products, including infant formula | UHPLC-ESI-TOF-MS (+) | 0.01–0.05 g/100 g cow’s whey and whole-milk powder in goat’s or sheep’s milk products, including infant formula | [66] | |
| EEYINELNR (Donkey) | Camel, Donkey, Goat, Sheep, Yak, Cow | Identify specific peptide markers of seven milk species and assess the impact of processing treatments for accurate quantification of cow milk adulteration in non-cow milk samples. | HPLC-QTOF-MS (DIA) | 1% cow’s milk | [112] | |
| HQGLPQEVLNENLLR (cow) | Camel, Cow, Water Buffalo, Donkey, Goat, Horse, Sheep | Use signature peptides to measure αs2-caseins, β-caseins, and κ-caseins from eight milk species, enabling accurate detection and evaluation of milk adulteration. | UHPLC-ESI-Orbitrap-MS (+) | LOD: 5 μg/L | [66] | |
| HQGLPQEVLNENLLR (1759.9449 m/z) (cow) | Cow, Goat | Inspect adulteration in goat milk, characteristic peptides of caseins from cow milk were screened out | MALDI-TOF/TOF (+) | 1% cow’s milk in goat milk | [117] | |
| αs1-casein HQGLPQEVLNENLLR (8–22) (cow) | Goat | Detect cow milk contamination in goat milk | nanoLC-ESI-IT-MS/MS (+) DDA | 1% of cow’s milk in goat milk | [116] | |
| YNQLQLQAIYAQEQLIR (Donkey) | Buffalo, Cow, Donkey, Goat, Sheep, Yak | Identify specific peptide markers of seven milk species and assess the impact of processing treatments for accurate quantification of cow milk adulteration in non-cow milk samples | HPLC-QTOF-MS (DIA) | 1% cow’s milk | [112] | |
| YNQLQLQAIYAQEQLIR (Donkey) | Cow, Water Buffalo, Yak, Goat, sheep, donkey, horse, camel | Use signature peptides to measure αs2-caseins, β-caseins, and κ-caseins from eight milk species, enabling accurate detection and evaluation of milk adulteration | UHPLC-ESI-Orbitrap-MS (+) | LOD: 10 μg/L; LOQ: 20 μg/L | [66] | |
| as2-casein | NMAINPSK (Cow) | Camel, Donkey, Goat, Sheep, Yak, Cow | Identify specific peptide markers of seven milk species and assess the impact of processing treatments for accurate quantification of cow milk adulteration in non-cow milk samples | HPLC-QTOF-MS (DIA) | 1% cow’s milk | [112] |
| NMAIHPSK (Buffalo) | Camel, Donkey, Goat, Sheep, Yak, Cow | Identify specific peptide markers of seven milk species and assess the impact of processing treatments for accurate quantification of cow milk adulteration in non-cow milk samples | HPLC-QTOF-MS (DIA) | 1% cow’s milk | [112] | |
| NHLNFLK (Sheep) | Camel, Donkey, Goat, Sheep, Yak, Cow | Identify specific peptide markers of seven milk species and assess the impact of processing treatments for accurate quantification of cow milk adulteration in non-cow milk samples | HPLC-QTOF-MS (DIA) | 1% cow’s milk | [112] | |
| IVLTPWDQTK (Donkey) | Camel, Donkey, Goat, Sheep, Yak, Cow | Identify specific peptide markers of seven milk species and assess the impact of processing treatments for accurate quantification of cow milk adulteration in non-cow milk samples | HPLC-QTOF-MS (DIA) | 1% cow’s milk | [112] | |
| TNSYQIIPVLR (Donkey) | Camel, Donkey, Goat, Sheep, Yak, Cow | Identify specific peptide markers of seven milk species and assess the impact of processing treatments for accurate quantification of cow milk adulteration in non-cow milk samples | HPLC-QTOF-MS (DIA) | 1% cow’s milk | [112] | |
| LNFLQYLQALR (Donkey) | Camel, Donkey, Goat, Sheep, Yak, Cow | Identify specific peptide markers of seven milk species and assess the impact of processing treatments for accurate quantification of cow milk adulteration in non-cow milk samples | HPLC-QTOF-MS (DIA) | 1% cow’s milk | [112] | |
| ISQHYQK (Buffalo) | Camel, Donkey, Goat, Sheep, Yak, Cow | Identify specific peptide markers of seven milk species and assess the impact of processing treatments for accurate quantification of cow milk adulteration in non-cow milk samples | HPLC-QTOF-MS (DIA) | 1% cow’s milk | [112] | |
| TNVIPYVR (Buffalo) | Camel, Donkey, Goat, Sheep, Yak, Cow | Identify specific peptide markers of seven milk species and assess the impact of processing treatments for accurate quantification of cow milk adulteration in non-cow milk samples | HPLC-QTOF-MS (DIA) | 1% cow’s milk | [112] | |
| AMKPWIQPK (Cow) | Camel, Donkey, Goat, Sheep, Yak, Cow | Identify specific peptide markers of seven milk species and assess the impact of processing treatments for accurate quantification of cow milk adulteration in non-cow milk samples | HPLC-QTOF-MS (DIA) | 1% cow’s milk | [112] | |
| LCTTSCEEVVR (51–61) (Goat and Sheep) | Cow, Goat, Sheep | Quantify cow’s whey and whole-milk powder percentage in goat or sheep milk products, including infant formula | UHPLC-ESI-TOF-MS (+) | 0.01–0.05 g/100 g cow’s whey and whole-milk powder in goat’s or sheep’s milk products, including infant formula | [115] | |
| NAVPITPTLNR (1195.6793 m/z) (cow) | Cow, Goat | Inspect adulteration in goat milk; screen out characteristic peptides of caseins from cow milk | MALDI-TOF/TOF (+) | 1% cow’s milk in goat milk | [117] | |
| NAVPITPTLNR (131–141) (cow) | Cow, Goat, Sheep | Quantify cow’s whey and whole-milk powder percentage in goat or sheep milk products, including infant formula | UHPLC-ESI-TOF-MS (+) | 0.01–0.05 g/100 g cow’s whey and whole-milk powder in goat’s or sheep’s milk products, including infant formula | [115] | |
| NAGPFTPTVNR (131–141) (Sheep and Goat) | Cow, Goat, Sheep | Quantify cow’s whey and whole-milk powder percentage in goat or sheep milk products, including infant formula | UHPLC-ESI-TOF-MS (+) | 0.01–0.05 g/100 g cow’s whey and whole-milk powder in goat’s or sheep’s milk products, including infant formula | [115] | |
| ENLCSTFCK (49–57) (cow) | Cow, Goat, Sheep | Quantification of cow’s whey and whole-milk powder percentage in goat or sheep milk products, including infant formula | UHPLC-ESI-TOF-MS (+) | 0.01–0.05 g/100 g cow’s whey and whole-milk powder in goat’s or sheep’s milk products, including infant formula | [115] | |
| TVYQHQK (198–204) (cow) | Cow, Goat, Sheep | Quantification of cow’s whey and whole-milk powder percentage in goat or sheep milk products, including infant formula | UHPLC-ESI-TOF-MS (+) | 0.01–0.05 g/100 g cow’s whey and whole-milk powder in goat’s or sheep’s milk products, including infant formula | [115] | |
| TVYQHQK (198–204) (Goat and Sheep) | Cow, Goat, Sheep | Quantification of cow’s whey and whole-milk powder percentage in goat or sheep milk products, including infant formula | UHPLC-ESI-TOF-MS (+) | 0.01–0.05 g/100 g cow’s whey and whole-milk powder in goat’s or sheep’s milk products, including infant formula | [115] | |
| FALPQYLK (cow) | Camel | Develop and validate a method using mass spectrometry to quantitatively analyze major camel and cow milk proteins through selected stable marker peptides and detect adulteration with cow milk | Identification of digested peptides: UPLC-ESI-TOF-MS (+); Quantitative analysis of peptides: UPLC-ESI-TQMS (MRM) | 0.045 ng/mL | [114] | |
| FALPQYLK (190–197) (cow) | Cow, Goat, Sheep | Quantify cow’s whey and whole-milk powder percentage in goat or sheep milk products, including infant formula | UHPLC-ESI-TOF-MS (+) | 0.01–0.05 g/100 g cow’s whey and whole-milk powder in goat’s or sheep’s milk products including infant formula | [115] | |
| FALPQYLK (190–197) (Goat and Sheep) | Cow, Goat, Sheep | Quantify cow’s whey and whole-milk powder percentage in goat or sheep milk products, including infant formula | UHPLC-ESI-TOF-MS (+) | 0.01–0.05 g/100 g cow’s whey and whole-milk powder in goat’s or sheep’s milk products, including infant formula | [115] | |
| FPQYLQYPYQGPIVLNPWDQVK (Goat) | Camel, donkey, Yak, goat, cow, sheep | Identify specific peptide markers of seven milk species and assess the impact of processing treatments for accurate quantification of cow milk adulteration in non-cow milk samples | HPLC-QTOF-MS (DIA) | 1% cow’s milk | [112] | |
| VLPVPQK (Cow) | Camel | Develop and validate a method using mass spectrometry to quantitatively analyze major camel and cow milk proteins through selected stable marker peptides and detect adulteration with cow milk | Identification of digested peptides: UPLC-ESI-TOF-MS (+;) Quantitative analysis of peptides: UPLC-ESI-TQMS (MRM) | 0.004 ng/mL | [114] | |
| β-casein | AVPYPQR (830.4519 m/z) (Cow) | Cow, Goat | Inspect adulteration in goat milk; screen out characteristic peptides of caseins from cow milk | MALDI-TOF/TOF (+) | 1% cow’s milk in goat milk | [117] |
| YPVEPFTER (Cow) | Camel, Donkey, Goat, Sheep, Yak, Cow | Identify specific peptide markers of seven milk species and assess the impact of processing treatments for accurate quantification of cow milk adulteration in non-cow milk samples | HPLC-QTOF-MS (DIA) | 1% cow’s milk | [112] | |
| IEEQQQTEDEQQDK (Camel) | Camel, Donkey, Goat, Sheep, Yak, Cow | Identify specific peptide markers of seven milk species and assess the impact of processing treatments for accurate quantification of cow milk adulteration in non-cow milk samples | HPLC-QTOF-MS (DIA) | 1% cow’s milk | [112] | |
| FQSEEQQQMEDELQDK (Buffalo) | Camel, Donkey, Goat, Sheep, Yak, Cow | Identify specific peptide markers of seven milk species and assess the impact of processing treatments for accurate quantification of cow milk adulteration in non-cow milk samples | HPLC-QTOF-MS (DIA) | 1% cow’s milk | [112] | |
| IHPFAQTQSLVYPFPGPIPK (Buffalo) | Camel, Donkey, Goat, Sheep, Yak, Cow | Identify specific peptide markers of seven milk species and assess the impact of processing treatments for accurate quantification of cow milk adulteration in non-cow milk samples | HPLC-QTOF-MS (DIA) | 1% cow’s milk | [112] | |
| AIPVQAVLPFQEPVPDPVR (Camel) | Camel, Donkey, Goat, Sheep, Yak, Cow | Identify specific peptide markers of seven milk species and assess the impact of processing treatments for accurate quantification of cow milk adulteration in non-cow milk samples | HPLC-QTOF-MS (DIA) | 1% cow’s milk | [112] | |
| VAPFPQPVVPYPQR (Donkey) | Camel, Donkey, Goat, Sheep, Yak, Cow | Identify specific peptide markers of seven milk species and assess the impact of processing treatments for accurate quantification of cow milk adulteration in non-cow milk samples | HPLC-QTOF-MS (DIA) | 1% cow’s milk | [112] | |
| GPFPIIV (217–224) (Goat and Sheep) | Cow, Goat, Sheep | Quantify cow’s whey and whole-milk powder percentage in goat or sheep milk products, including infant formula | UHPLC-ESI-TOF-MS (+) | 0.01–0.05 g/100 g cow’s whey and whole-milk powder in goat’s or sheep’s milk products, including infant formula | [115] | |
| YIPIQYVLSR | Camel | Develop and validate a method using mass spectrometry to quantitatively analyze major camel and cow milk proteins through selected stable marker peptides and detect adulteration with cow milk | Identification of digested peptides: UPLC-ESI-TOF-MS (+); Quantitative analysis of peptides: UPLC-ESI-TQMS (MRM) | 0.103 ng/mL | [114] | |
| κ-casein | FFSDK (38–42) (Cow) | Cow, Goat, Sheep | Quantify cow’s whey and whole-milk powder percentage in goat or sheep milk products, including infant formula | UHPLC-ESI-TOF-MS (+) | 0.01–0.05 g/100 g cow’s whey and whole-milk powder in goat’s or sheep’s milk products, including infant formula | [115] |
| FFDDK (38–42) (Goat and Sheep) | Cow, Goat, Sheep | Quantify cow’s whey and whole-milk powder percentage in goat or sheep milk products including infant formula | UHPLC-ESI-TOF-MS (+) | 0.01–0.05 g/100 g cow’s whey and whole-milk powder in goat’s or sheep’s milk products, including infant formula | [115] | |
| SPAQTLQWQVLPNTVPAK (Goat) | Camel, Donkey, Goat, Sheep, Yak, Cow | Identify specific peptide markers of seven milk species and assess the impact of processing treatments for accurate quantification of cow milk adulteration in non-cow milk samples | HPLC-QTOF-MS (DIA) | 1% cow’s milk | [112] | |
| SPAQTLQWQVLPNTVPAK (Goat) | Cow, Water Buffalo, Yak, Goat, sheep, donkey, horse, camel | Use signature peptides to measure αs2-caseins, β-caseins, and κ-caseins from eight milk species, enabling accurate detection and evaluation of milk adulteration | UHPLC-ESI-Orbitrap-MS (+) | LOD: 10 μg/L; LOQ: 30 μg/L | [66] | |
| SPAQTLQWQVLPNAVPAK (Sheep) | Camel, Donkey, Goat, Sheep, Yak, Cow | Identify specific peptide markers of seven milk species and assess the impact of processing treatments for accurate quantification of cow milk adulteration in non-cow milk samples | HPLC-QTOF-MS (DIA) | 1% cow’s milk | [112] | |
| SPAQTLQWQVLPNAVPAK (Sheep) | Cow, Water Buffalo, Yak, Goat, sheep, donkey, horse, camel | Use signature peptides to measure αs2-caseins, β-caseins, and κ-caseins from eight milk species, enabling accurate detection and evaluation of milk adulteration | UHPLC-ESI-Orbitrap-MS (+) | LOD: 10 μg/L; LOQ: 30 μg/L | [3] | |
| SPAQILQWQVLPNTVPAK (Buffalo) | Camel, Donkey, Goat, Sheep, Yak, Cow | Identify specific peptide markers of seven milk species and assess the impact of processing treatments for accurate quantification of cow milk adulteration in non-cow milk samples | HPLC-QTOF-MS (DIA) | 1% cow’s milk | [112] | |
| SCQAQPTTMAR (Cow) | Camel, Donkey, Goat, Sheep, Yak, Cow | Identify specific peptide markers of seven milk species and assess the impact of processing treatments for accurate quantification of cow milk adulteration in non-cow milk samples | HPLC-QTOF-MS (DIA) | 1% cow’s milk | [112] | |
| SCQDQPTAMAR (Sheep) | Camel, Donkey, Goat, Sheep, Yak, Cow | Identify specific peptide markers of seven milk species and assess the impact of processing treatments for accurate quantification of cow milk adulteration in non-cow milk samples | HPLC-QTOF-MS (DIA) | 1% cow’s milk | [112] | |
| YFPIQFVQSR (Camel) | Camel, Donkey, Goat, Sheep, Yak, Cow | Identify specific peptide markers of seven milk species and assess the impact of processing treatments for accurate quantification of cow milk adulteration in non-cow milk samples | HPLC-QTOF-MS (DIA) | 1% cow’s milk | [112] | |
| IDALNENK (Cow) | Camel | Analyze major camel and cow milk proteins through selected stable marker peptides and detect adulteration with cow milk | Identification of digested peptides: UPLC-ESI-TOF-MS (+); Quantitative analysis of peptides: UPLC-ESI-TQMS (MRM) | 0.066 ng/mL | [114] | |
| LSFNPTQLEEQCHI (167–180) (cow) | Cow, Goat, Sheep | Quantify cow’s whey and whole-milk powder percentage in goat or sheep milk products, including infant formula | UHPLC-ESI-TOF-MS (+) | 0.01–0.05 g/100 g cow’s whey and whole-milk powder in goat’s or sheep’s milk products, including infant formula | [115] | |
| β-Lactoglobulin LSFNPTQLEEQCHI (149–162) | Goat | Detect cow milk contamination in goat milk | nanoLC-ESI-IT-MS/MS (+) DDA | 1% of cow’s milk in goat milk | [116] | |
| β-lactoglobulin | LAFNPTQLEGQCHV (167–180) (Goat and Sheep) | Cow, Goat, Sheep | Quantify cow’s whey and whole-milk powder percentage in goat or sheep milk products, including infant formula | UHPLC-ESI-TOF-MS (+) | 0.01–0.05 g/100 g cow’s whey and whole-milk powder in goat’s or sheep’s milk products, including infant formula | [115] |
| LSFNPTQLEGQCHI (Yak) | Camel, Donkey, Goat, Sheep, Yak, Cow | Identify specific peptide markers of seven milk species and assess the impact of processing treatments for accurate quantification of cow milk adulteration in non-cow milk samples | HPLC-QTOF-MS (DIA) | 1% cow’s milk | [112] | |
| WENDECAQK (Cow) | Camel, Donkey, Goat, Sheep, Yak, Cow | Identify specific peptide markers of seven milk species and assess the impact of processing treatments for accurate quantification of cow milk adulteration in non-cow milk samples | HPLC-QTOF-MS (DIA) | 1% cow’s milk | [112] | |
| LSFNPTQLEGQCHI (Yak) | Camel, Donkey, Goat, Sheep, Yak, Cow | Identify specific peptide markers of seven milk species and assess the impact of processing treatments for accurate quantification of cow milk adulteration in non-cow milk samples | HPLC-QTOF-MS (DIA) | 1% cow’s milk | [112] | |
| NICNISCDK (90–98) (Goat and Sheep) | Cow, Goat, Sheep | Quantify cow’s whey and whole-milk powder percentage in goat or sheep milk products, including infant formula | UHPLC-ESI-TOF-MS (+) | 0.01–0.05 g/100 g cow’s whey and whole-milk powder in goat’s or sheep’s milk products, including infant formula | [115] | |
| CEVFR (25–29) (Cow) | Cow, Goat, Sheep | Quantify cow’s whey and whole-milk powder percentage in goat or sheep milk products, including infant formula | UHPLC-ESI-TOF-MS (+) | 0.01–0.05 g/100 g cow’s whey and whole-milk powder in goat’s or sheep’s milk products, including infant formula | [115] | |
| α-lactalbumin | NICNISCDK (90–98) (Goat and Sheep) | Cow, Goat, Sheep | Quantify cow’s whey and whole-milk powder percentage in goat or sheep milk products, including infant formula | UHPLC-ESI-TOF-MS (+) | 0.01–0.05 g/100 g cow’s whey and whole-milk powder in goat’s or sheep’s milk products, including infant formula | [115] |
| CEVFR (25–29) (Cow) | Cow, Goat, Sheep | Quantify cow’s whey and whole-milk powder percentage in goat or sheep milk products, including infant formula | UHPLC-ESI-TOF-MS (+) | 0.01–0.05 g/100 g cow’s whey and whole-milk powder in goat’s or sheep’s milk products, including infant formula | [115] |
| Marker Proteins | Marker Peptide Sequence | Milk Origin | Ref. |
|---|---|---|---|
| αs1-casein | FFVAPFPEVFGK (38–49) | Cow | [114,115] |
| FVVAPFPEVFR (38–48) | Goat and Sheep | [115] | |
| VNELSK (52–57) | Cow | [115] | |
| ENINELSK (50–57) | Goat and Sheep | [66] | |
| EEYINELNR | Donkey | [112] | |
| HQGLPQEVLNENLLR (1759.9449 m/z) | Cow | [117] | |
| YNQLQLQAIYAQEQLIR | Donkey | [66,112] | |
| as2-casein | NMAINPSK | Cow | [112] |
| NMAIHPSK | Buffalo | [112] | |
| NHLNFLK | Sheep | [112] | |
| IVLTPWDQTK | Donkey | [112] | |
| TNSYQIIPVLR | Donkey | [112] | |
| LNFLQYLQALR | Donkey | [112] | |
| ISQHYQK | Buffalo | [112] | |
| TNVIPYVR | Buffalo | [112] | |
| AMKPWIQPK | Cow | [112] | |
| LCTTSCEEVVR (51–61) | Goat and Sheep | [115] | |
| NAVPITPTLNR (131–141) (1195.6793 m/z) | Cow | [115,117] | |
| NAGPFTPTVNR (131–141) | Sheep and Goat | [115] | |
| ENLCSTFCK (49–57) | Cow | [115] | |
| TVYQHQK (198–204) | Cow | [115] | |
| FALPQYLK (190–197) | Cow | [114,115] | |
| FPQYLQYPYQGPIVLNPWDQVK | Goat | [112] | |
| VLPVPQK | Cow | [114] | |
| β-casein | AVPYPQR (830.4519 m/z) | Cow | [117] |
| YPVEPFTER | Cow | [112] | |
| IEEQQQTEDEQQDK | Camel | [112] | |
| FQSEEQQQMEDELQDK | Buffalo | [112] | |
| IHPFAQTQSLVYPFPGPIPK | Buffalo | [112] | |
| AIPVQAVLPFQEPVPDPVR | Camel | [112] | |
| VAPFPQPVVPYPQR | Donkey | [112] | |
| GPFPIIV (217–224) | Goat and Sheep | [115] | |
| YIPIQYVLSR | Cow | [114] | |
| κ-casein | FFSDK (38–42) | Cow | [115] |
| SPAQTLQWQVLPNTVPAK | Goat | [66,112] | |
| SPAQTLQWQVLPNAVPAK | Sheep | [112] | |
| SPAQILQWQVLPNTVPAK | Buffalo | [112] | |
| SCQAQPTTMAR | Cow | [112] | |
| SCQDQPTAMAR | Sheep | [112] | |
| YFPIQFVQSR | Camel | [112] | |
| IDALNENK | Cow | [114] | |
| LSFNPTQLEEQCHI (167–180) | Cow | [115] | |
| β-lactoglobulin | LAFNPTQLEGQCHV (167–180) | Goat and Sheep | [115] |
| LSFNPTQLEGQCHI | Yak | [112] | |
| WENDECAQK | Cow | [112] | |
| α-lactalbumin | NICNISCDK (90–98) | Goat and Sheep | [115] |
| CEVFR (25–29) | Cow | [115] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Karamoutsios, A.; Lekka, P.; Voidarou, C.C.; Dasenaki, M.; Thomaidis, N.S.; Skoufos, I.; Tzora, A. Assessing Milk Authenticity Using Protein and Peptide Biomarkers: A Decade of Progress in Species Differentiation and Fraud Detection. Foods 2025, 14, 2588. https://doi.org/10.3390/foods14152588
Karamoutsios A, Lekka P, Voidarou CC, Dasenaki M, Thomaidis NS, Skoufos I, Tzora A. Assessing Milk Authenticity Using Protein and Peptide Biomarkers: A Decade of Progress in Species Differentiation and Fraud Detection. Foods. 2025; 14(15):2588. https://doi.org/10.3390/foods14152588
Chicago/Turabian StyleKaramoutsios, Achilleas, Pelagia Lekka, Chrysoula Chrysa Voidarou, Marilena Dasenaki, Nikolaos S. Thomaidis, Ioannis Skoufos, and Athina Tzora. 2025. "Assessing Milk Authenticity Using Protein and Peptide Biomarkers: A Decade of Progress in Species Differentiation and Fraud Detection" Foods 14, no. 15: 2588. https://doi.org/10.3390/foods14152588
APA StyleKaramoutsios, A., Lekka, P., Voidarou, C. C., Dasenaki, M., Thomaidis, N. S., Skoufos, I., & Tzora, A. (2025). Assessing Milk Authenticity Using Protein and Peptide Biomarkers: A Decade of Progress in Species Differentiation and Fraud Detection. Foods, 14(15), 2588. https://doi.org/10.3390/foods14152588









