Abstract
Apples are one of the most popular fruits in Germany, valued for their regional availability and health benefits. When deciding which apple to buy, several characteristics are important to consumers, including the taxonomic variety, organic cultivation and regional production. To verify that these characteristics are correctly declared, powerful analytical methods are required. In this study, ultra-high performance liquid chromatography quadrupole time-of-flight mass spectrometry (UHPLC-Q-ToF-MS) is applied in combination with random forest to 193 apple samples for the analysis of various authentication issues. Accuracies of 93.3, 85.5, 85.6 and 90% were achieved for distinguishing between German and non-German, North and South German, organic and conventional apples and for six different taxonomic varieties. Since the classification models largely use different parts of the data, which is shown by variable selection, this method is very well suited to answer different authentication issues with one analytical approach.
1. Introduction
According to regulation (EU) No 1169/2011 of 25 October 2011, it should be ensured that consumers are appropriately informed about the food they consume to make choices influenced by, inter alia, health, economics, environmental, social and ethical considerations. Lately, environmental awareness, e.g., for species extinction and resource depletion, has increased and consumers are more and more interested in sustainably produced food with short transport distances [1,2], and the demand for local and organic food is rising in European countries [3]. This also applies to apples, Malus × domestica borkh., a fruit that is grown in many parts of the world and is the most popular fruit in Germany [4]. In addition to local and organic production, the taxonomic variety is also of interest for the consumer, as this strongly influences the taste of the apple. This interest is even more present if the consumer is allergic to apples, as the allergenicity of different varieties is different [5,6]. It is therefore important to develop reliable approaches for apple authentication that enable identification of the geographical origin, production method and taxonomic variety.
Various methods have been developed to analyze apples for these different authentication issues individually. The discrimination of apple origins using inductively coupled plasma mass spectrometry (ICP-MS), for example, achieved classification accuracies of 91%, 83% and 92% for the differentiation of European and non-European, German and non-German and North and South German samples, respectively [7]. To differentiate organic from conventional apples, Song et al. developed a low-cost and non-destructive sensor system for in-line food authentication using a smartphone and a diffraction grating sheet [8]. In addition, Barberis et al. developed a non-invasive method to differentiate the varieties of apple samples using a functionalized strip in combination with gas chromatography mass spectrometry (GC-MS) [9].
To develop a method that can analyze several of these authentication issues simultaneously, untargeted approaches that examine the metabolome are promising, as it reflects the phenotype of the sample and is influenced by external factors such as the geographical and taxonomic origin as well as the production method [10,11,12]. For this analysis, GC-MS and liquid chromatography mass spectrometry (LC-MS) as well as Proton Nuclear Magnetic Resonance (1H-NMR) methods are suitable. Wenck et al. applied 1H-NMR spectroscopy to analyze several authentication issues of apples and the discrimination of seven taxonomic varieties, German and non-German samples, as well as organically and conventionally grown apples was possible with classification accuracies of 73%, 89 and 80%, respectively [13].
Compared to 1H-NMR, LC-MS shows higher sensitivity [14], which is why it has already been applied for the authentication of various food [15,16,17,18,19,20,21], including apple juice [22,23]. However, due to the comparatively low robustness of LC-MS [12,14], only spectra of samples measured at the same time and on the same device are compared in these applications. When processing this data, a correspondence step is usually applied in which the detected and aligned chromatographic peaks are matched between the different spectra to generate a common peak list that is unique for each processing batch. This means that the peak lists generated in different batches cannot be used for joint data analysis, preventing the measurement and evaluation of data over a longer period of time, e.g., different harvest years. To make this possible, we have developed a data processing approach called bucketing of LC-MS spectra (BOULS) [16,24] that applies a bucketing step in which the intensities of all signals in a defined area are summed up. In this way, a standardized data structure is generated independent of the processing batch. We have shown that this approach enables the prediction and addition of data from new samples that have been processed individually to an existing RF model. In addition, generating buckets compensates for technical variances, thereby reducing the need for batch correction such as Locally Estimated Scatterplot Smoothing [25], which is difficult to implement in routine analyses. For the analysis of honey, the application of BOULS enabled long-term use of LC-MS to determine the geographical origin with a classification accuracy of 94% for 126 test samples from six different countries [24].
In order to harness the analytical data and obtain classification models for the analysis of authentication issues, machine learning approaches such as artificial neural networks, support vector machines and random forests (RF) are applied. RF is a non-parametric ensemble learning algorithm based on multiple binary decision trees. Each of these trees is trained on a subset of the samples, called bootstrap samples, and the remaining samples, called out-of-the-bag samples, can be used to generate an independent out-of-bag-error (OOB-error) [26,27]. This error is equivalent to that obtained by the use of independent validation data, when no parameter optimization is conducted, which is particularly advantageous when analyzing small data sets. This and other features, such as suitability for high-dimensional data and the ability to adjust the bootstrap sample for imbalanced training data, make RF advantageous for use in food authentication based on LC-MS data [17,18,20,21,28].
In this study, we combined untargeted ultra-high performance liquid chromatography quadrupole time-of-flight mass spectrometry (UHPLC-Q-ToF-MS) with BOULS and RF for the authentication of apple samples that were measured over a period of 8 months. In addition to analyzing whether it is possible to successfully classify apples with respect to the different authentication issues of geographical origin, variety and production method using the same approach, we also analyzed whether BOULS is generally applicable in other laboratories using different LC-MS instruments than those on which it was developed. This is particularly important because this approach promises that data can be evaluated independently of these parameters, which should be verified.
2. Materials and Methods
2.1. Sample Preparation and LC-MS Data Acquisition
In this study, 193 apple samples were analyzed once, of which 57, 124 and 12 were harvested in the years 2020, 2021 and 2022, respectively. The sample preparation and LC-MS data acquisition procedures of the routine laboratory were adopted in order to provide realistic conditions for testing the general applicability of the BOULS approach. First, the apples were washed with deionized water and juiced using a centrifugal juicer (Nutri Juicer Cold Juicer) as soon as they were completely dry, and leaves and stems were removed. The samples were stored at −20 °C until the measurement. The standard operating procedure of the laboratory was followed to create realistic conditions and, accordingly, 1 mL of the defrosted samples and 1 mL of a stock solution with caffeine (CAS-no. 58-08-2), 2,5-dihydroxybenzoic acid (CAS-no. 490-79-9) and rosmarinic acid (CAS-no. 20283-92-5) in methanol (hypergrade, Merck-no. 1.06035) were homogenized and centrifuged (20 min, 4000 g, room temperature), and the upper part was filtered (IC Millex LG 0.20 µm Low Protein Binding Hydrophilic LCR (PTFE) Merck Millipore) into HPLC vials.
A 1220 Infinity II LC system in combination with 6545 ESI-Q-TOF-MS from Agilent was used as UHPLC-Q-ToF-MS-system with an endcapped-C18 column (Waters, Acquity HSS T3, 1.8 µm, 100 x 2.1 mm). The mobile phases of the UHPLC method were water (Eluent A, H2O bidest.) with 0.1% formic acid (Honeywell-Fluka no. 09676) and acetonitrile (Eluent B, hypergrade, Merck-no. 1.00029) with 0.1% formic acid). The flow rate was set to 0.4 mL/minute and the start gradient was 95/5 Eluent A/Eluent B. At 3 min, 9 min, 11 min and 13 min, the mobile phase was shifted to 80/20, 55/45, 0/100 and 95/5 Eluent A/Eluent B, respectively. Stoptime was 13.5 min plus 2.5 min postrun. The injection volume was 2 µL, the column temperature was 40 °C and electrospray ionization was performed with VCap 3500 V, Nozzle 300 V and 200 °C gas temperature. The samples were measured in a randomized order.
2.2. Data Used for the Different Authentication Issues
Depending on the information available, different data sets were built for the different authentication issues. The data set for the differentiation between German and non-German samples comprised 193 samples, from which 117 samples originated from Schleswig-Holstein (59), Hamburg (6), Lower Saxony (14) and Baden-Württemberg (38) in Germany. The 76 non-German samples originated from Chile (15), Italy (26), New Zealand (18) and South Africa (17) and were purchased in supermarkets. The data set for the differentiation of the regional origin within Germany comprised the 117 German samples, with the 79 samples originating from Schleswig-Holstein, Hamburg and Lower Saxony categorized as North German samples and the 38 samples from Baden-Württemberg categorized as South German samples. For the differentiation according to the production method, 153 samples were used. The 113 conventionally grown apple samples originated from Chile (7), Italy (22), New Zealand (16), South Africa (15), and Germany, more precisely from the federal states Schleswig-Holstein (16), Lower Saxony (4) and Baden-Württemberg (33). The 40 organically grown samples originated from Italy (1), New Zealand (1), Schleswig-Holstein (29) and Lower Saxony (9).
The data set for the determination of the taxonomic variety comprised 80 samples. The 11 samples of the variety Braeburn originated from South Africa (3), New Zealand (4), Baden Württemberg (3) and Schleswig-Holstein (1). The 12 samples of the variety Cripps Pink originated from South Africa (5), New Zealand (2), Chile (3) and Italy (2). The 12 samples of the variety Elstar originated from Baden Württemberg (8), Schleswig-Holstein (3) and Hamburg (1). The 9 Boskoop apples originated from Schleswig-Holstein (2), Lower Saxony (2), Baden Württemberg (4) and Hamburg (1). The 28 Gala apples originated from Italy (5), New Zealand (6), South Africa (4), Chile (3), Baden Württemberg (9) and Schleswig-Holstein (1). The 8 Jonagold apples originated from Lower Saxony (1), Baden Württemberg (5) and Schleswig-Holstein (2).
2.3. Data Processing and Analysis
Data processing and analysis were carried out in R (version 4.4.1) [29] using the BOULS approach [24] (https://github.com/AGSeifert/BOULS, accessed on 9 January 2025, requires Linux OS). BOULS is based on the xcms workflow [30] and uses the same functions for data import and retention time alignment.
Before importing the data into R, the Agilent-specific .d files were converted to open-format mzXML files using MSConvert, which is part of the ProteoWizard software package [31] (version 3.0.21078-7da1f1136 (developer build)), and the filter peakPicking was used to convert the profile data into centroided data. The Bioconductor package mzR (version 2.26.0) was used for data import into R [31,32,33,34] and the package MSnbase (version 2.18.0) [35,36] was used to load and store the data in an object compatible with those of the xcms package (version 3.14.0) [37]. Retention time alignment was conducted using the obiwarp method [38] with a bin size of 0.1 and localAlignment set to TRUE, after peak detection using the centWave algorithm [39] with the parameters peakwidth of 15 s and ppm of 5 Da. The same sample was used as the center spectrum for each processing batch.
Subsequently, the data were processed using BOULS with a bucket size of 20 s in the retention time dimension and 2 Da in the mass dimension and the data were normalized by dividing the summarized intensities of each bucket by the sum of intensities of all buckets. This parameter combination was previously determined during optimization of data processing with BOULS.
The stats package [40,41,42] (version 4.4.1) was used for PCA and the scores plots were visualized using the ggplot2 package [43] (version 3.5.1). For classification, RF was applied using the ranger package [44] (version 0.16.0) with the parameter ntree set to 5000 and the respective default settings for mtry and min.node.size (133, which is the square root of the total number of variables and 1, respectively). To compensate for class imbalance, the parameter case.weights was chosen according to the size of the respective classes. The performance of the models was evaluated by the out-of-bag (OOB) error of the obtained random forests. The Pomona package (version 1.0.1) was used for Boruta variable selection with the value “impurity_corrected” for the importance parameter, 0.01 for the p-value and 100 for maxRuns [45,46].
3. Results and Discussion
The apple samples were analyzed using LC-MS, and Figure S1 shows representative chromatograms, which were then processed and evaluated using PCA and RF.
3.1. Principal Component Analysis
In order to analyze the main variances of the processed data of the samples, PCA was applied, and the scores of the first two principal components labeled according to the country of origin and taxonomic variety, as well as to the region within Germany and the production method, are shown in Figure 1, Figures S2 and S3, respectively. It can be observed that the groupings are not based on the respective authentication issue but on the days on which the data was obtained. Similar results were achieved when analyzing honey using this approach [16]. They can be explained by the varying age of the columns and degree of contamination of the devices [47], which influence the data even after processing with BOULS. Similarly to the analysis of honey in the first implementation of BOULS, unsupervised data analysis cannot be applied to the analysis of apples, which is why random forest was used as a supervised method to analyze the different authentication issues.
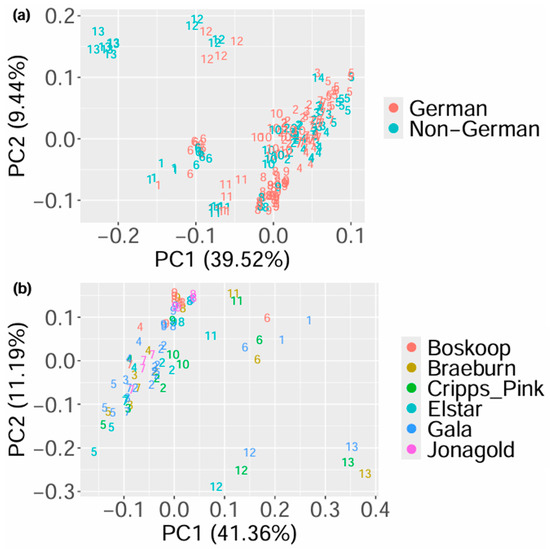
Figure 1.
Results of the PCA showing the scores of the first and second principal component with colors according to the geographical origin (a) and to the taxonomic variety (b). The numbers correspond to the days on which the data was obtained, meaning that data points with the same number were measured on the same day.
3.2. Differentiation Between German and Non-German Samples
The question whether the samples originated from Germany or not arose from the assumption that it is of most importance for the consumer whether the apple originates from the country in which it is sold. Therefore, we trained a corresponding RF model and the results are shown in Table 1. In total, 8 non-German samples were misclassified as German, and 2, 5 and 1 of them originated from New Zealand, Italy and South Africa, respectively (see Table S1). The misclassification of the Italian samples could be attributed to a similarity between Italian and German apples, which has presumably arisen from an adaptation to consumer expectations, given that a large number of apples are imported from Italy to Germany every year [48]. Another possible reason for the incorrect classification is that the non-German samples were purchased in supermarkets, meaning the origin information cannot be guaranteed with certainty. However, it is highly unlikely that German samples were mislabeled, so this has negligible influence on the classification results. The overall accuracy achieved is 93.3%, which is higher than that obtained for the same authentication issue by ICP-MS (83.2%) [7] and 1H-NMR (88.5%) [13]. Just as in these studies, non-German samples are misclassified more frequently than German samples, which could be due to the higher within-class variance of these samples and the generally smaller number of samples. In general, the results show that LC-MS in combination with BOULS and RF is a promising approach for the identification of German apple samples.

Table 1.
Classification results of the apple data set according to the geographical origin reaching a total accuracy of 93.3% with a sensitivity of 89.5% and 95.7% for the non-German and German samples, respectively.
3.3. Differentiation of the Regional Origin Within Germany
The regionality of food is becoming increasingly important for the consumer due to increasing environmental awareness, with a view to shorting transport distances. Therefore, it was also investigated whether North and South German samples can be distinguished, and the results are shown in Table 2. The overall accuracy is 85.5%, and 2 (of 59) and 3 (of 14) samples from Schleswig-Holstein and Lower Saxony, respectively, are misclassified (see Table S1). This could be explained by similar soil compositions for North and South German samples [49]. The reason for the much higher sensitivity of the North German class (93.7%) compared to the South German class (68.4%) could be that a smaller number of samples were used here. This may not be sufficient to adequately represent the variance of this class in the model. A similar conclusion about the influence of different sample sizes on the classification was drawn for the analysis of apple samples by 1H-NMR and for the analysis of honey by the approach used here [13,16,24].

Table 2.
Classification results of the apple data set according to the regionality within Germany, reaching a total accuracy of 85.5% with a sensitivity of 93.7% and 68.4% for the North and South German samples, respectively.
The accuracy for distinguishing apples from North and South Germany is higher for LC-MS (about 85%) than for 1H-NMR (80.7%), but lower than for ICP-MS (92.3%). This better performance of ICP-MS could be due to soil differences, that are mainly reflected in the different element composition [50], which could also be used for the regional authentication of asparagus, almonds, walnuts, hazelnuts and truffles [51,52,53,54,55,56].
3.4. Differentiation Between Organically and Conventionally Produced Apples
Since the production method is also relevant for consumers, a classification model was also trained to differentiate between organically and conventionally grown apples. The results are shown in Table 3 and the model achieved an overall accuracy of 85.6%. Similarly to the distinction between apples from different regions within Germany, the sensitivity of the larger class of conventionally produced apples is much higher (97.3%) than that of the smaller class of organically produced apples, which has a sensitivity of 52.5%. As before, the difference here could be explained by the different sample sizes of the classes. However, the very high sensitivity for conventionally produced apples, which is approximately 10% higher than for the analysis of this issue with 1H-NMR [13], suggests that this method is very promising for application. This is because an authentication method is only needed to detect deliberate mislabeling of conventionally produced apples as organic, not vice versa, as conventional apples have lower production costs.

Table 3.
Classification results of the apple data set according to the production method, reaching a total accuracy of 85.6% with a sensitivity of 97.3% and 52.5% for the conventional and biologically produced apple samples, respectively.
3.5. Differentiation by Taxonomic Variety
Since it is also relevant to identify the taxonomic variety of apples, especially with regard to the different allergenicity, a corresponding model was subsequently trained. The results are shown in Table 4, reaching a total accuracy of 90% for the differentiation of the six different varieties. The reason for individually occurring misclassifications could be that many of the apple varieties share common ancestry, which results in close genetic relationships [57].

Table 4.
Classification results of the apple data set according to the taxonomic variety, reaching a total accuracy of 90% and sensitivities of 88.9%, 72.7%, 100%, 83.3%, 96.4% and 87.5% for the classes Boskoop, Braeburn, Cripps Pink, Elstar, Gala and Jonagold, respectively.
Although the individual classes here are smaller than for the analyses shown above, there is no discernible influence of the corresponding class sizes on performance. The results here are therefore very different from those of the NMR analysis, where a clear influence was observed in this respect [13]. It can be concluded that the differences in the LC-MS data between the varieties are comparatively large, resulting in a fairly clear distinction.
3.6. Analysis of the Intersection Between the Important Variables for Different Authentication Issues
In order to analyze the variables on which the respective classifications are based on, variable selection was carried out. The results are shown in Tables S2–S5 and the overlap of the selected variables is shown in the UpSet plot in Figure 2. In total, 114, 55, 20 and 18 variables are individually relevant for the classification according to taxonomic variety, geographical origin, cultivation method and regional origin, respectively. The fact that most of the variables are selected for the taxonomic variety is probably due to the fact that, comparatively, many classes are distinguished there, namely six. In general, there are hardly any variables that are relevant for several classification models. However, 20 variables are selected for both the taxonomic variety and the identification of German samples. The reason for this is probably that the German and non-German samples are not evenly distributed in terms of taxonomic variety. For example, all Jonagold samples originate from Germany, while the Gala samples originate from Germany, Italy, New Zealand, South Africa and Chile (see Table S1).
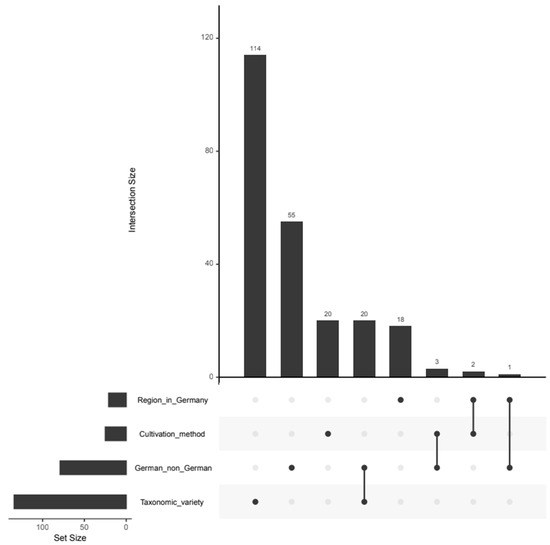
Figure 2.
UpSet Plot showing the intersection of the selected variables for the differentiation of apples regarding the regional origin within Germany, the cultivation method, the geographical origin (German or non-German) and the taxonomic variety. The bars show the number of variables selected for a single authentication issue for single dots or multiple authentication issues for connected dots.
4. Conclusions and Outlook
In this study UHPLC-Q-ToF-MS combined with BOULS data processing and RF analysis was used to differentiate apples in terms of production method, taxonomic variety, and geographical and local origin. Since high classification accuracies were achieved, especially in comparison with previously developed methods of apple authentication, and different parts of the complex data are used for the different classifications; this approach is very promising for the simultaneous analysis of different authentication issues of apples. However, as the main focus was on testing general applicability, some of the investigated groups were relatively small. In order to ensure that the full range of variation within each group is represented in the model, a larger sample size will be required for future applications. The results of this study demonstrate the general applicability of the BOULS approach to data processing, as the algorithm has been applied here in a different laboratory and to data from a different instrument manufacturer than when the approach was developed. Whether a model transfer, i.e., the application of a model developed in another laboratory, is also possible should be analyzed in a future study.
Nevertheless, this work provides the basis for the long-term application of LC-MS, e.g., in commercial or public laboratories, for the authentication of apples at different levels and other food.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/foods14152655/s1, Figure S1: Representative chromatograms of samples with the properties (a) German, Jonagold and organically produced, (b) non-German, Cripps Pink and conventionally produced, (c) non-German, Braeburn and conventionally produced, (d) non-German, Gala and conventionally produced, (e) German, Elstar and unknown production method, (f) German, Boskoop and conventionally produced; Figure S2: Results of the PCA showing the scores of the first and second principal component with colors according to the regional origin in Germany. The numbers correspond to the days on which the data was obtained; Figure S3: Results of the PCA showing the scores of the first and second principal component with colors according to the production method. The numbers correspond to the days on which the data was obtained; Table S1: Results of the RF model for the classification regarding the geographical origin of the apple samples; Table S2: Selected variables for the differentiation of German and non-German apple samples. The variable names indicate the start position of the buckets in retention time/mass level dimension; Table S3: Selected variables for the differentiation of apple samples from different origins within Germany. The variable names indicate the start position of the buckets in retention time/mass level dimension; Table S4: Selected variables for the differentiation of the different production methods. The variable names indicate the start position of the buckets in retention time/mass level dimension; Table S5: Selected variables for the differentiation according to the taxonomic variety of the apple samples. The variable names indicate the start position of the buckets in retention time/mass level dimension.
Author Contributions
Conceptualization, I.F., C.K. and S.S.; methodology, S.S., C.K., J.H. and I.F.; software, J.H.; validation, J.H.; formal analysis, I.F. and J.H.; investigation, I.F. and R.S.; resources, S.S. and I.F.; data curation, J.H.; writing—original draft preparation, J.H.; writing—review and editing, S.S.; visualization, J.H.; supervision, S.S.; project administration, S.S. and I.F.; funding acquisition, S.S. All authors have read and agreed to the published version of the manuscript.
Funding
We acknowledge financial support from the Open Access Publication Fund of Universität Hamburg.
Data Availability Statement
The data sets analyzed during the current study are not publicly available. However, the BOULS approach is published in an R package here: https://github.com/AGSeifert/BOULS (accessed on 9 January 2025, requires Linux OS) and example data are provided here: https://www.fdr.uni-hamburg.de/record/13583 (accessed on 17 July 2025).
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Schaffner, D.; Demarmels, S.; Juettner, U. Promoting Biodiversity: Do Consumers Prefer Feelings, Facts, Advice or Appeals? J. Consum. Mark. 2015, 32, 266–277. [Google Scholar] [CrossRef]
- Tulloch, A.I.T.; Miller, A.; Dean, A.J. Does Scientific Interest in the Nature Impacts of Food Align with Consumer Information-Seeking Behavior? Sustain. Sci. 2021, 16, 1029–1043. [Google Scholar] [CrossRef]
- Denver, S.; Jensen, J.D. Consumer Preferences for Organically and Locally Produced Apples. Food Qual. Prefer. 2014, 31, 129–134. [Google Scholar] [CrossRef]
- SINUS-Instritut; YouGov. Studie zum Apfel: Die Hälfte Hat Schon Einmal Äpfel vom Nachbarsbaum Gepflückt; SINUS Markt- und Sozialforschung GmbH: Heidelberg, Germany, 2017. [Google Scholar]
- Kaeswurm, J.A.H.; Neuwald, D.A.; Straub, L.V.; Buchweitz, M. Impact of Cultivation and Storage Conditions on Total Mal d 1 Content and Isoallergen Profile in Apples. J. Agric. Food Chem. 2023, 71, 12975–12985. [Google Scholar] [CrossRef]
- Kaeswurm, J.A.H.; Straub, L.V.; Siegele, A.; Brockmeyer, J.; Buchweitz, M. Characterization and Quantification of Mal d 1 Isoallergen Profiles and Contents in Traditional and Commercial Apple Varieties by Mass Spectrometry. J. Agric. Food Chem. 2023, 71, 2554–2565. [Google Scholar] [CrossRef] [PubMed]
- Müller, M.-S.; Oest, M.; Scheffler, S.; Horns, A.L.; Paasch, N.; Bachmann, R.; Fischer, M. Food Authentication Goes Green: Method Optimization for Origin Discrimination of Apples Using Apple Juice and ICP-MS. Foods 2024, 13, 3783. [Google Scholar] [CrossRef] [PubMed]
- Song, W.; Jiang, N.; Wang, H.; Guo, G. Evaluation of Machine Learning Methods for Organic Apple Authentication Based on Diffraction Grating and Image Processing. J. Food Compos. Anal. 2020, 88, 103437. [Google Scholar] [CrossRef]
- Barberis, E.; Amede, E.; Dondero, F.; Marengo, E.; Manfredi, M. New Non-Invasive Method for the Authentication of Apple Cultivars. Foods 2021, 11, 89. [Google Scholar] [CrossRef]
- Dettmer, K.; Aronov, P.A.; Hammock, B.D. Mass Spectrometry-Based Metabolomics: MASS SPECTROMETRY-BASED METABOLOMICS. Mass Spectrom. Rev. 2007, 26, 51–78. [Google Scholar] [CrossRef]
- Cubero-Leon, E.; Peñalver, R.; Maquet, A. Review on Metabolomics for Food Authentication. Food Res. Int. 2014, 60, 95–107. [Google Scholar] [CrossRef]
- Creydt, M.; Fischer, M. Food Phenotyping: Recording and Processing of Non-Targeted Liquid Chromatography Mass Spectrometry Data for Verifying Food Authenticity. Molecules 2020, 25, 3972. [Google Scholar] [CrossRef] [PubMed]
- Wenck, S.; Bachmann, R.; Barmbold, S.-M.; Horns, A.L.; Paasch, N.; Seifert, S. Authentication of Apples (Malus × Domestica Borkh.) According to Geographical Origin, Variety and Production Method Using 1H NMR Spectroscopy and Random Forest. Food Control 2025, 167, 110817. [Google Scholar] [CrossRef]
- Emwas, A.-H.; Roy, R.; McKay, R.T.; Tenori, L.; Saccenti, E.; Gowda, G.A.N.; Raftery, D.; Alahmari, F.; Jaremko, L.; Jaremko, M.; et al. NMR Spectroscopy for Metabolomics Research. Metabolites 2019, 9, 123. [Google Scholar] [CrossRef] [PubMed]
- Martinello, M.; Stella, R.; Baggio, A.; Biancotto, G.; Mutinelli, F. LC-HRMS-Based Non-Targeted Metabolomics for the Assessment of Honey Adulteration with Sugar Syrups: A Preliminary Study. Metabolites 2022, 12, 985. [Google Scholar] [CrossRef]
- Hansen, J.; Kunert, C.; Raezke, K.-P.; Seifert, S. Detection of Sugar Syrups in Honey Using Untargeted Liquid Chromatography–Mass Spectrometry and Chemometrics. Metabolites 2024, 14, 633. [Google Scholar] [CrossRef]
- Schütz, D.; Achten, E.; Creydt, M.; Riedl, J.; Fischer, M. Non-Targeted LC-MS Metabolomics Approach towards an Authentication of the Geographical Origin of Grain Maize (Zea mays L.) Samples. Foods 2021, 10, 2160. [Google Scholar] [CrossRef]
- Creydt, M.; Hudzik, D.; Rurik, M.; Kohlbacher, O.; Fischer, M. Food Authentication: Small-Molecule Profiling as a Tool for the Geographic Discrimination of German White Asparagus. J. Agric. Food Chem. 2018, 66, 13328–13339. [Google Scholar] [CrossRef]
- Klockmann, S.; Reiner, E.; Bachmann, R.; Hackl, T.; Fischer, M. Food Fingerprinting: Metabolomic Approaches for Geographical Origin Discrimination of Hazelnuts (Corylus Avellana) by UPLC-QTOF-MS. J. Agric. Food Chem. 2016, 64, 9253–9262. [Google Scholar] [CrossRef]
- Lösel, H.; Brockelt, J.; Gärber, F.; Teipel, J.; Kuballa, T.; Seifert, S.; Fischer, M. Comparative Analysis of LC-ESI-IM-qToF-MS and FT-NIR Spectroscopy Approaches for the Authentication of Organic and Conventional Eggs. Metabolites 2023, 13, 882. [Google Scholar] [CrossRef]
- Wenck, S.; Creydt, M.; Hansen, J.; Gärber, F.; Fischer, M.; Seifert, S. Opening the Random Forest Black Box of the Metabolome by the Application of Surrogate Minimal Depth. Metabolites 2022, 12, 5. [Google Scholar] [CrossRef]
- Dinis, K.; Tsamba, L.; Thomas, F.; Jamin, E.; Camel, V. Preliminary Authentication of Apple Juices Using Untargeted UHPLC-HRMS Analysis Combined to Chemometrics. Food Control 2022, 139, 109098. [Google Scholar] [CrossRef]
- Wang, J.; Chow, W. Study of Ultrahigh-Performance Liquid Chromatography Electrospray Ionization Q-Orbitrap Mass Spectrometry and Various Extraction Methods for Fingerprinting and Identification of Molecular Authenticity Markers in Apple and Grape Juices. ACS Food Sci. Technol. 2022, 2, 1326–1338. [Google Scholar] [CrossRef]
- Hansen, J.; Kunert, C.; Münstermann, H.; Raezke, K.-P.; Seifert, S. Application of Untargeted Liquid Chromatography-Mass Spectrometry to Routine Analysis of Food Using Three-Dimensional Bucketing and Machine Learning. Sci. Rep. 2024, 14, 16594. [Google Scholar] [CrossRef] [PubMed]
- Han, W.; Li, L. Evaluating and Minimizing Batch Effects in Metabolomics. Mass Spectrom. Rev. 2022, 41, 421–442. [Google Scholar] [CrossRef] [PubMed]
- Breiman, L. Random Forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef]
- Zhang, C.; Ma, Y. (Eds.) Ensemble Machine Learning: Methods and Applications; Springer: New York, NY, USA, 2012; ISBN 978-1-4419-9325-0. [Google Scholar]
- Lim, D.K.; Long, N.P.; Mo, C.; Dong, Z.; Lim, J.; Kwon, S.W. Optimized Mass Spectrometry-Based Metabolite Extraction and Analysis for the Geographical Discrimination of White Rice (Oryza sativa L.): A Method Comparison Study. J. AOAC Int. 2018, 101, 498–506. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2023. [Google Scholar]
- Rainer, J. Metabolomics Data Pre-Processing Using Xcms. 2020. Available online: https://jorainer.github.io/metabolomics2018/xcms-preprocessing.html (accessed on 9 January 2025).
- Chambers, M.C.; Maclean, B.; Burke, R.; Amodei, D.; Ruderman, D.L.; Neumann, S.; Gatto, L.; Fischer, B.; Pratt, B.; Egertson, J.; et al. A Cross-Platform Toolkit for Mass Spectrometry and Proteomics. Nat. Biotechnol. 2012, 30, 918–920. [Google Scholar] [CrossRef]
- Martens, L.; Chambers, M.; Sturm, M.; Kessner, D.; Levander, F.; Shofstahl, J.; Tang, W.H.; Römpp, A.; Neumann, S.; Pizarro, A.D.; et al. mzML—A Community Standard for Mass Spectrometry Data. Mol. Cell. Proteom. 2011, 10, R110.000133. [Google Scholar] [CrossRef]
- Pedrioli, P.G.A.; Eng, J.K.; Hubley, R.; Vogelzang, M.; Deutsch, E.W.; Raught, B.; Pratt, B.; Nilsson, E.; Angeletti, R.H.; Apweiler, R.; et al. A Common Open Representation of Mass Spectrometry Data and Its Application to Proteomics Research. Nat. Biotechnol. 2004, 22, 1459–1466. [Google Scholar] [CrossRef]
- Keller, A.; Eng, J.; Zhang, N.; Li, X.; Aebersold, R. A Uniform Proteomics MS/MS Analysis Platform Utilizing Open XML File Formats. Mol. Syst. Biol. 2005, 1, 1-8. [Google Scholar] [CrossRef]
- Gatto, L.; Lilley, K.S. MSnbase-an R/Bioconductor Package for Isobaric Tagged Mass Spectrometry Data Visualization, Processing and Quantitation. Bioinformatics 2012, 28, 288–289. [Google Scholar] [CrossRef]
- Gatto, L.; Gibb, S.; Rainer, J. MSnbase, Efficient and Elegant R-Based Processing and Visualization of Raw Mass Spectrometry Data. J. Proteome Res. 2021, 20, 1063–1069. [Google Scholar] [CrossRef]
- Benton, H.P.; Want, E.J.; Ebbels, T.M.D. Correction of Mass Calibration Gaps in Liquid Chromatography–Mass Spectrometry Metabolomics Data. Bioinformatics 2010, 26, 2488–2489. [Google Scholar] [CrossRef]
- Prince, J.T.; Marcotte, E.M. Chromatographic Alignment of ESI-LC-MS Proteomics Data Sets by Ordered Bijective Interpolated Warping. Anal. Chem. 2006, 78, 6140–6152. [Google Scholar] [CrossRef]
- Tautenhahn, R.; Böttcher, C.; Neumann, S. Highly Sensitive Feature Detection for High Resolution LC/MS. BMC Bioinform. 2008, 9, 504. [Google Scholar] [CrossRef] [PubMed]
- Sánchez, J.; Mardia, K.V.; Kent, J.M. BIBBY: Multivariate Analysis. Academic Press, London-New York-Toronto-Sydney-San Francisco 1979. xv, 518 pp., $ 61.00. Biom. J 1982, 24, 502. [Google Scholar] [CrossRef]
- Becker, R.M.; Chambers, J.M.; Wilks, A.R. The New S Language Data Analysis: A Programming Environment for Data Analysis and Graphics; The Wadsworth & Brooks/Cole statistics, probability series; Wadsworth & Brooks/Cole: Pacific Grove, CA, USA, 1988; ISBN 978-0-534-09193-4. [Google Scholar]
- Venables, W.N.; Ripley, B.D. Modern Applied Statistics with S, 4th ed.; Springer: New York, NY, USA, 2002. [Google Scholar]
- Wickham, H. Ggplot2; Use R! Springer International Publishing: Cham, Switzerland, 2016; ISBN 978-3-319-24275-0. [Google Scholar]
- Wright, M.N.; Ziegler, A. Ranger: A Fast Implementation of Random Forests for High Dimensional Data in C++ and R. J. Stat. Soft. 2017, 77, 1–17. [Google Scholar] [CrossRef]
- Degenhardt, F.; Seifert, S.; Szymczak, S. Evaluation of Variable Selection Methods for Random Forests and Omics Data Sets. Brief. Bioinform. 2019, 20, 492–503. [Google Scholar] [CrossRef]
- Kursa, M.B.; Rudnicki, W.R. Feature Selection with the Boruta Package. J. Stat. Soft. 2010, 36, 1–13. [Google Scholar] [CrossRef]
- Zelena, E.; Dunn, W.B.; Broadhurst, D.; Francis-McIntyre, S.; Carroll, K.M.; Begley, P.; O’Hagan, S.; Knowles, J.D.; Halsall, A.; HUSERMET Consortium; et al. Development of a Robust and Repeatable UPLC−MS Method for the Long-Term Metabolomic Study of Human Serum. Anal. Chem. 2009, 81, 1357–1364. [Google Scholar] [CrossRef]
- UN Comtrade. UN Comtrade Database. Available online: https://comtradeplus.un.org/TradeFlow?Frequency=A&Flows=M&CommodityCodes=080810&Partners=276&Reporters=all&period=2024&AggregateBy=none&BreakdownMode=plus (accessed on 6 April 2025).
- Schimmel, J.; Gentsch, N.; Boy, J.; Uteau, D.; Rohr, A.-D.; Winkelmann, T.; Busnena, B.; Liu, B.; Krueger, J.; Kaufhold, S.; et al. Alleviation of Apple Replant Disease in Sandy Soils by Clay Amendments. Silicon 2024, 16, 4343–4360. [Google Scholar] [CrossRef]
- Creydt, M.; Fischer, M. Omics Approaches for Food Authentication. Electrophoresis 2018, 39, 1569–1581. [Google Scholar] [CrossRef]
- Richter, B.; Gurk, S.; Wagner, D.; Bockmayr, M.; Fischer, M. Food Authentication: Multi-Elemental Analysis of White Asparagus for Provenance Discrimination. Food Chem. 2019, 286, 475–482. [Google Scholar] [CrossRef]
- Von Wuthenau, K.; Segelke, T.; Müller, M.-S.; Behlok, H.; Fischer, M. Food Authentication of Almonds (Prunus Dulcis Mill.). Origin Analysis with Inductively Coupled Plasma Mass Spectrometry (ICP-MS) and Chemometrics. Food Control 2022, 134, 108689. [Google Scholar] [CrossRef]
- Von Wuthenau, K.; Müller, M.-S.; Cvancar, L.; Oest, M.; Fischer, M. Food Authentication of Almonds (Prunus Dulcis Mill.). Fast Origin Analysis with Laser Ablation Inductively Coupled Plasma Mass Spectrometry and Chemometrics. J. Agric. Food Chem. 2022, 70, 5237–5244. [Google Scholar] [CrossRef]
- Segelke, T.; Von Wuthenau, K.; Kuschnereit, A.; Müller, M.-S.; Fischer, M. Origin Determination of Walnuts (Juglans regia L.) on a Worldwide and Regional Level by Inductively Coupled Plasma Mass Spectrometry and Chemometrics. Foods 2020, 9, 1708. [Google Scholar] [CrossRef] [PubMed]
- Müller, M.-S.; Springer, C.; Middendorf, E.; Cvancar, L.; Oest, M.; Fischer, M. Food Authentication Goes Green: Development of a Fast and Resource-Saving Method for Determining the Geographical Origin of Hazelnuts Using ICP-MS and Laser Ablation. J. Food Compos. Anal. 2025, 139, 107168. [Google Scholar] [CrossRef]
- Segelke, T.; Von Wuthenau, K.; Neitzke, G.; Müller, M.-S.; Fischer, M. Food Authentication: Species and Origin Determination of Truffles (Tuber spp.) by Inductively Coupled Plasma Mass Spectrometry and Chemometrics. J. Agric. Food Chem. 2020, 68, 14374–14385. [Google Scholar] [CrossRef] [PubMed]
- Bannier, H.-J. Moderne Apfelzüchtung: Genetische Verarmung und Tendenzen zur Inzucht: Vitalitätsverluste erst bei Verzicht auf Fungizideinsatz sichtbar. Erwerbs-Obstbau 2011, 52, 85–110. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).