Classification Prediction of Jujube Variety Based on Hyperspectral Imaging: A Comparative Study of Intelligent Optimization Algorithms
Abstract
1. Introduction
2. Materials and Methods
2.1. Jujube Samples
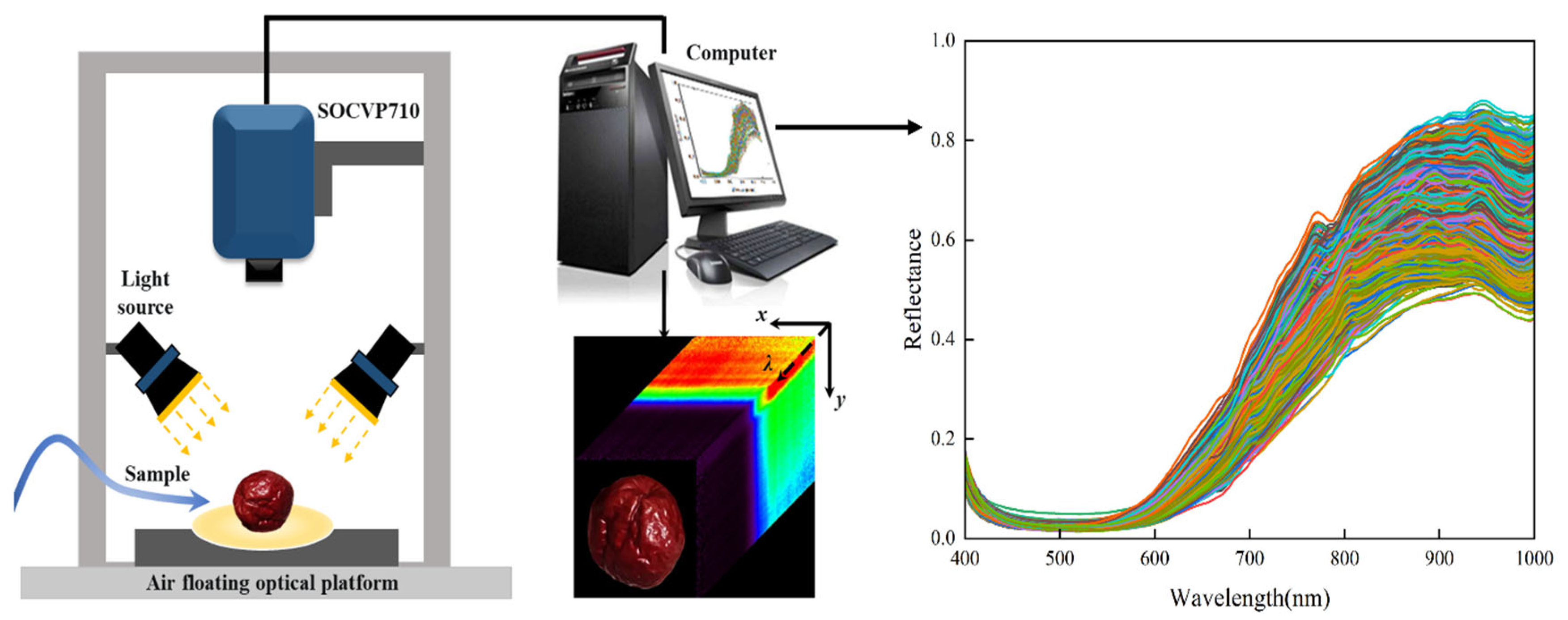
2.2. Hyperspectral Imaging System and Data Acquisition
2.3. Data Processing
2.3.1. Isolation Forest (IF) algorithm
2.3.2. Spectral Data Preprocessing
2.3.3. Characteristic Variable Selection Based on CARS
2.4. Principle and Implementation of the Algorithm
2.4.1. Support Vector Machine (SVM)
2.4.2. Dimensionality Reduction and Visualization Analysis
2.4.3. Zebra Optimization Algorithm (ZOA)
2.4.4. Genetic Algorithm (GA)
2.4.5. Particle Swarm Optimization (PSO)
2.4.6. Grey Wolf Optimizer (GWO)
2.5. Sample Splitting and Optimizer Parameter Settings
3. Results
3.1. Removal of Abnormal Data
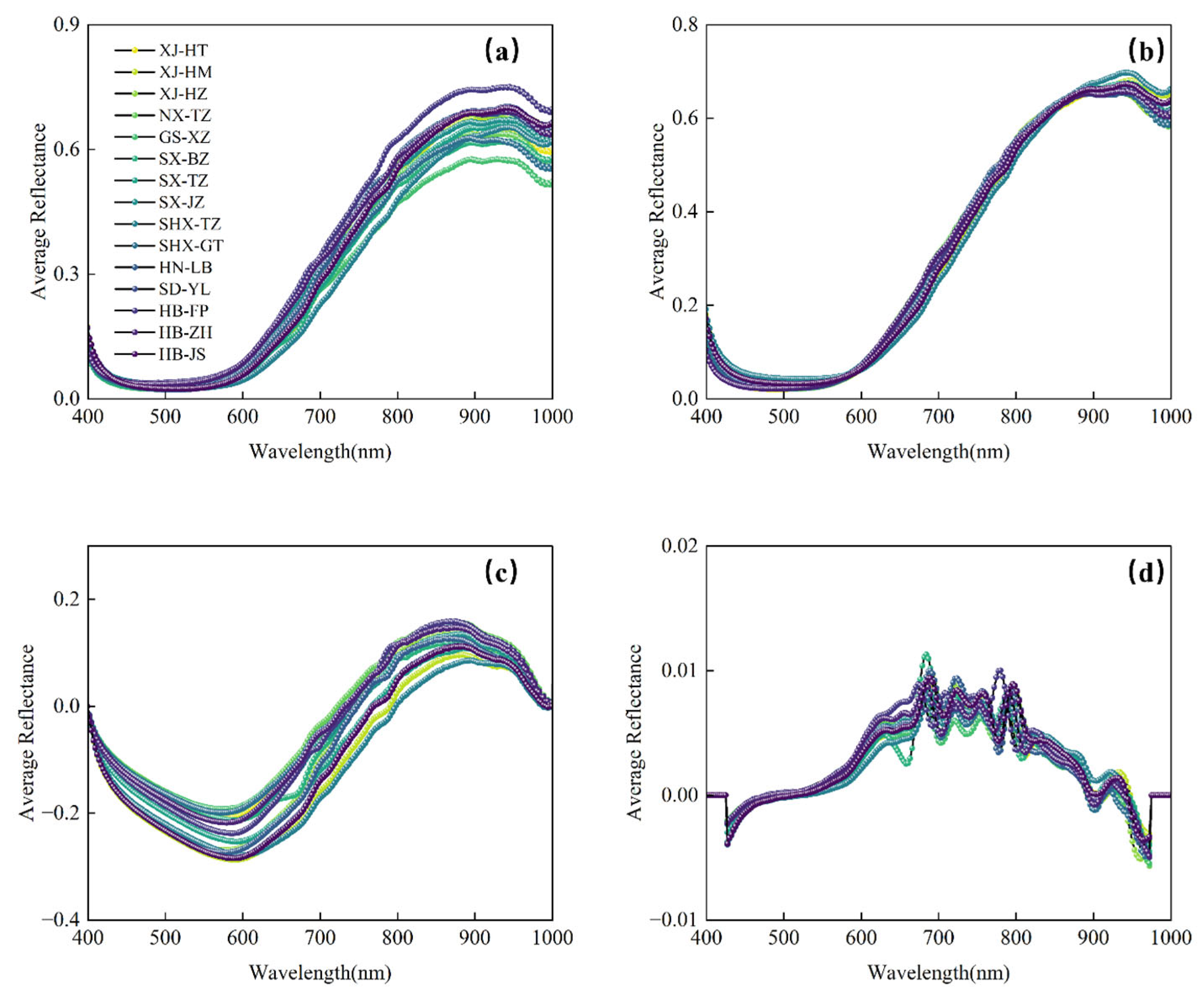
3.2. Spectral Characteristics
3.3. PCA, t-SNE, and UMAP Visualization Analysis
3.4. Results of CARS Feature Wavelength Selection
3.5. Results of Intelligent Optimization Algorithm Classification
3.5.1. Fitness Curve Analysis of Intelligent Optimization Algorithms
3.5.2. Classification Accuracy of Optimized SVM Models with Intelligent Algorithms
3.6. Confusion Matrix Analysis of SG1st and SG1st-CARS Under the GWO-SVM Model
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Liu, Y.; Liao, Y.; Guo, M.; Zhang, W.; Sang, Y.; Wang, H.; Cheng, S.; Chen, G. Comparative elucidation of bioactive and volatile components in dry mature jujube fruit (Ziziphus jujuba Mill.) subjected to different drying methods. Food Chem. X 2022, 14, 100311. [Google Scholar] [CrossRef] [PubMed]
- Wei, Y.; Liu, Q.; Fan, S.; Jiang, X.; Chen, Y.; Wang, F.; Cao, X.; Yan, L. Development of a predictive model for assessing quality of winter jujube during storage utilizing hyperspectral imaging technology. J. Food Process Eng. 2024, 47, e14688. [Google Scholar] [CrossRef]
- Zhang, L.; Wei, Y.; Liu, J.; Wu, J.; An, D. A hyperspectral band selection method based on sparse band attention network for maize seed variety identification. Expert Syst. Appl. 2024, 238, 122273. [Google Scholar] [CrossRef]
- Dong, Z.; Yang, F.; Du, J.; Wang, K.; Lv, L.; Long, W. Identification of varieties in Camellia oleifera leaf based on deep learning technology. Ind. Crops Prod. 2024, 216, 118635. [Google Scholar] [CrossRef]
- Kong, X.; Xu, M.; Wang, K.; Chen, Q.; Han, L.; Li, Q.; Guo, Q.; Wan, H.; Nie, J. Development of a comprehensive evaluation system for the sensory and nutritional quality of winter jujube (Ziziphus jujuba Mill. cv. Dongzao). LWT 2024, 194, 115777. [Google Scholar] [CrossRef]
- Wang, C.; He, W.-Z.; Zhao, D.-Y.; Liu, Z.; Fan, Y.; Tian, W.-N.; Wu, W.-L.; Rogers, K.M. Modeling of stable isotope and multi-element compositions of jujube (Ziziphus jujuba Mill.) for origin traceability of protected geographical indication (PGI) products in Xinjiang, China. J. Food Compos. Anal. 2020, 92, 103577. [Google Scholar] [CrossRef]
- Hu, Y.; Chen, C.; Liu, H.; Bai, B.; He, W.; Tian, J.; Mei, X.; Li, B.; Yu, S.; Shen, Q.; et al. Geographical origin traceability of Xiaobai apricot (Prunus armeniaca L.cv.) using stable isotope and multi-element analysis. J. Food Compos. Anal. 2025, 145, 107814. [Google Scholar] [CrossRef]
- Li, X.; Zhao, X.; Wang, S.; Wu, B.; Du, Y.; Dong, C.; Wu, Z.; Sun, F.; Wu, J.; Wang, L.; et al. Qualities and bitter substances profile of Chinese winter jujube (Ziziphus jujuba Mill. cv. Dong zao) under the ultra-low oxygen environment. Postharvest Biol. Technol. 2025, 222, 113416. [Google Scholar] [CrossRef]
- Cai, W.; Fei, L.; Zhang, D.; Ni, H.; Peng, B.; Zhao, X.; Zhang, Q.; Tang, F.; Zhang, Y.; Shan, C. Impact of ultra-high-pressure treatment on microbial community composition and flavor quality of jujube juice: Insights from high-throughput sequencing technology, intelligent bionic sensory system, and metabolomics approach. Food Res. Int. 2024, 191, 114688. [Google Scholar] [CrossRef] [PubMed]
- Yu, K.; Zhao, Y.; Li, X.; Shao, Y.; Zhu, F.; He, Y. Identification of crack features in fresh jujube using Vis/NIR hyperspectral imaging combined with image processing. Comput. Electron. Agric. 2014, 103, 1–10. [Google Scholar] [CrossRef]
- Mohd Ali, M.; Hashim, N.; Aziz, S.A.; Lasekan, O. Emerging non-destructive thermal imaging technique coupled with chemometrics on quality and safety inspection in food and agriculture. Trends Food Sci. Technol. 2020, 105, 176–185. [Google Scholar] [CrossRef]
- Guo, Z.; Zhang, Y.; Xiao, H.; Jayan, H.; Majeed, U.; Ashiagbor, K.; Jiang, S.; Zou, X. Multi-sensor fusion and deep learning for batch monitoring and real-time warning of apple spoilage. Food Control 2025, 172, 111174. [Google Scholar] [CrossRef]
- Chen, B.; Shi, B.; Gong, J.; Shi, G.; Jin, H.; Qin, T.; Yang, Z.; Lim, K.-J.; Liu, W.; Zhang, J.; et al. Quality detection and variety classification of pecan seeds using hyperspectral imaging technology combined with machine learning. J. Food Compos. Anal. 2024, 131, 106248. [Google Scholar] [CrossRef]
- Gong, J.; Dou, X.; Wang, D.; Fang, M.; Yu, L.; Ma, F.; Wang, X.; Xu, B.; Li, P.; Zhang, L. Authentication of rapeseed variety based on hyperspectral imaging and chemometrics. Appl. Food Res. 2025, 5, 100941. [Google Scholar] [CrossRef]
- Ouyang, Q.; Wang, L.; Park, B.; Kang, R.; Wang, Z.; Chen, Q.; Guo, Z. Assessment of matcha sensory quality using hyperspectral microscope imaging technology. LWT 2020, 125, 109254. [Google Scholar] [CrossRef]
- Pham, Q.T.; Lu, S.-E.; Liou, N.-S. Development of sorting and grading methodology of jujubes using hyperspectral image data. Postharvest Biol. Technol. 2025, 222, 113406. [Google Scholar] [CrossRef]
- Liu, Q.; Jiang, X.; Wang, F.; Zhu, B.; Yan, L.; Wei, Y.; Chen, Y. Detection of dried jujube from fresh jujube with different variety and maturity after hot air drying based on hyperspectral imaging technology. J. Food Compos. Anal. 2024, 133, 106378. [Google Scholar] [CrossRef]
- Liu, Z.; Kou, J.; Yan, Z.; Wang, P.; Liu, C.; Sun, C.; Shao, A.; Klein, B. Enhancing XRF sensor-based sorting of porphyritic copper ore using particle swarm optimization-support vector machine (PSO-SVM) algorithm. Int. J. Min. Sci. Technol. 2024, 34, 545–556. [Google Scholar] [CrossRef]
- Zhang, S.; Yin, Y.; Liu, C.; Li, J.; Sun, X.; Wu, J. Discrimination of wheat flour grade based on PSO-SVM of hyperspectral technique. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2023, 302, 123050. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Yu, C.; Ma, Y.; Zhang, H.; Yan, L.; Fan, S. Prediction of Key Quality Parameters in Hot Air-Dried Jujubes Based on Hyperspectral Imaging. Foods 2025, 14, 1855. [Google Scholar] [CrossRef] [PubMed]
- Jiang, X.; Liu, Q.; Yan, L.; Cao, X.; Chen, Y.; Wei, Y.; Wang, F.; Xing, H. Hyperspectral imaging combined with spectral-imagery feature fusion convolutional neural network to discriminate different geographical origins of wolfberries. J. Food Compos. Anal. 2024, 132, 106259. [Google Scholar] [CrossRef]
- Liu, F.T.; Ting, K.M.; Zhou, Z.-H. Isolation Forest. In Proceedings of the 2008 Eighth IEEE International Conference on Data Mining (ICDM), Pisa, Italy, 15–19 December 2008; pp. 413–422. [Google Scholar] [CrossRef]
- Marcelli, E.; Barbariol, T.; Sartor, D.; Susto, G.A. Active Learning-based Isolation Forest (ALIF): Enhancing anomaly detection with expert feedback. Inf. Sci. 2024, 678, 121012. [Google Scholar] [CrossRef]
- Li, J.; Luo, W.; Han, L.; Cai, Z.; Guo, Z. Two-wavelength image detection of early decayed oranges by coupling spectral classification with image processing. J. Food Compos. Anal. 2022, 111, 104642. [Google Scholar] [CrossRef]
- Li, X.; Wu, J.; Bai, T.; Wu, C.; He, Y.; Huang, J.; Li, X.; Shi, Z.; Hou, K. Variety classification and identification of jujube based on near-infrared spectroscopy and 1D-CNN. Comput. Electron. Agric. 2024, 223, 109122. [Google Scholar] [CrossRef]
- Xi, Q.; Chen, Q.; Ahmad, W.; Pan, J.; Zhao, S.; Xia, Y.; Ouyang, Q.; Chen, Q. Quantitative analysis and visualization of chemical compositions during shrimp flesh deterioration using hyperspectral imaging: A comparative study of machine learning and deep learning models. Food Chem. 2025, 481, 143997. [Google Scholar] [CrossRef] [PubMed]
- Guo, Z.; Wang, M.; Barimah, A.O.; Chen, Q.; Li, H.; Shi, J.; El-Seedi, H.R.; Zou, X. Label-free surface enhanced Raman scattering spectroscopy for discrimination and detection of dominant apple spoilage fungus. Int. J. Food Microbiol. 2021, 338, 108990. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Jiang, X.; Wang, F.; Fan, S.; Zhu, B.; Yan, L.; Chen, Y.; Wei, Y.; Chen, W. Evaluation and process monitoring of jujube hot air drying using hyperspectral imaging technology and deep learning for quality parameters. Food Chem. 2025, 467, 141999. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Ouyang, Q.; Park, B.; Kang, R.; Wang, Z.; Wang, L.; Chen, Q. Physicochemical indicators coupled with multivariate analysis for comprehensive evaluation of matcha sensory quality. Food Chem. 2022, 371, 131100. [Google Scholar] [CrossRef] [PubMed]
- Tang, Y.; Wang, F.; Zhao, X.; Yang, G.; Xu, B.; Zhang, Y.; Xu, Z.; Yang, H.; Yan, L.; Li, L. A nondestructive method for determination of green tea quality by hyperspectral imaging. J. Food Compos. Anal. 2023, 123, 105621. [Google Scholar] [CrossRef]
- Li, S.; Zhao, Z.; Miao, T.; Li, X.; Yu, S.; Zhang, Y.; Wei, J.; Zou, K. Discrimination of Pb-Zn deposit types using the trace element data of galena based on deep learning. Ore Geol. Rev. 2024, 170, 106133. [Google Scholar] [CrossRef]
- Trojovska, E.; Dehghani, M.; Trojovsky, P. Zebra Optimization Algorithm: A New Bio-Inspired Optimization Algorithm for Solving Optimization Algorithm. IEEE Access 2022, 10, 49445–49473. [Google Scholar] [CrossRef]
- Ma, Z.; Li, X.; Sun, J. A data-driven fault detection approach for unknown large-scale systems based on GA-SVM. Inf. Sci. 2024, 658, 120023. [Google Scholar] [CrossRef]
- Priya, G.V.; Ganguly, S. Multi-swarm surrogate model assisted PSO algorithm to minimize distribution network energy losses. Appl. Soft Comput. 2024, 159, 111616. [Google Scholar] [CrossRef]
- Mirjalili, S.; Mirjalili, S.M.; Lewis, A. Grey Wolf Optimizer. Adv. Eng. Softw. 2014, 69, 46–61. [Google Scholar] [CrossRef]
- Ouyang, Q.; Chang, H.; Fan, Z.; Ma, S.; Chen, Q.; Liu, Z. Monitoring changes in constituents during black tea fermentation using snapshot multispectral imaging and 1D-CNN enhanced with data augmentation. Comput. Electron. Agric. 2025, 237, 110643. [Google Scholar] [CrossRef]
- Wei, Y.; Yuan, M.; Hu, H.; Xu, H.; Mao, X. Estimation for soluble solid content in Hetian jujube using hyperspectral imaging with fused spectral and textural Features. J. Food Compos. Anal. 2024, 128, 106079. [Google Scholar] [CrossRef]
- Li, Y.; Ma, B.; Li, C.; Yu, G. Accurate prediction of soluble solid content in dried Hami jujube using SWIR hyperspectral imaging with comparative analysis of models. Comput. Electron. Agric. 2022, 193, 106655. [Google Scholar] [CrossRef]
- Qi, Z.; Wu, X.; Yang, Y.; Wu, B.; Fu, H. Discrimination of the Red Jujube Varieties Using a Portable NIR Spectrometer and Fuzzy Improved Linear Discriminant Analysis. Foods 2022, 11, 763. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Yang, Z.; Yang, Y.; Wu, B.; Sun, J. Geographical Origin Identification of Chinese Red Jujube Using Near-Infrared Spectroscopy and Adaboost-CLDA. Foods 2025, 14, 803. [Google Scholar] [CrossRef] [PubMed]
- Cui, T.; Chen, H.; Li, J.; Zhou, J.; Han, L.; Tian, X.; He, F.; Chen, X.; Wang, H. A novel strategy for rapid quantification of multiple quality markers and authenticity identification based on near-infrared spectroscopy and machine learning algorithms, Fructus Gardeniae as a case study. Microchem. J. 2025, 209, 112697. [Google Scholar] [CrossRef]
- Zhang, H.; Li, G.; Qin, C.; Xu, C.; Li, Z.; Yin, X.; Chen, T.; Wang, Y.; Wang, K. Rapid detection of Pu-erh tea vintage by data fusion strategy based on Terahertz and Raman Spectral technology. Infrared Phys. Technol. 2025, 147, 105803. [Google Scholar] [CrossRef]







Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, Q.; Zhou, J.; Wu, Z.; Ma, D.; Ma, Y.; Fan, S.; Yan, L. Classification Prediction of Jujube Variety Based on Hyperspectral Imaging: A Comparative Study of Intelligent Optimization Algorithms. Foods 2025, 14, 2527. https://doi.org/10.3390/foods14142527
Liu Q, Zhou J, Wu Z, Ma D, Ma Y, Fan S, Yan L. Classification Prediction of Jujube Variety Based on Hyperspectral Imaging: A Comparative Study of Intelligent Optimization Algorithms. Foods. 2025; 14(14):2527. https://doi.org/10.3390/foods14142527
Chicago/Turabian StyleLiu, Quancheng, Jun Zhou, Zhaoyi Wu, Didi Ma, Yuxuan Ma, Shuxiang Fan, and Lei Yan. 2025. "Classification Prediction of Jujube Variety Based on Hyperspectral Imaging: A Comparative Study of Intelligent Optimization Algorithms" Foods 14, no. 14: 2527. https://doi.org/10.3390/foods14142527
APA StyleLiu, Q., Zhou, J., Wu, Z., Ma, D., Ma, Y., Fan, S., & Yan, L. (2025). Classification Prediction of Jujube Variety Based on Hyperspectral Imaging: A Comparative Study of Intelligent Optimization Algorithms. Foods, 14(14), 2527. https://doi.org/10.3390/foods14142527





