Investigating the Microbial Dynamics of Hermetia illucens Powder Throughout Rearing and Processing: An Integrated Approach Using Cultural and Metabarcoding Methods
Abstract
1. Introduction
2. Materials and Methods
2.1. Systematic Review of the Microbial Diversity of BSF Larvae, Substrate, and Frass
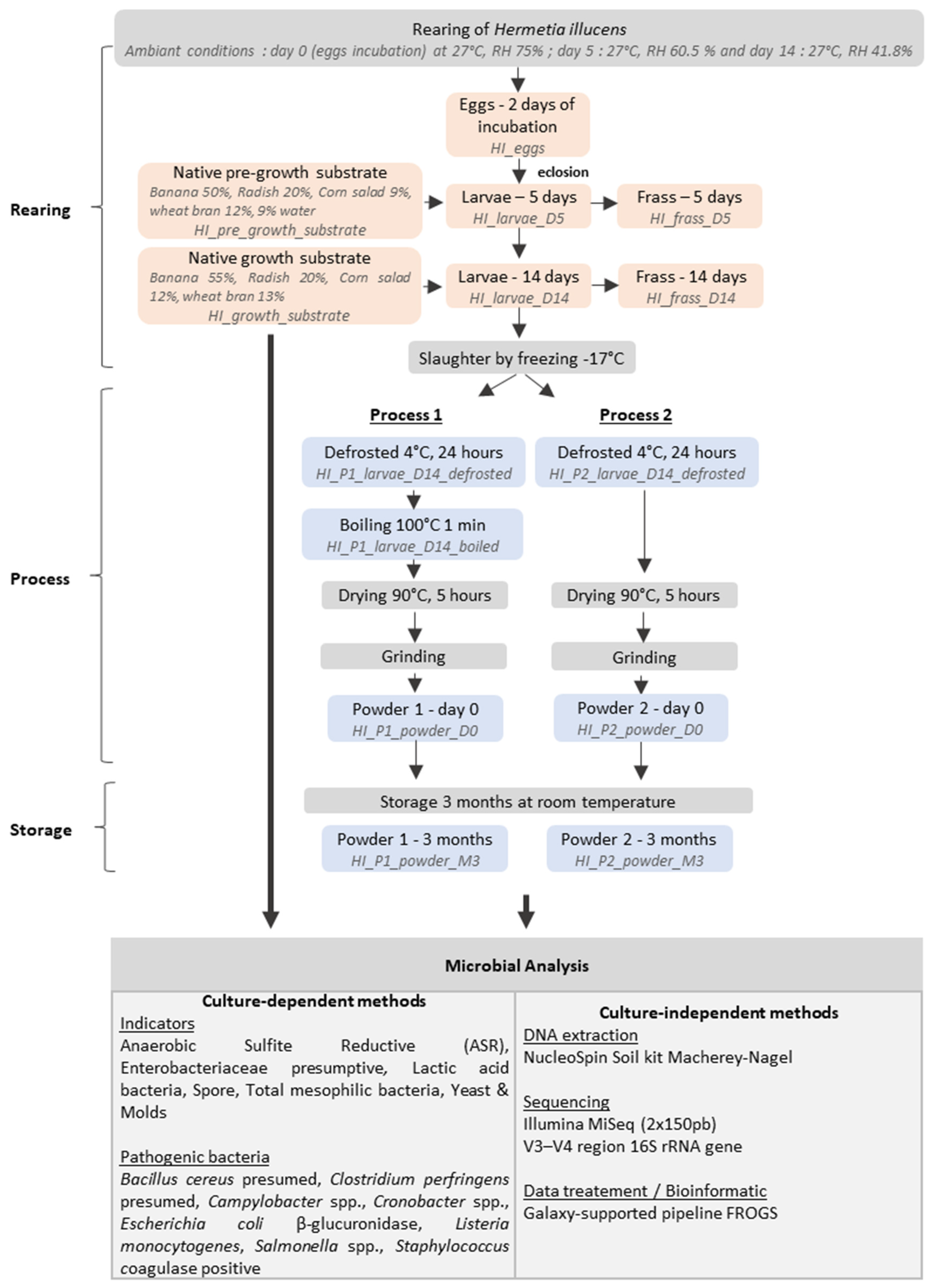
2.2. Experimental Design
2.3. Rearing Conditions Measurement
2.4. Microbial Enumeration
2.5. DNA Extraction and Sequencing of Bacterial rRNA 16S Amplicons
2.6. Bioinformatic and Statistical Analyses
3. Results
3.1. Systematic Review on the Microbial Diversity of BSF
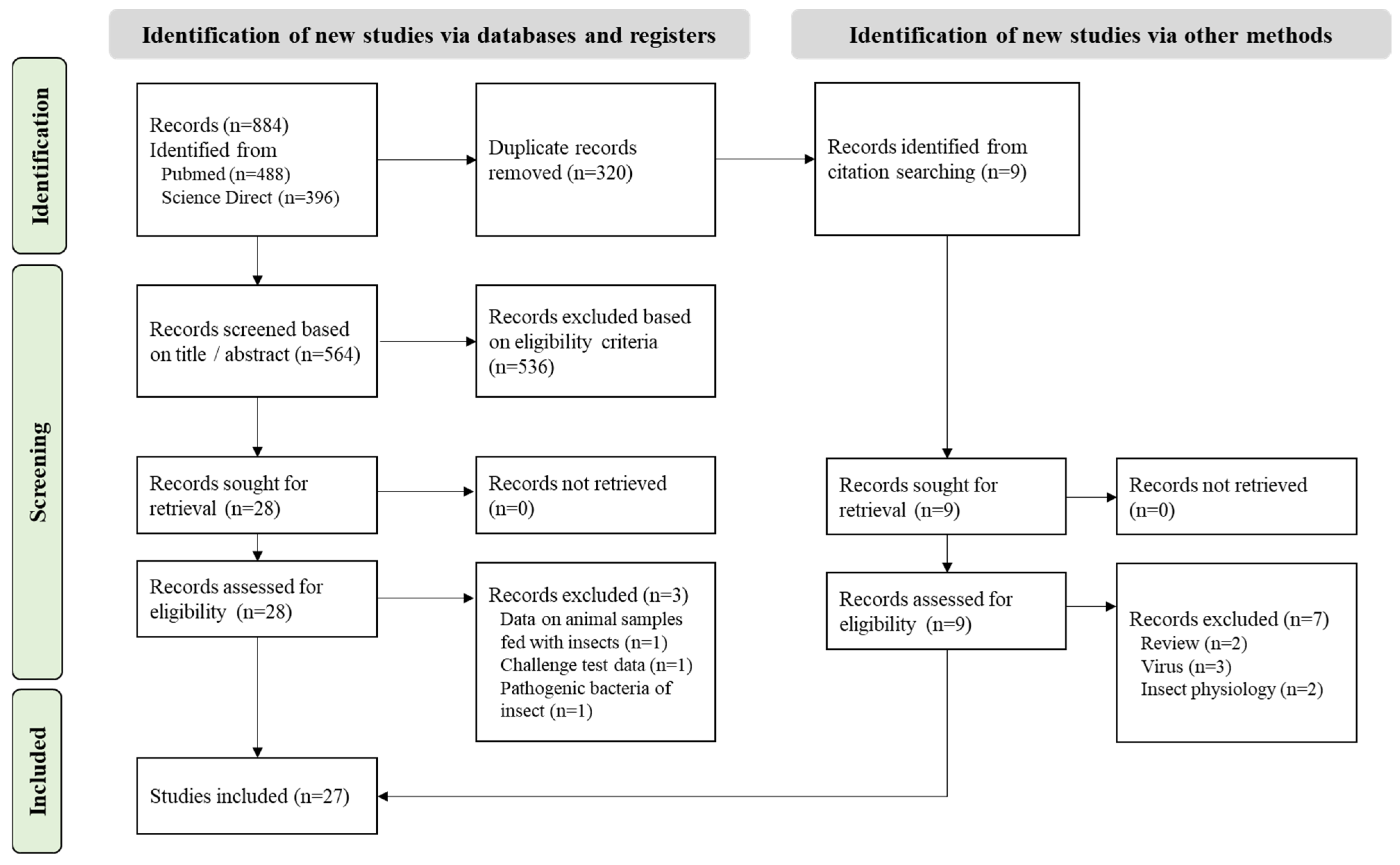
3.1.1. Study Selected
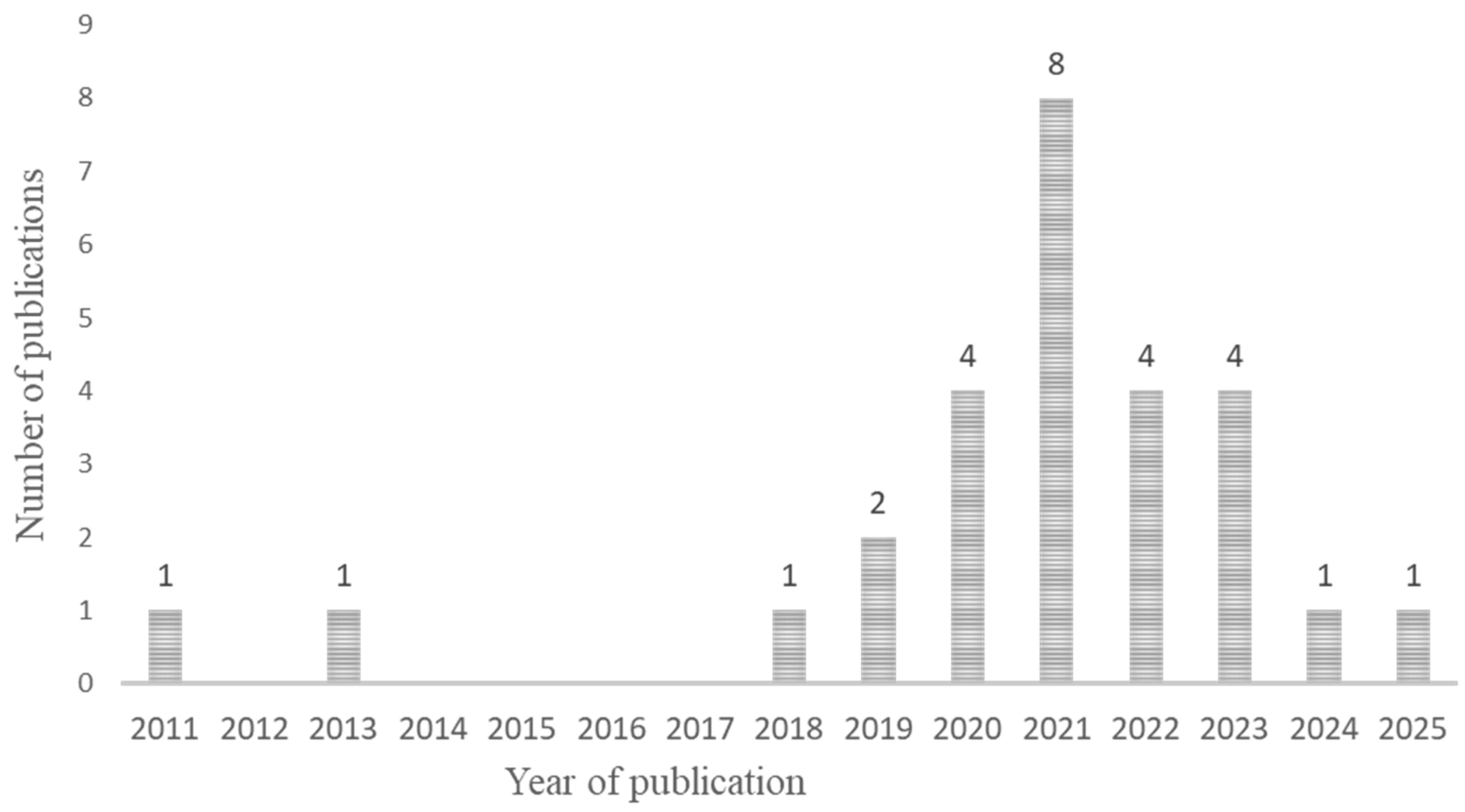
3.1.2. Study Description
3.1.3. Microbial Profile of BSF Already Published
| Acinetobacter | Actinomyces | Bacillus | Bacteroides | Burkholderia | Campylobacter | Clostridium | Corynebacterium | Dysgonomonas | Enterobacter | Enterobacteriaceae | Enterococcus | Escherichia | Escherichia-Shigella | Ignatzschineria | Klebsiella | Listeria | Lysinibacillus | Morganella | Paenalcaligenes | Proteus | Providencia | Pseudomonas | Rhizobiales | Salmonella | Staphylococcus | Vagococcus | Weissella | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Ao et al., 2021 [48] | X | X | X | |||||||||||||||||||||||||
| Auger et al., 2023 [52] | X | X | X | X | X | |||||||||||||||||||||||
| Bruno et al., 2019 [51] | X | X | X | X | X | X | X | X | ||||||||||||||||||||
| Chen et al., 2023 [50] | X | X | X | X | X | |||||||||||||||||||||||
| Cifuentes et al., 2020 [53] | X | X | X | X | X | |||||||||||||||||||||||
| Cifuentes et al., 2022 [8] | X | X | X | X | X | X | X | |||||||||||||||||||||
| Gorrens et al., 2021 [30] | X | X | X | X | X | |||||||||||||||||||||||
| Gorrens et al., 2022 [47] | X | X | X | X | X | X | X | |||||||||||||||||||||
| Jeon et al., 2011 [54] | X | X | ||||||||||||||||||||||||||
| Jiang et al., 2019 [55] | X | X | X | |||||||||||||||||||||||||
| Klammsteiner et al., 2020 [56] | X | X | X | |||||||||||||||||||||||||
| Klammsteiner et al., 2021 [57] | X | X | X | X | ||||||||||||||||||||||||
| Li et al., 2023 [34] | X | X | X | X | ||||||||||||||||||||||||
| Liu et al., 2020 [58] | X | X | X | X | ||||||||||||||||||||||||
| Mašková et al., 2025 [59] | X | X | X | X | X | X | X | X | ||||||||||||||||||||
| Pei et al., 2023 [60] | X | X | X | |||||||||||||||||||||||||
| Querejeta et al., 2023 [61] | X | X | X | X | X | |||||||||||||||||||||||
| Raimondi et al., 2020 [21] | X | X | X | X | X | |||||||||||||||||||||||
| Schreven et al., 2021 [62] | X | X | X | X | ||||||||||||||||||||||||
| Schreven et al., 2022 [63] | X | X | X | X | X | X | ||||||||||||||||||||||
| Tanga et al., 2021 [64] | X | X | X | X | X | X | ||||||||||||||||||||||
| Tegtmeier et al., 2021 [35] | X | X | X | X | X | |||||||||||||||||||||||
| Van Looveren et al., 2024 [65] | X | X | X | X | X | X | X | |||||||||||||||||||||
| Wu et al., 2022 [49] | X | X | X | X | X | |||||||||||||||||||||||
| Wynants et al., 2018 [23] | X | X | X | X | X | |||||||||||||||||||||||
| Yang et al., 2021 [66] | X | X | X | X | X | X | ||||||||||||||||||||||
| Zheng et al., 2013 [67] | X | X |
3.2. Physico-Chemical Analysis of Rearing Conditions
3.3. Quantification of Microbial Indicators and Pathogens
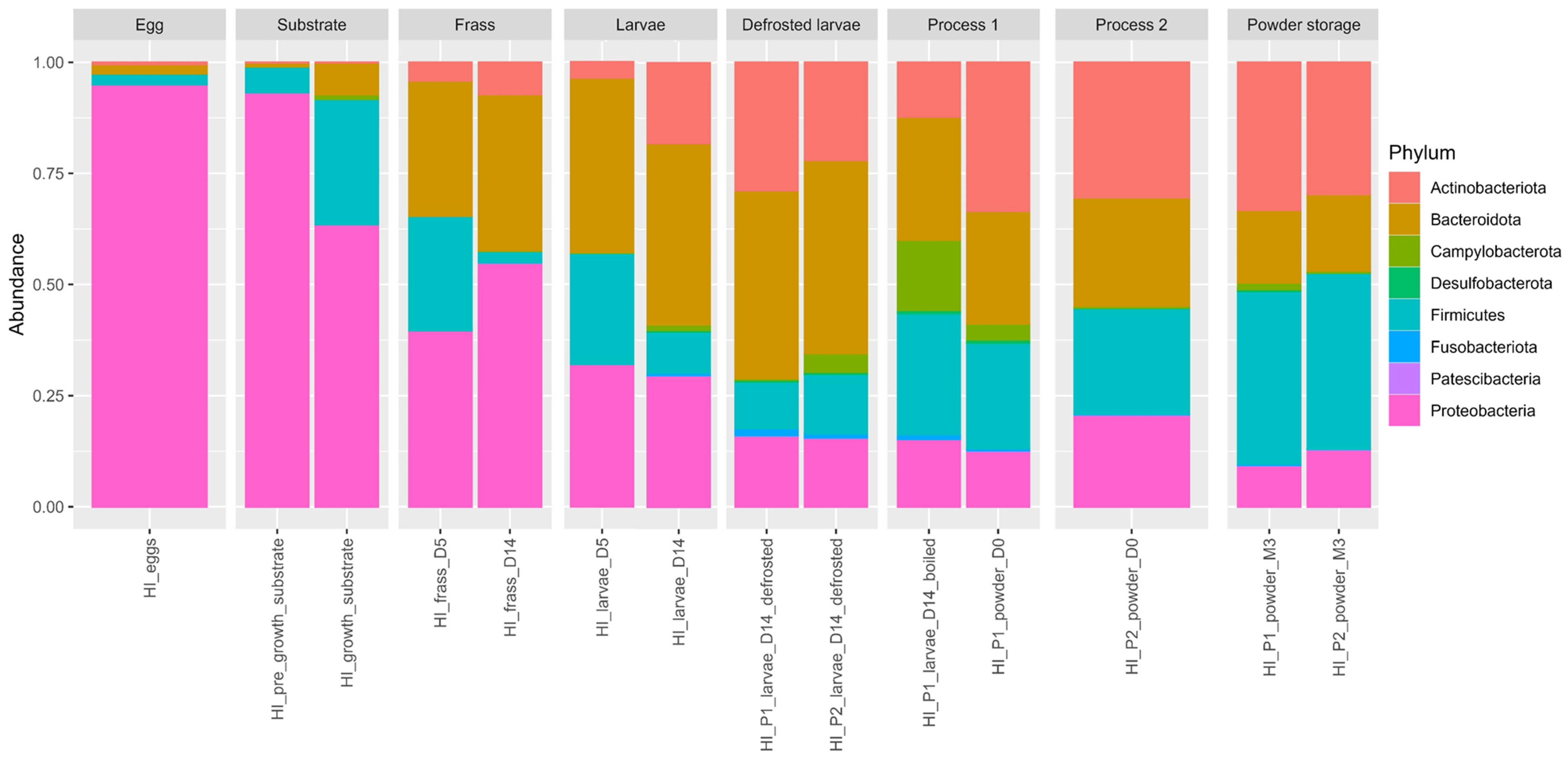
3.4. Distribution of Global Phylum Taxonomic
3.5. Taxonomic Identification of Potential Pathogenic Bacteria
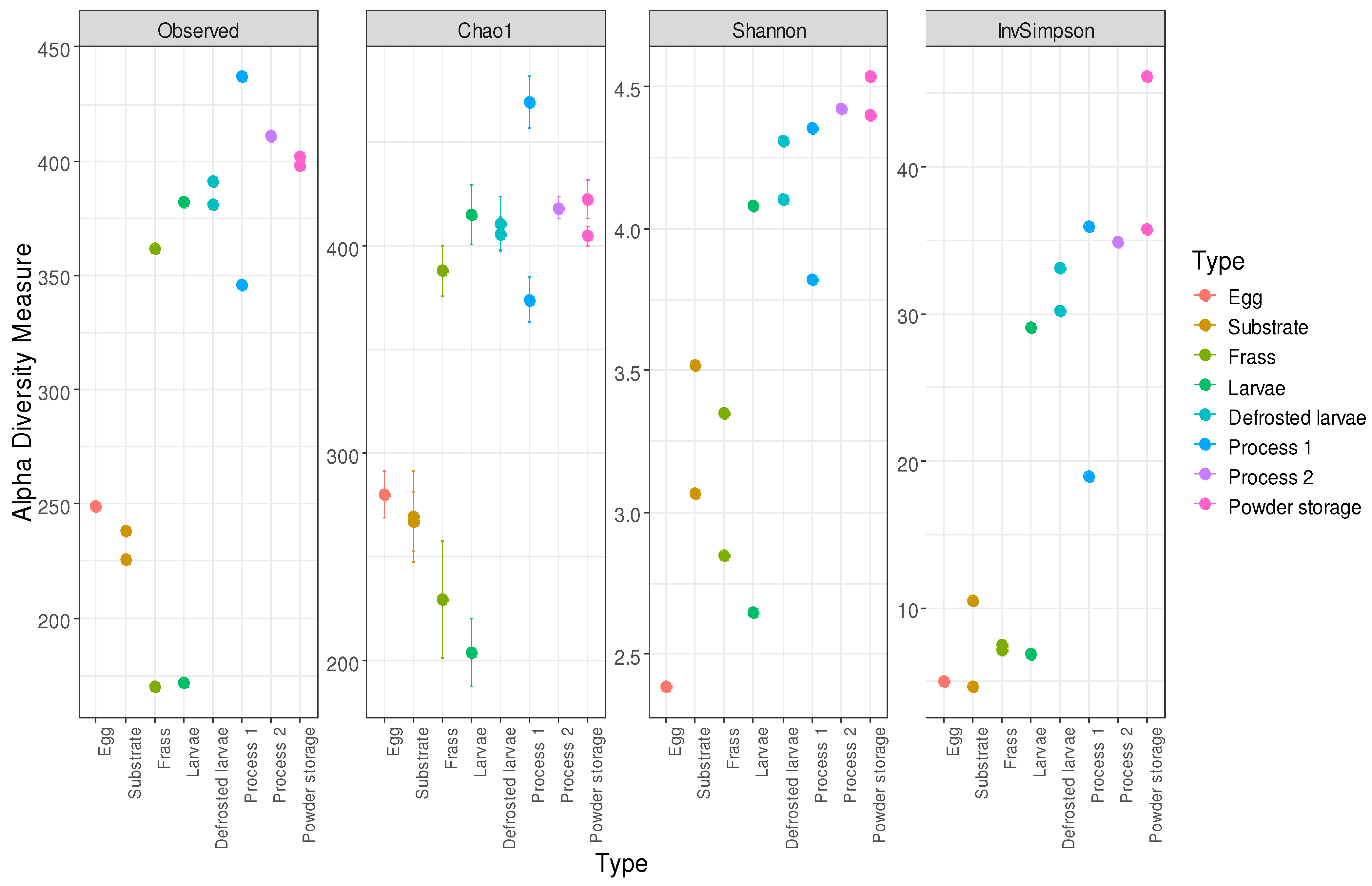
3.6. Alpha Diversity Analysis
3.7. Abundance of Microbial Communities
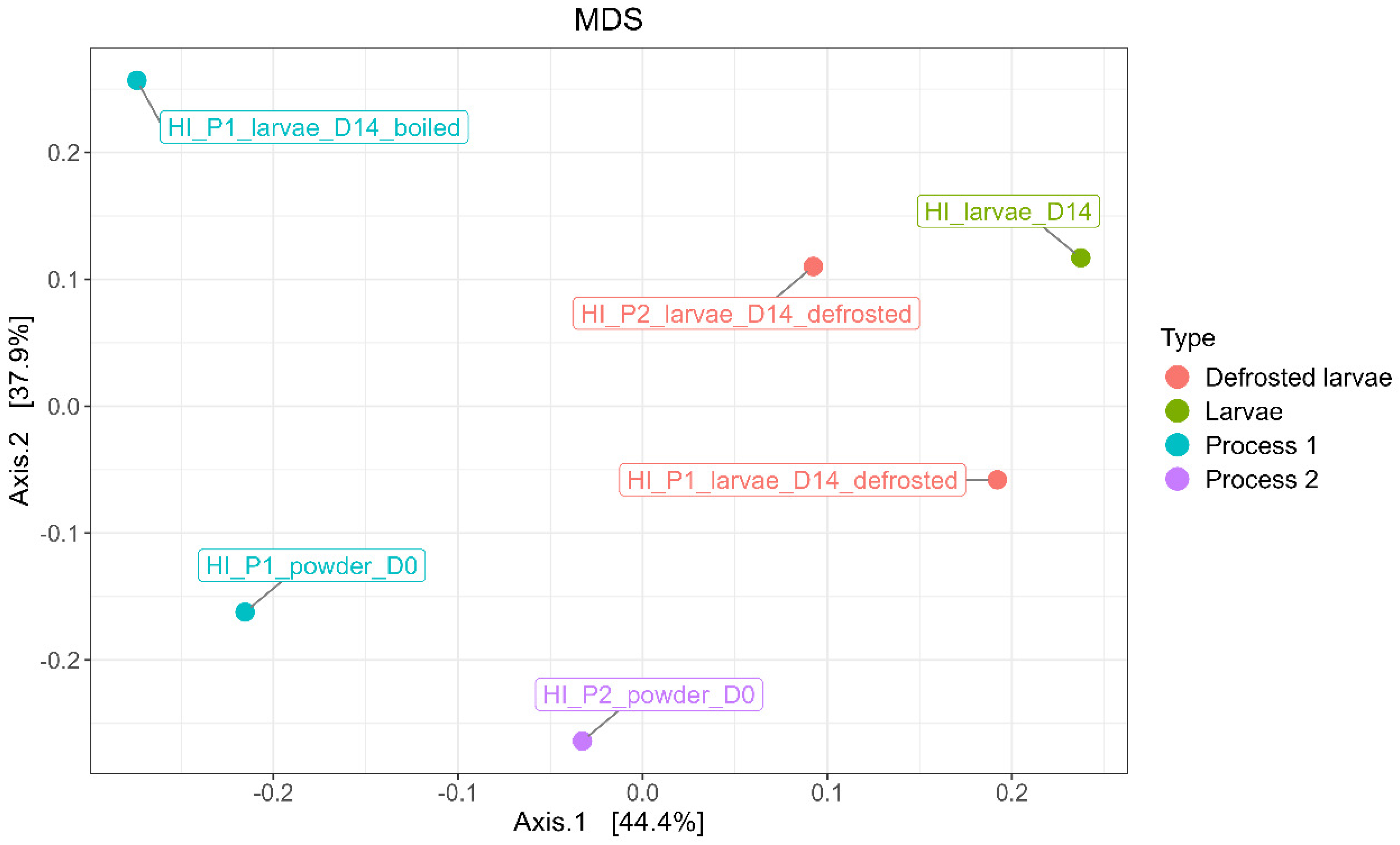
3.8. Comparison of Microbial Diversity (Process/Rearing)
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Godfray, H.C.J.; Crute, I.R.; Haddad, L.; Lawrence, D.; Muir, J.F.; Nisbett, N.; Pretty, J.; Robinson, S.; Toulmin, C.; Whiteley, R. The Future of the Global Food System. Phil. Trans. R. Soc. B 2010, 365, 2769–2777. [Google Scholar] [CrossRef] [PubMed]
- Van Der Spiegel, M.; Noordam, M.Y.; Van Der Fels-Klerx, H.J. Safety of Novel Protein Sources (Insects, Microalgae, Seaweed, Duckweed, and Rapeseed) and Legislative Aspects for Their Application in Food and Feed Production. Comp. Rev. Food Sci. Food Safe 2013, 12, 662–678. [Google Scholar] [CrossRef] [PubMed]
- Van Huis, A. Potential of Insects as Food and Feed in Assuring Food Security. Annu. Rev. Entomol. 2013, 58, 563–583. [Google Scholar] [CrossRef] [PubMed]
- Makkar, H.P.S.; Tran, G.; Heuzé, V.; Ankers, P. State-of-the-Art on Use of Insects as Animal Feed. Anim. Feed. Sci. Technol. 2014, 197, 1–33. [Google Scholar] [CrossRef]
- Gill, K.; Chenier, K.A.; Free, A.; Goff, J.; Pitchford, J.L.; Cressman, K.; Posten, M.; Brunden, E.; Shelton, M.; Swanson, K.; et al. Research Needs, Environmental Concerns, and Logistical Considerations for Incorporating Livestock Grazing into Coastal Upland Habitat Management. J. Environ. Manag. 2023, 329, 117119. [Google Scholar] [CrossRef]
- Dobermann, D.; Swift, J.A.; Field, L.M. Opportunities and Hurdles of Edible Insects for Food and Feed. Nutr. Bull. 2017, 42, 293–308. [Google Scholar] [CrossRef]
- Fernandez-Cassi, X.; Söderqvist, K.; Bakeeva, A.; Vaga, M.; Dicksved, J.; Vagsholm, I.; Jansson, A.; Boqvist, S. Microbial Communities and Food Safety Aspects of Crickets (Acheta Domesticus) Reared under Controlled Conditions. J. Insects Food Feed. 2020, 6, 429–440. [Google Scholar] [CrossRef]
- Cifuentes, Y.; Vilcinskas, A.; Kämpfer, P.; Glaeser, S.P. Isolation of Hermetia illucens Larvae Core Gut Microbiota by Two Different Cultivation Strategies. Antonie Van. Leeuwenhoek 2022, 115, 821–837. [Google Scholar] [CrossRef]
- Vandeweyer, D.; Crauwels, S.; Lievens, B.; Van Campenhout, L. Metagenetic Analysis of the Bacterial Communities of Edible Insects from Diverse Production Cycles at Industrial Rearing Companies. Int. J. Food Microbiol. 2017, 261, 11–18. [Google Scholar] [CrossRef]
- Vandeweyer, D.; Crauwels, S.; Lievens, B.; Van Campenhout, L. Microbial Counts of Mealworm Larvae (Tenebrio molitor) and Crickets (Acheta domesticus and Gryllodes sigillatus) from Different Rearing Companies and Different Production Batches. Int. J. Food Microbiol. 2017, 242, 13–18. [Google Scholar] [CrossRef]
- Stoops, J.; Crauwels, S.; Waud, M.; Claes, J.; Lievens, B.; Van Campenhout, L. Microbial Community Assessment of Mealworm Larvae (Tenebrio molitor) and Grasshoppers (Locusta Migratoria Migratorioides) Sold for Human Consumption. Food Microbiol. 2016, 53, 122–127. [Google Scholar] [CrossRef] [PubMed]
- Kooh, P.; Ververis, E.; Tesson, V.; Boué, G.; Federighi, M. Entomophagy and Public Health: A Review of Microbiological Hazards. Health 2019, 11, 1272–1290. [Google Scholar] [CrossRef]
- Garofalo, C.; Milanović, V.; Cardinali, F.; Aquilanti, L.; Clementi, F.; Osimani, A. Current Knowledge on the Microbiota of Edible Insects Intended for Human Consumption: A State-of-the-Art Review. Food Res. Int. 2019, 125, 108527. [Google Scholar] [CrossRef] [PubMed]
- Garofalo, C.; Osimani, A.; Milanović, V.; Taccari, M.; Cardinali, F.; Aquilanti, L.; Riolo, P.; Ruschioni, S.; Isidoro, N.; Clementi, F. The Microbiota of Marketed Processed Edible Insects as Revealed by High-Throughput Sequencing. Food Microbiol. 2017, 62, 15–22. [Google Scholar] [CrossRef]
- Grabowski, N.T.; Klein, G. Microbiology of Processed Edible Insect Products – Results of a Preliminary Survey. Int. J. Food Microbiol. 2017, 243, 103–107. [Google Scholar] [CrossRef]
- Fasolato, L.; Cardazzo, B.; Carraro, L.; Fontana, F.; Novelli, E.; Balzan, S. Edible Processed Insects from E-Commerce: Food Safety with a Focus on the Bacillus Cereus Group. Food Microbiol. 2018, 76, 296–303. [Google Scholar] [CrossRef]
- Osimani, A.; Milanović, V.; Cardinali, F.; Garofalo, C.; Clementi, F.; Pasquini, M.; Riolo, P.; Ruschioni, S.; Isidoro, N.; Loreto, N.; et al. The Bacterial Biota of Laboratory-Reared Edible Mealworms (Tenebrio molitor L.): From Feed to Frass. Int. J. Food Microbiol. 2018, 272, 49–60. [Google Scholar] [CrossRef]
- Greenhalgh, J.P.; Amund, D. Examining the Presence of Cronobacter Spp. in Ready-to-Eat Edible Insects. Food Saf. 2019, 7, 74–78. [Google Scholar] [CrossRef]
- Mancini, S.; Paci, G.; Ciardelli, V.; Turchi, B.; Pedonese, F.; Fratini, F. Listeria Monocytogenes Contamination of Tenebrio molitor Larvae Rearing Substrate: Preliminary Evaluations. Food Microbiol. 2019, 83, 104–108. [Google Scholar] [CrossRef]
- Wynants, E.; Frooninckx, L.; Van Miert, S.; Geeraerd, A.; Claes, J.; Van Campenhout, L. Risks Related to the Presence of Salmonella Sp. during Rearing of Mealworms (Tenebrio molitor) for Food or Feed: Survival in the Substrate and Transmission to the Larvae. Food Control 2019, 100, 227–234. [Google Scholar] [CrossRef]
- Raimondi, S.; Spampinato, G.; Macavei, L.I.; Lugli, L.; Candeliere, F.; Rossi, M.; Maistrello, L.; Amaretti, A. Effect of Rearing Temperature on Growth and Microbiota Composition of Hermetia illucens. Microorganisms 2020, 8. [Google Scholar] [CrossRef] [PubMed]
- De Smet, J.; Wynants, E.; Cos, P.; Van Campenhout, L. Microbial Community Dynamics during Rearing of Black Soldier Fly Larvae (Hermetia illucens) and Impact on Exploitation Potential. Appl. Env. Microbiol. 2018, 84. [Google Scholar] [CrossRef] [PubMed]
- Wynants, E.; Frooninckx, L.; Crauwels, S.; Verreth, C.; De Smet, J.; Sandrock, C.; Wohlfahrt, J.; Van Schelt, J.; Depraetere, S.; Lievens, B.; et al. Assessing the Microbiota of Black Soldier Fly Larvae (Hermetia illucens) Reared on Organic Waste Streams on Four Different Locations at Laboratory and Large Scale. Microb. Ecol. 2019, 77, 913–930. [Google Scholar] [CrossRef] [PubMed]
- Klunder, H.C.; Wolkers-Rooijackers, J.; Korpela, J.M.; Nout, M.J.R. Microbiological Aspects of Processing and Storage of Edible Insects. Food Control 2012, 26, 628–631. [Google Scholar] [CrossRef]
- Osimani, A.; Ferrocino, I.; Corvaglia, M.R.; Roncolini, A.; Milanović, V.; Garofalo, C.; Aquilanti, L.; Riolo, P.; Ruschioni, S.; Jamshidi, E.; et al. Microbial Dynamics in Rearing Trials of Hermetia illucens Larvae Fed Coffee Silverskin and Microalgae. Food Res. Int. 2021, 140, 110028. [Google Scholar] [CrossRef]
- Cacchiarelli, C.; Fratini, F.; Puccini, M.; Vitolo, S.; Paci, G.; Mancini, S. Effects of Different Blanching Treatments on Colour and Microbiological Profile of Tenebrio molitor and Zophobas Morio Larvae. LWT Food Sci. Technol. 2022, 157, 113112. [Google Scholar] [CrossRef]
- Mancini, S.; Fratini, F.; Tuccinardi, T.; Turchi, B.; Nuvoloni, R.; Paci, G. Effects of Different Blanching Treatments on Microbiological Profile and Quality of the Mealworm (Tenebrio molitor). J. Insects Food Feed. 2019, 5, 225–234. [Google Scholar] [CrossRef]
- Ribeiro, J.C.; Marques, J.P.; Fernandes, T.R.; Pintado, M.E.; Carvalho, S.M.P.; Cunha, L.M. Effect of Blanching, Storage and Drying Conditions on the Macro-Composition, Color and Safety of Mealworm Tenebrio molitor Larvae. LWT 2024, 191, 115646. [Google Scholar] [CrossRef]
- Adamek, M.; Mlček, J.; Adámková, A.; Suchánková, J.; Janalíková, M.; Borkovcová, M.; Bednářová, M. Effect of Different Storage Conditions on the Microbiological Characteristics of Insect. Potravinarstvo Slovak J. Food Sci. 2018, 12, 248–253. [Google Scholar] [CrossRef]
- Gorrens, E.; Van Moll, L.; Frooninckx, L.; De Smet, J.; Van Campenhout, L. Isolation and Identification of Dominant Bacteria From Black Soldier Fly Larvae (Hermetia illucens) Envisaging Practical Applications. Front. Microbiol. 2021, 12, 665546. [Google Scholar] [CrossRef]
- Juste, A.; Thomma, B.; Lievens, B. Recent Advances in Molecular Techniques to Study Microbial Communities in Food-Associated Matrices and Processes. Food Microbiol. 2008, 25, 745–761. [Google Scholar] [CrossRef] [PubMed]
- Staats, M.; Arulandhu, A.J.; Gravendeel, B.; Holst-Jensen, A.; Scholtens, I.; Peelen, T.; Prins, T.W.; Kok, E. Advances in DNA Metabarcoding for Food and Wildlife Forensic Species Identification. Anal. Bioanal. Chem. 2016, 408, 4615–4630. [Google Scholar] [CrossRef] [PubMed]
- Kariuki, E.G.; Kibet, C.; Paredes, J.C.; Mboowa, G.; Mwaura, O.; Njogu, J.; Masiga, D.; Bugg, T.D.H.; Tanga, C.M. Metatranscriptomic Analysis of the Gut Microbiome of Black Soldier Fly Larvae Reared on Lignocellulose-Rich Fiber Diets Unveils Key Lignocellulolytic Enzymes. Front. Microbiol. 2023, 14, 1120224. [Google Scholar] [CrossRef] [PubMed]
- Li, X.-Y.; Mei, C.; Luo, X.-Y.; Wulamu, D.; Zhan, S.; Huang, Y.-P.; Yang, H. Dynamics of the Intestinal Bacterial Community in Black Soldier Fly Larval Guts and Its Influence on Insect Growth and Development. Insect Sci. 2023, 30, 947–963. [Google Scholar] [CrossRef]
- Tegtmeier, D.; Hurka, S.; Mihajlovic, S.; Bodenschatz, M.; Schlimbach, S.; Vilcinskas, A. Culture-Independent and Culture-Dependent Characterization of the Black Soldier Fly Gut Microbiome Reveals a Large Proportion of Culturable Bacteria with Potential for Industrial Applications. Microorganisms 2021, 9, 1642. [Google Scholar] [CrossRef]
- Brulé, L.; Misery, B.; Baudouin, G.; Yan, X.; Guidou, C.; Trespeuch, C.; Foltyn, C.; Anthoine, V.; Moriceau, N.; Federighi, M.; et al. Evaluation of the Microbial Quality of Hermetia illucens Larvae for Animal Feed and Human Consumption: Study of Different Type of Rearing Substrates. Foods 2024, 13, 1587. [Google Scholar] [CrossRef]
- Page, M.J.; Moher, D.; Bossuyt, P.M.; Boutron, I.; Hoffmann, T.C.; Mulrow, C.D.; Shamseer, L.; Tetzlaff, J.M.; Akl, E.A.; Brennan, S.E.; et al. PRISMA 2020 Explanation and Elaboration: Updated Guidance and Exemplars for Reporting Systematic Reviews. BMJ 2021, 372, n160. [Google Scholar] [CrossRef]
- Kooh, P.; Jury, V.; Laurent, S.; Audiat-Perrin, F.; Sanaa, M.; Tesson, V.; Federighi, M.; Boué, G. Control of Biological Hazards in Insect Processing: Application of HACCP Method for Yellow Mealworm (Tenebrio molitor) Powders. Foods 2020, 9, 1528. [Google Scholar] [CrossRef]
- Vandeweyer, D.; Lenaerts, S.; Callens, A.; Van Campenhout, L. Effect of Blanching Followed by Refrigerated Storage or Industrial Microwave Drying on the Microbial Load of Yellow Mealworm Larvae (Tenebrio molitor). Food Control 2017, 71, 311–314. [Google Scholar] [CrossRef]
- Turner, S.; Pryer, K.M.; Miao, V.P.W.; Palmer, J.D. Investigating Deep Phylogenetic Relationships among Cyanobacteria and Plastids by Small Subunit rRNA Sequence Analysis1. J. Eukaryot. Microbiol. 1999, 46, 327–338. [Google Scholar] [CrossRef]
- Kisand, V.; Cuadros, R.; Wikner, J. Phylogeny of Culturable Estuarine Bacteria Catabolizing Riverine Organic Matter in the Northern Baltic Sea. Appl. Environ. Microbiol. 2002, 68, 379–388. [Google Scholar] [CrossRef] [PubMed]
- Escudié, F.; Auer, L.; Bernard, M.; Mariadassou, M.; Cauquil, L.; Vidal, K.; Maman, S.; Hernandez-Raquet, G.; Combes, S.; Pascal, G. FROGS: Find, Rapidly, OTUs with Galaxy Solution. Bioinformatics 2018, 34, 1287–1294. [Google Scholar] [CrossRef] [PubMed]
- McMurdie, P.J.; Holmes, S. Phyloseq: An R Package for Reproducible Interactive Analysis and Graphics of Microbiome Census Data. PLoS ONE 2013, 8, e61217. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Huber, W.; Anders, S. Moderated Estimation of Fold Change and Dispersion for RNA-Seq Data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Gómez-Rubio, V. Ggplot2—Elegant Graphics for Data Analysis (2nd Edition). J. Stat. Soft. 2017, 77. [Google Scholar] [CrossRef]
- Plotly Technologies Inc. Collaborative Data Science; Plotly Technologies Inc.: Montreal, QC, Canada, 2015. [Google Scholar]
- Gorrens, E.; Smet, J.D.; Vandeweyer, D.; Bossaert, S.; Crauwels, S.; Lievens, B.; Campenhout, L.V. The Bacterial Communities of Black Soldier Fly Larvae (Hermetia illucens) during Consecutive, Industrial Rearing Cycles. J. Insects Food Feed. 2022, 8, 1061–1076. [Google Scholar] [CrossRef]
- Ao, Y.; Yang, C.; Wang, S.; Hu, Q.; Yi, L.; Zhang, J.; Yu, Z.; Cai, M.; Yu, C. Characteristics and Nutrient Function of Intestinal Bacterial Communities in Black Soldier Fly (Hermetia illucens L.) Larvae in Livestock Manure Conversion. Microb. Biotechnol. 2021, 14, 886–896. [Google Scholar] [CrossRef]
- Wu, N.; Wang, X.; Mao, Z.; Liang, J.; Liu, X.; Xu, X. Bioconversion of Chicken Meat and Bone Meal by Black Soldier Fly Larvae: Effects of Straw Addition on the Quality and Microbial Profile of Larval Frass. J. Environ. Manag. 2022, 307, 114579. [Google Scholar] [CrossRef]
- Chen, G.; Zhang, K.; Tang, W.; Li, Y.; Pang, J.; Yuan, X.; Song, X.; Jiang, L.; Yu, X.; Zhu, H.; et al. Feed Nutritional Composition Affects the Intestinal Microbiota and Digestive Enzyme Activity of Black Soldier Fly Larvae. Front. Microbiol. 2023, 14, 1184139. [Google Scholar] [CrossRef]
- Bruno, D.; Bonelli, M.; De Filippis, F.; Di Lelio, I.; Tettamanti, G.; Casartelli, M.; Ercolini, D.; Caccia, S. The Intestinal Microbiota of Hermetia illucens Larvae Is Affected by Diet and Shows a Diverse Composition in the Different Midgut Regions. Appl. Env. Microbiol. 2019, 85, e01864-18. [Google Scholar] [CrossRef]
- Auger, L.; Deschamps, M.-H.; Vandenberg, G.; Derome, N. Microbiota Is Structured by Gut Regions, Life Stage, and Diet in the Black Soldier Fly (Hermetia illucens). Front. Microbiol. 2023, 14, 1221728. [Google Scholar] [CrossRef] [PubMed]
- Cifuentes, Y.; Glaeser, S.P.; Mvie, J.; Bartz, J.-O.; Müller, A.; Gutzeit, H.O.; Vilcinskas, A.; Kämpfer, P. The Gut and Feed Residue Microbiota Changing during the Rearing of Hermetia illucens Larvae. Antonie Van. Leeuwenhoek 2020, 113, 1323–1344. [Google Scholar] [CrossRef] [PubMed]
- Jeon, H.; Park, S.; Choi, J.; Jeong, G.; Lee, S.-B.; Choi, Y.; Lee, S.-J. The Intestinal Bacterial Community in the Food Waste-Reducing Larvae of Hermetia illucens. Curr. Microbiol. 2011, 62, 1390–1399. [Google Scholar] [CrossRef] [PubMed]
- Jiang, C.-L.; Jin, W.-Z.; Tao, X.-H.; Zhang, Q.; Zhu, J.; Feng, S.-Y.; Xu, X.-H.; Li, H.-Y.; Wang, Z.-H.; Zhang, Z.-J. Black Soldier Fly Larvae (Hermetia illucens) Strengthen the Metabolic Function of Food Waste Biodegradation by Gut Microbiome. Microb. Biotechnol. 2019, 12, 528–543. [Google Scholar] [CrossRef]
- Klammsteiner, T.; Walter, A.; Bogataj, T.; Heussler, C.D.; Stres, B.; Steiner, F.M.; Schlick-Steiner, B.C.; Arthofer, W.; Insam, H. The Core Gut Microbiome of Black Soldier Fly (Hermetia illucens) Larvae Raised on Low-Bioburden Diets. Front. Microbiol. 2020, 11, 993. [Google Scholar] [CrossRef]
- Klammsteiner, T.; Walter, A.; Bogataj, T.; Heussler, C.D.; Stres, B.; Steiner, F.M.; Schlick-Steiner, B.C.; Insam, H. Impact of Processed Food (Canteen and Oil Wastes) on the Development of Black Soldier Fly (Hermetia illucens) Larvae and Their Gut Microbiome Functions. Front. Microbiol. 2021, 12, 619112. [Google Scholar] [CrossRef]
- Liu, C.; Yao, H.; Chapman, S.J.; Su, J.; Wang, C. Changes in Gut Bacterial Communities and the Incidence of Antibiotic Resistance Genes during Degradation of Antibiotics by Black Soldier Fly Larvae. Environ. Int. 2020, 142, 105834. [Google Scholar] [CrossRef]
- Mašková, Z.; Medo, J.; Kolesár, E.; Tančinová, D.; Ivanišová, E.; Urminská, D.; Hleba, L.; Urminská, J.; Mrvová, M.; Barboráková, Z. Hermetia illucens in the Process of Kitchen Waste Biodegradation: The Effect of Different Approaches to Waste Storage on the Microbiological Profile and Nutritional Parameters of the Larvae. Insects 2025, 16. [Google Scholar] [CrossRef]
- Pei, Y.; Sun, M.; Zhang, J.; Lei, A.; Chen, H.; Kang, X.; Ni, H.; Yang, S. Comparative Metagenomic and Metatranscriptomic Analyses Reveal the Response of Black Soldier Fly (Hermetia illucens) Larvae Intestinal Microbes and Reduction Mechanisms to High Concentrations of Tetracycline. Toxics 2023, 11. [Google Scholar] [CrossRef]
- Querejeta, M.; Hervé, V.; Perdereau, E.; Marchal, L.; Herniou, E.A.; Boyer, S.; Giron, D. Changes in Bacterial Community Structure Across the Different Life Stages of Black Soldier Fly (Hermetia illucens). Microb. Ecol. 2023, 86, 1254–1267. [Google Scholar] [CrossRef]
- Schreven, S.J.J.; de Vries, H.; Hermes, G.D.A.; Smidt, H.; Dicke, M.; Jvan Loon, J.J.A. Relative Contributions of Egg-Associated and Substrate-Associated Microorganisms to Black Soldier Fly Larval Performance and Microbiota. FEMS Microbiol. Ecol. 2021, 97. [Google Scholar] [CrossRef]
- Schreven, S.J.J.; de Vries, H.; Hermes, G.D.A.; Zeni, G.; Smidt, H.; Dicke, M.; van Loon, J.J.A. Black Soldier Fly Larvae Influence Internal and Substrate Bacterial Community Composition Depending on Substrate Type and Larval Density. Appl. Env. Microbiol. 2022, 88, e0008422. [Google Scholar] [CrossRef] [PubMed]
- Tanga, C.M.; Waweru, J.W.; Tola, Y.H.; Onyoni, A.A.; Khamis, F.M.; Ekesi, S.; Paredes, J.C. Organic Waste Substrates Induce Important Shifts in Gut Microbiota of Black Soldier Fly (Hermetia illucens L.): Coexistence of Conserved, Variable, and Potential Pathogenic Microbes. Front. Microbiol. 2021, 12, 635881. [Google Scholar] [CrossRef] [PubMed]
- Van Looveren, N.; IJdema, F.; van der Heijden, N.; Van Der Borght, M.; Vandeweyer, D. Microbial Dynamics and Vertical Transmission of Escherichia Coli across Consecutive Life Stages of the Black Soldier Fly (Hermetia illucens). Anim. Microbiome 2024, 6, 29. [Google Scholar] [CrossRef]
- Yang, F.; Tomberlin, J.K.; Jordan, H.R. Starvation Alters Gut Microbiome in Black Soldier Fly (Diptera: Stratiomyidae) Larvae. Front. Microbiol. 2021, 12, 601253. [Google Scholar] [CrossRef]
- Zheng, L.; Crippen, T.L.; Singh, B.; Tarone, A.M.; Dowd, S.; Yu, Z.; Wood, T.K.; Tomberlin, J.K. A Survey of Bacterial Diversity from Successive Life Stages of Black Soldier Fly (Diptera: Stratiomyidae) by Using 16S rDNA Pyrosequencing. J. Med. Entomol. 2013, 50, 647–658. [Google Scholar] [CrossRef]
- Eke, M.; Tougeron, K.; Hamidovic, A.; Tinkeu, L.S.N.; Hance, T.; Renoz, F. Deciphering the Functional Diversity of the Gut Microbiota of the Black Soldier Fly (Hermetia illucens): Recent Advances and Future Challenges. Anim. Microbiome 2023, 5, 40. [Google Scholar] [CrossRef]
- Bruno, D.; Bonelli, M.; Cadamuro, A.G.; Reguzzoni, M.; Grimaldi, A.; Casartelli, M.; Tettamanti, G. The Digestive System of the Adult Hermetia illucens (Diptera: Stratiomyidae): Morphological Features and Functional Properties. Cell Tissue Res. 2019, 378, 221–238. [Google Scholar] [CrossRef]
- Osimani, A.; Aquilanti, L. Spore-Forming Bacteria in Insect-Based Foods. Curr. Opin. Food Sci. 2021, 37, 112–117. [Google Scholar] [CrossRef]
- Chaisowwong, W.; Kusumoto, A.; Hashimoto, M.; Harada, T.; Maklon, K.; Kawamoto, K. Physiological Characterization of Campylobacter Jejuni under Cold Stresses Conditions: Its Potential for Public Threat. J. Vet. Med. Sci. 2012, 74, 43–50. [Google Scholar] [CrossRef]
- İzgördü, Ö.K.; Darcan, C.; Kariptaş, E. Overview of VBNC, a Survival Strategy for Microorganisms. 3 Biotech. 2022, 12, 307. [Google Scholar] [CrossRef] [PubMed]
- Gill, C.; Bahrndorff, S.; Lowenberger, C. Campylobacter Jejuni in Musca Domestica: An Examination of Survival and Transmission Potential in Light of the Innate Immune Responses of the House Flies. Insect Sci. 2017, 24, 584–598. [Google Scholar] [CrossRef] [PubMed]
- Strother, K.O.; Steelman, C.D.; Gbur, E.E. Reservoir Competence of Lesser Mealworm (Coleoptera: Tenebrionidae) for Campylobacter Jejuni (Campylobacterales: Campylobacteraceae). J. Med. Entomol. 2005, 42, 42–47. [Google Scholar] [CrossRef] [PubMed]
- Bahrndorff, S.; Gill, C.; Lowenberger, C.; Skovgård, H.; Hald, B. The Effects of Temperature and Innate Immunity on Transmission of Campylobacter Jejuni (Campylobacterales: Campylobacteraceae) Between Life Stages of Musca Domestica (Diptera: Muscidae). J. Med. Entomol. 2014, 51, 670–677. [Google Scholar] [CrossRef]
- Jensen, A.N.; Hald, B. Campylobacter Contamination Level in Houseflies after Exposure to Materials Containing Campylobacter. J. Insects Food Feed. 2018, 4, 179–186. [Google Scholar] [CrossRef]
- Carrillo, C.D.; Plante, D.; Iugovaz, I.; Kenwell, R.; Bélanger, G.; Boucher, F.; Poulin, N.; Trottier, Y.-L. Method-Dependent Variability in Determination of Prevalence of Campylobacter Jejuni and Campylobacter Coli in Canadian Retail Poultry. J. Food Prot. 2014, 77, 1682–1688. [Google Scholar] [CrossRef]
- Magajna, B.; Schraft, H. Evaluation of Propidium Monoazide and Quantitative PCR To Quantify Viable Campylobacter Jejuni Biofilm and Planktonic Cells in Log Phase and in a Viable but Nonculturable State. J. Food Prot. 2015, 78, 1303–1311. [Google Scholar] [CrossRef]
- Nicholson, W.L. Roles of Bacillus Endospores in the Environment. Cell. Mol. Life Sci. (CMLS) 2002, 59, 410–416. [Google Scholar] [CrossRef]
- Heussler, C.D.; Klammsteiner, T.; Stonig, K.T.; Insam, H.; Schlick-Steiner, B.C.; Steiner, F.M. Microbial Influences on Black Soldier Fly Reproduction: A Focus on Egg Surface Colonization. Res. Sq. 2023. [Google Scholar] [CrossRef]
- Wells, C.; Wilkins, T. Clostridia: Sporeforming Anaerobic Bacilli. In Medical Microbiology, 4th ed.; University of Texas Medical Branch at Galveston: Galveston, TX, USA, 1996; Chapter 18. [Google Scholar]
- Zeng, T.; Jaffar, S.; Xu, Y.; Qi, Y. The Intestinal Immune Defense System in Insects. Int. J. Mol. Sci. 2022, 23, 15132. [Google Scholar] [CrossRef]
- Vogel, M.; Shah, P.N.; Voulgari-Kokota, A.; Maistrou, S.; Aartsma, Y.; Beukeboom, L.W.; Salles, J.F.; Van Loon, J.J.A.; Dicke, M.; Wertheim, B. Health of the Black Soldier Fly and House Fly under Mass-Rearing Conditions: Innate Immunity and the Role of the Microbiome. J. Insects Food Feed. 2022, 8, 857–878. [Google Scholar] [CrossRef]
- Van Huis, A.; Rumpold, B.A.; Van Der Fels-Klerx, H.J.; Tomberlin, J.K. Advancing Edible Insects as Food and Feed in a Circular Economy. J. Insects Food Feed. 2021, 7, 935–948. [Google Scholar] [CrossRef]
- Nakagawa, A.; Sakamoto, T.; Kanost, M.R.; Tabunoki, H. Development of New Methods to Stimulate the Production of Antimicrobial Peptides in the Larvae of the Black Soldier Fly Hermetia illucens. Int. J. Mol. Sci. 2023, 24, 15765. [Google Scholar] [CrossRef] [PubMed]
- Moretta, A.; Salvia, R.; Scieuzo, C.; Di Somma, A.; Vogel, H.; Pucci, P.; Sgambato, A.; Wolff, M.; Falabella, P. A Bioinformatic Study of Antimicrobial Peptides Identified in the Black Soldier Fly (BSF) Hermetia illucens (Diptera: Stratiomyidae). Sci. Rep. 2020, 10, 16875. [Google Scholar] [CrossRef]
- Van Moll, L.; De Smet, J.; Paas, A.; Tegtmeier, D.; Vilcinskas, A.; Cos, P.; Van Campenhout, L. In Vitro Evaluation of Antimicrobial Peptides from the Black Soldier Fly (Hermetia illucens) against a Selection of Human Pathogens. Microbiol. Spectr. 2022, 10, e01664-21. [Google Scholar] [CrossRef]
- Johnson, J.S.; Spakowicz, D.J.; Hong, B.-Y.; Petersen, L.M.; Demkowicz, P.; Chen, L.; Leopold, S.R.; Hanson, B.M.; Agresta, H.O.; Gerstein, M.; et al. Evaluation of 16S rRNA Gene Sequencing for Species and Strain-Level Microbiome Analysis. Nat. Commun. 2019, 10, 5029. [Google Scholar] [CrossRef]
- Parniakov, O.; Mikhrovska, M.; Wiktor, A.; Alles, M.; Ristic, D.; Bogusz, R.; Nowacka, M.; Devahastin, S.; Mujumdar, A.; Heinz, V.; et al. Insect Processing for Food and Feed: A Review of Drying Methods. Dry. Technol. 2022, 40, 1500–1513. [Google Scholar] [CrossRef]
- Kewuyemi, Y.O.; Kesa, H.; Chinma, C.E.; Adebo, O.A. Fermented Edible Insects for Promoting Food Security in Africa. Insects 2020, 11, 283. [Google Scholar] [CrossRef]
- Pérez-Rodríguez, E.; Ibarra-Herrera, C.C.; Pérez-Carrillo, E. Effect of Incorporation of Solid-State Fermented Edible Insects Tenebrio molitor and Sphenarium Purpurascens with Aspergillus Oryzae in the Elaboration of Bread. LWT 2023, 184, 115003. [Google Scholar] [CrossRef]
- Rubiola, S.; Macori, G.; Civera, T.; Fanning, S.; Mitchell, M.; Chiesa, F. Comparison Between Full-Length 16S rRNA Metabarcoding and Whole Metagenome Sequencing Suggests the Use of Either Is Suitable for Large-Scale Microbiome Studies. Foodborne Pathog. Dis. 2022, 19, 495–504. [Google Scholar] [CrossRef]
- Olivier, S.A.; Bull, M.K.; Strube, M.L.; Murphy, R.; Ross, T.; Bowman, J.P.; Chapman, B. Long-Read MinIONTM Sequencing of 16S and 16S-ITS-23S rRNA Genes Provides Species-Level Resolution of Lactobacillaceae in Mixed Communities. Front. Microbiol. 2023, 14, 1290756. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Li, H.; Ma, S.; Cao, J.; Liao, H.; Huang, Q.; Chen, W. The Newest Oxford Nanopore R10.4.1 Full-Length 16S rRNA Sequencing Enables the Accurate Resolution of Species-Level Microbial Community Profiling. Appl. Environ. Microbiol. 2023, 89, e00605-23. [Google Scholar] [CrossRef] [PubMed]
- Matsuo, Y.; Komiya, S.; Yasumizu, Y.; Yasuoka, Y.; Mizushima, K.; Takagi, T.; Kryukov, K.; Fukuda, A.; Morimoto, Y.; Naito, Y.; et al. Full-Length 16S rRNA Gene Amplicon Analysis of Human Gut Microbiota Using MinIONTM Nanopore Sequencing Confers Species-Level Resolution. BMC Microbiol. 2021, 21, 35. [Google Scholar] [CrossRef] [PubMed]
- Bertolo, A.; Valido, E.; Stoyanov, J. Optimized Bacterial Community Characterization through Full-Length 16S rRNA Gene Sequencing Utilizing MinION Nanopore Technology. BMC Microbiol. 2024, 24, 58. [Google Scholar] [CrossRef]
- De Filippis, F.; Parente, E.; Ercolini, D. Metagenomics Insights into Food Fermentations. Microb. Biotechnol. 2017, 10, 91–102. [Google Scholar] [CrossRef]
- Butowski, C.F.; Dixit, Y.; Reis, M.M.; Mu, C. Metatranscriptomics for Understanding the Microbiome in Food and Nutrition Science. Metabolites 2025, 15, 185. [Google Scholar] [CrossRef]




| Indicators | Methods |
|---|---|
| ASR, Anaerobic Sulfite Reductive (46 °C) | NF V08-061 boxes |
| Bacillus cereus presumed | NF EN ISO 7932 |
| Campylobacter spp. | NF EN ISO 10272-2/A1 |
| Clostridium perfringens presumed | NF EN ISO 7937 |
| Cronobacter spp. | NF EN ISO 22964 |
| Enterobacteriaceae presumed | NF V08-054 |
| Escherichia coli β-glucuronidase positive | NF ISO 16649-2 |
| Lactic acid bacteria | NF ISO 15214 |
| Listeria monocytogenes | BKR 23/02-11/02 |
| Molds | Internal method adapted (DG18, 120 h ± 3 h) |
| Salmonella spp. | BKR 23/07-10/11 |
| Staphylococcus coagulase-positive | Internal method adapted of NF EN ISO 6888-2 |
| Yeasts | Internal method adapted (NF V 08-036) |
| Rearing Data Sample | Environmental Rearing Data (Room) | |||||
|---|---|---|---|---|---|---|
| Date | pH | T°C | RH% | T°C | RH% | |
| Incubation (eggs) | Day 0 | - | - | - | 27.0 ± 0.1 | 75.0 ± 0.3 |
| Pre-Growth Batch * | Day 2 | 5.2 | 24.7 ± 0.09 | 52.7 ± 8.4 | 26.9 ± 0.8 | 52.7 ± 8.4 |
| Day 5 | 7.3 | 42.7 ± 0.97 | 63.5 ± 8.8 | 26.8 ± 0.5 | 63.5 ± 8.8 | |
| Growth batch * | Day 6 | 4.1 | 24.64 ± 0.43 | 43.7 ± 2.5 | 27.9 ± 1.7 | 43.7 ± 2.5 |
| Day 8 | 8.5 | 31.84 ± 0.64 | 44.2 ± 2.8 | 27.7 ± 1.5 | 44.2 ± 2.8 | |
| Day 12 | 8.9 | 32.08 ± 0.81 | 36.2 ± 2.1 | 28.0 ± 1.2 | 36.2 ± 2.1 | |
| Day 14 | 9.0 | 30.2 ± 0.47 | 43.6 ± 2.6 | 27.8 ± 2.0 | 43.6 ± 2.6 | |
| Anaerobic Sulfite-Reductive | Lactic Acid Bacteria | Mold | Spores | Total Mesophilic Bacteria | Yeast | |
|---|---|---|---|---|---|---|
| Substrate | 2.70 a ± 0.99 | 8.48 ± 0.00 | 3.10 ± 0.71 | 5.18 ± 0.30 | 7.53 ± 0.48 | 3.58 ± 1.45 |
| Frass | 4.72 ± 0.51 | 6.24 b ± 3.17 | 3.43 ± 0.61 | 7.82 ± 0.29 | 9.29 ± 0.61 | 3.36 ± 1.11 |
| Sample Code | Bacillus cereus Presumed | Clostridium perfringens Presumed | Cronobacter spp. | Enterobacteriaceae Presumed | Escherichia coli β-Glucuronidase-Positive | Listeria monocytogenes | Salmonella spp. | Staphylococci Coagulase-Positive | |
|---|---|---|---|---|---|---|---|---|---|
| Substrate | HI_pre_growth_substrate HI_growth_substrate | <1 | <1 | Not detected | 3.08 ± 0.18 | <1 | Not detected | Not detected | <1 |
| Frass | HI_frass_D5, HI_frass_D14 | <1 | <1 | Not detected | 2.78 ± 1.11 | <1, 2.51 * | Not detected | Not detected | <1 |
| Larvae | HI_larvae_D5, HI_larvae_D14, HI_P1_larvae_D14_defrosted, HI_P2_larvae_D14_defrosted | <1 | <1, <1, <1, 4.28 * | Not detected | 3.61 ± 0.77 | 2.62 ± 0.26 | Not detected | Not detected | <1 |
| Boiled larvae (applied for Process 1) | HI_P1_larvae_D14_boiled | <1 | <1 | Not detected | <2 | <1 | Not detected | Not detected | <1 |
| Powder Process 1 | HI_P1_powder_D0, HI_P1_powder_M3 | <1 | <1 | Not detected | <2 | <1 | Not detected | Not detected | <1 |
| Powder Process 2 | HI_P2_powder_D0, HI_P2_powder_M3 | <1 | <1 | Not detected | <2 | <1 | Not detected | Not detected | <1 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Misery, B.; Brulé, L.; Djema, R.; Yan, X.; Le Cozic, V.; Baudouin, G.; Federighi, M.; Boué, G. Investigating the Microbial Dynamics of Hermetia illucens Powder Throughout Rearing and Processing: An Integrated Approach Using Cultural and Metabarcoding Methods. Foods 2025, 14, 2161. https://doi.org/10.3390/foods14132161
Misery B, Brulé L, Djema R, Yan X, Le Cozic V, Baudouin G, Federighi M, Boué G. Investigating the Microbial Dynamics of Hermetia illucens Powder Throughout Rearing and Processing: An Integrated Approach Using Cultural and Metabarcoding Methods. Foods. 2025; 14(13):2161. https://doi.org/10.3390/foods14132161
Chicago/Turabian StyleMisery, Boris, Lenaïg Brulé, Rima Djema, Xin Yan, Victoire Le Cozic, Guillaume Baudouin, Michel Federighi, and Géraldine Boué. 2025. "Investigating the Microbial Dynamics of Hermetia illucens Powder Throughout Rearing and Processing: An Integrated Approach Using Cultural and Metabarcoding Methods" Foods 14, no. 13: 2161. https://doi.org/10.3390/foods14132161
APA StyleMisery, B., Brulé, L., Djema, R., Yan, X., Le Cozic, V., Baudouin, G., Federighi, M., & Boué, G. (2025). Investigating the Microbial Dynamics of Hermetia illucens Powder Throughout Rearing and Processing: An Integrated Approach Using Cultural and Metabarcoding Methods. Foods, 14(13), 2161. https://doi.org/10.3390/foods14132161









