Effects of Heterologous Expression of Genes Related L–Malic acid Metabolism in Saccharomyces uvarum on Flavor Substances Production in Wine
Abstract
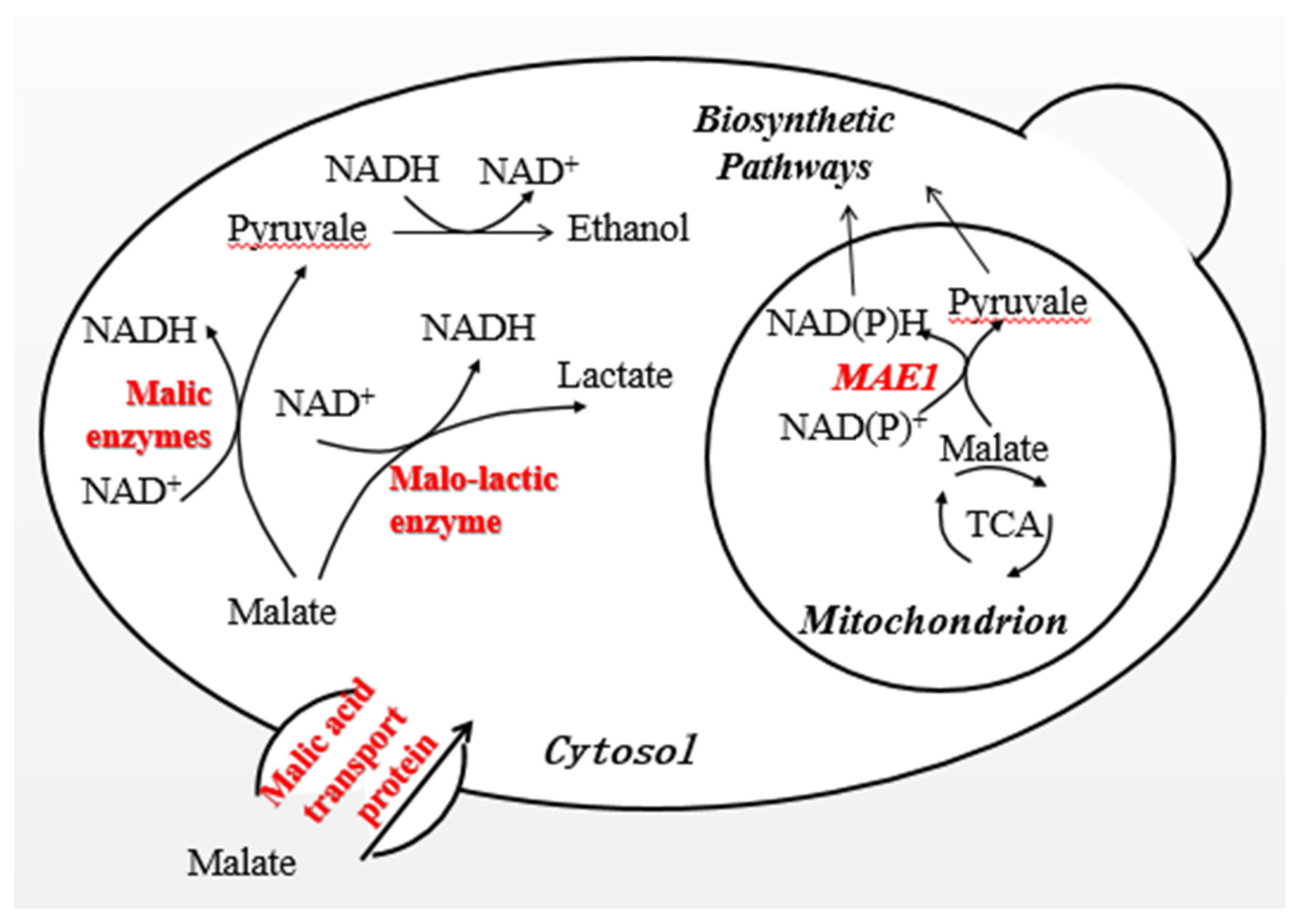
1. Introduction
2. Materials and Methods
2.1. Strains and Media
2.2. Construction of Plasmids
2.3. Transformation Strategy
2.4. Fermentation Conditions
2.5. Chemical Analysis
2.6. Real-Time Quantitative PCR (RT-qPCR)
2.7. Statistical Analysis
3. Results and Discussion
3.1. Effect on L–Malic acid Degradation of Single-Gene Heterologous Expression in S. uvarum
3.2. Effects of Coexpression of Malate Permease mleP from L. lactis with Different Malolactic Enzyme or Malic Enzyme Genes in S. uvarum on the Degradation of L–Malic Acid
3.3. Effects of Coexpression of Malate Permease MAE1 from S. pombe with Different Malolactic Enzyme or Malic Enzyme Genes in S. uvarum on the Degradation of L–Malic Acid
3.4. Effects of the Gene Expressions in S. uvarum on Higher Alcohols and Other Flavor Substances Production
3.5. Effects of the Gene Expressions in S. uvarum on Growth and Fermentation Performance
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Ruiz-De-Villa, C.; Poblet, M.; Cordero-Otero, R.; Bordons, A.; Reguant, C.; Rozès, N. Screening of Saccharomyces cerevisiae and Torulaspora delbrueckii strains in relation to their effect on malolactic fermentation. Food Microbiol. 2023, 112, 104212. [Google Scholar] [CrossRef]
- Balmaseda, A.; Rozès, N.; Bordons, A.; Reguant, C. Modulation of a defined community of Oenococcus oeni strains by Torulaspora delbrueckii and its impact on malolactic fermentation. Aust. J. Grape Wine Res. 2022, 28, 374–382. [Google Scholar] [CrossRef]
- Dimopoulou, M.; Troianou, V.; Paramithiotis, S.; Proksenia, N.; Kotseridis, Y. Evaluation of malolactic starters in white and rosé winemaking of moschofilero wines. Appl. Sci. 2022, 12, 5722–5736. [Google Scholar] [CrossRef]
- Umino, M.; Onozato, M.; Sakamoto, T.; Koishi, M.; Fukushima, T. Analyzing citramalic acid enantiomers in apples and commercial fruit juice by liquid Chromatography–Tandem mass spectrometry with pre-column derivatization. Molecules 2023, 28, 1556. [Google Scholar] [CrossRef]
- Fu, J.; Wang, L.; Sun, J.; Ju, N.; Jin, G. Malolactic fermentation: New approaches to old problems. Microorganisms 2022, 10, 2363–2378. [Google Scholar] [CrossRef]
- Liszt, K.I.; Walker, J.; Somoza, V. Identification of organic acids in wine that stimulate mechanisms of gastric acid secretion. J. Agric. Food Chem. 2012, 60, 7022–7030. [Google Scholar] [CrossRef]
- Krieger-Weber, S.; Heras, J.M.; Suarez, C. Lactobacillus plantarum, a new biological tool to control malolactic fermentation: A review and an outlook. Beverages 2020, 6, 23. [Google Scholar] [CrossRef]
- Gabler, A.M.; Ludwig, A.; Biener, F.; Waldner, M.; Dawid, C.; Oliver, F. Chemical characterization of red wine polymers and their interaction affinity with odorants. Foods 2024, 13, 526. [Google Scholar] [CrossRef]
- Jakabová, S.; Fikselová, M.; Mendelová, A.; Ševčík, M.; Jakab, I.; Aláčová, Z.; Ivanova-Petropulos, V. Chemical composition of white wines produced from different grape varieties and wine regions in slovakia. Appl. Sci. 2021, 11, 11059. [Google Scholar] [CrossRef]
- Tzamourani, A.; Paramithiotis, S.; Favier, M.; Coulon, J.; Moine, V.; Paraskevopoulos, I.; Dimopoulou, M. New insights into the production of assyrtiko wines from the volcanic terroir of santorini island using Lachancea thermotolerans. Microorganisms 2024, 12, 786. [Google Scholar] [CrossRef]
- Bauer, R.; Dicks, L.M.T. Control of malolactic fermentation in wine. A review. S. Afr. Soc. Enol. Vitic. 2017, 25, 74–88. [Google Scholar] [CrossRef]
- Prusova, B.; Licek, J.; Kumsta, M.; Baron, M.; Sochor, J. Effect of new methods for inhibiting malolactic fermentation on the analytical and sensory parameters of wines. Fermentation 2024, 10, 122. [Google Scholar] [CrossRef]
- Edwards, C.G.; Reynolds, A.G.; Rodriguez, A.V.; Semon, M.J.; Mills, J.M. Implication of acetic acid in the induction of slow/stuck grape juice fermentations and inhibition of yeast by Lactobacillus sp. Am. J. Enol. Vitic. 1999, 50, 204–210. [Google Scholar] [CrossRef]
- Sgouros, G.; Mallouchos, A.; Dourou, D.; Banilas, G.; Chalvantzi, I.; Kourkoutas, Y.; Nisiotou, A. Torulaspora delbrueckii may help manage total and volatile acidity of santorini-assyrtiko wine in view of global warming. Foods 2023, 12, 191. [Google Scholar] [CrossRef]
- Vicente, J.; Vladic, L.; Marquina, D.; Brezina, S.; Rauhut, D.; Santiago, B. The influence of chitosan on the chemical composition of wines fermented with Lachancea thermotolerans. Foods 2024, 13, 987. [Google Scholar] [CrossRef]
- Li, P.; Gao, Y.; Wang, C.; Zhang, C.Y.; Guo, X.; Xiao, D. Effect of ILV6 deletion and expression of aldB from Lactobacillus plantarum in Saccharomyces uvarum on diacetyl production and wine flavor. J. Agric. Food Chem. 2018, 66, 8556–8565. [Google Scholar] [CrossRef]
- Zelle, R.M.; Harrison, J.C.; Pronk, J.T.; Maris, A.J.A.V. Anaplerotic role for cytosolic malic enzyme in engineered Saccharomyces cerevisiae strains. Appl. Environ. Microbiol. 2011, 77, 732–738. [Google Scholar] [CrossRef][Green Version]
- Schümann, C.; Michlmayr, H.; Eder, R.; Del Hierro, A.M.; Kulbe, K.D.; Mathiesen, G.; Nguyen, T. Heterologous expression of Oenococcus oeni malolactic enzyme in Lactobacillus plantarum for improved malolactic fermentation. AMB Express 2012, 2, 19. [Google Scholar] [CrossRef]
- Landete, J.M.; Ferrer, S.; Monedero, V.; Zuniga, M. Malic enzyme and malolactic enzyme pathways are functionally linked but independently regulated in Lactobacillus casei BL23. Appl. Environ. Microbiol. 2013, 79, 5509–5518. [Google Scholar] [CrossRef] [PubMed]
- Taillandier, P.; Strehaiano, P. The role of malic acid in the metabolism of Schizosaccharomyces pombe: Substrate con2 sumption and cell growth. Appl. Microbiol. Biotechnol. 1991, 35, 541–543. [Google Scholar] [CrossRef]
- Camarasa, C.; Bidard, F.; Bony, M.; Barre, P.; Dequin, S. Characterization of Schizosaccharomyces pombe malate permease by expression in Saccharomyces cerevisiae. Appl. Environ. Microbiol. 2001, 67, 4144–4151. [Google Scholar] [CrossRef]
- Bony, M.; Bidart, F.; Camarasa, C.; Ansanay, V.; Dulau, L.; Barre, P.; Dequin, S. Metabolic analysis of S. cerevisiae strains engineered for malolactic fermentation. FEBS Lett. 1997, 410, 452–456. [Google Scholar] [CrossRef] [PubMed]
- Volschenk, H.; Viljoen-Bloom, M.; Subden, R.E.; van Vuuren, H.J.J. Malo-ethanolic fermentation in grape must by recombinant strains of Saccharomyces cerevisiae. Yeast 2001, 18, 963–970. [Google Scholar] [CrossRef] [PubMed]
- Husnik, J.I.; Volschenk, H.; Bauer, J.; Colavizza, D.; Luo, Z.; van Vuuren, H.J. Metabolic engineering of malolactic wine yeast. Metab. Eng. 2006, 8, 315–323. [Google Scholar] [CrossRef]
- Gamero, A.; Tronchoni, J.; Querol, A.; Belloch, C. Production of aroma compounds by cryotolerant Saccharomyces species and hybrids at low and moderate fermentation temperatures. J. Appl. Microbiol. 2013, 114, 1405–1414. [Google Scholar] [CrossRef]
- Almeida, P.; Gonçalves, C.; Teixeira, S.; Libkind, D.; Bontrager, M.; Masneuf-Pomarède, I.; Albertin, W.; Durrens, P.; Sherman, D.J.; Marullo, P.; et al. A Gondwanan imprint on global diversity and domestication of wine and cider yeast Saccharomyces uvarum. Nat. Commun. 2014, 5, 4044–4067. [Google Scholar] [CrossRef] [PubMed]
- Stribny, J.; Gamero, A.; Pérez-Torrado, R.; Querol, A. Saccharomyces kudriavzevii and Saccharomyces uvarum differ from Saccharomyces cerevisiae during the production of aroma-active higher alcohols and acetate esters using their amino acidic precursors. Int. J. Food Microbiol. 2015, 205, 41–46. [Google Scholar] [CrossRef]
- Solomon, M.; Schulkin, A.; Kolouchova, R.; Francis, I.L.; Schmidt, S.A. Aromatic higher alcohols in wine: Implication on aroma and palate attributes during chardonnay aging. Molecules 2021, 26, 4979. [Google Scholar] [CrossRef]
- Valero, E.; Moyano, L.; Millan, M.; Medina, M.; Ortega, J. Higher alcohols and esters production by Saccharomyces cerevisiae influence of the initial oxygenation of the grape must. Food Chem. 2002, 78, 57–61. [Google Scholar] [CrossRef]
- Henschke, P.A.; Jiranek, V. Yeast-Metabolism of Nitrogen Compounds in Wine Microbiology and Biotechnology; Harwood Academic Publishers: Reading, UK, 1993; pp. 77–164. [Google Scholar]
- Hazelwood, L.A.; Daran, J.M.; van Maris, A.J.; Pronk, J.T.; Dickinson, J.R. The Ehrlich pathway for fusel alcohol production: A century of research on Saccharomyces cerevisiae metabolism. Appl. Environ. Microbiol. 2008, 74, 2259–2266. [Google Scholar] [CrossRef]
- Cui, D.Y.; Ge, J.L.; Song, Y.M.; Feng, P.P.; Lin, L.C.; Guo, L.Y.; Zhang, C.Y. Regulating the ratio of higher alcohols to esters by simultaneously overexpressing ATF1 and deleting BAT2 in brewer’s yeast Saccharomyces pastorianus. Food Biosci. 2021, 3, 101231. [Google Scholar] [CrossRef]
- Teste, M.-A.; Duquenne, M.; François, J.M.; Parrou, J.-L. Validation of reference genes for quantitative expression analysis by real-time RT-PCR in Saccharomyces cerevisiae. BMC Mol. Biol. 2009, 10, 99. [Google Scholar] [CrossRef] [PubMed]
- Williams, S.A.; Hodges, R.A.; Strike, T.L.; Snow, R.; Kunkee, R.E. Cloning the gene for the malolactic fermentation of wine from lactobacillus delbrueckii in escherichia coli and yeasts. Appl. Environ. Microbiol. 1984, 47, 288–293. [Google Scholar] [CrossRef] [PubMed]
- Ansanay, V.; Dequin, S.; Camarasa, C.; Schaeffer, V.; Grivet, J.P.; Blondin, B.; Salmon, J.M.; Barre, P. Malolactic fermentation by engineered Saccharomyces cerevisiae as compared with engineered Schizosaccharomyces pombe. Yeast 1996, 12, 215–225. [Google Scholar] [CrossRef]
- Lemme, A.; Sztajer, H.; Wagner-Döbler, I. Characterization of mleR, a positive regulator of malolactic fermentation and part of the acid tolerance response in streptococcus mutans. BMC Microbiol. 2010, 10, 58. [Google Scholar] [CrossRef] [PubMed]
- Paramithiotis, S.; Stasinou, V.; Tzamourani, A.; Kotseridis, Y.; Dimopoulou, M. Malolactic fermentation—Theoretical advances and practical considerations. Fermentation 2022, 8, 521. [Google Scholar] [CrossRef]
- Styger, G.; Jacobson, D.; Bauer, F.F. Identifying genes that impact on aroma profiles produced by Saccharomyces cerevisiae and the production of higher alcohols. Appl. Microbiol. Biotechnol. 2001, 91, 713–730. [Google Scholar] [CrossRef] [PubMed]
- Eden, A.; Nedervelde, L.V.; Drukker, M.; Benvenisty, N.; Debourg, A. Involvement of branched-chain amino acid aminotransferases in the production of fusel alcohols during fermentation in yeast. Appl. Microbiol. Biotechnol. 2001, 55, 296–300. [Google Scholar] [CrossRef]
- Kim, B.; Cho, B.R.; Hahn, J.S. Metabolic engineering of Saccharomyces cerevisiae for the production of 2-phenylethanol via ehrlich pathway. Biotechnol. Bioeng. 2014, 111, 115–124. [Google Scholar] [CrossRef]









| Strains or Plasmids | Relevant Characteristic | Reference or Source |
|---|---|---|
| Strain | ||
| E. coli DH5α | supE44ΔlacU169(ϕ80lacZΔM15) hsdR17 recAl endAl gyrA96 thi-1 relA | This lab |
| WY1 | Wild-type industrial Saccharomyces uvarum | This lab |
| Lactococcus lactis | Wild-type industrial Lactococcus lactis | This lab |
| Lactobacillus plantarum | Wild-type industrial Lactobacillus plantarum | This lab |
| Oenococcus oeni | Wild-type industrial Oenococcus oeni | This lab |
| Schizosaccharomyces pombe | Wild-type industrial Schizosaccharomyces pombe | This lab |
| WY0 | PGK1p-PGK1t-loxP-KanMX-loxP | This study |
| WYE | PGK1p-MAE1-PGK1t-loxP-KanMX-loxP | This study |
| WYSN | PGK1p-mleSN (Lactococcus lactis)-PGK1t-loxP-KanMX-loxP | This study |
| WYSZ | PGK1p-mleSZ (Lactobacillus plantarum)-PGK1t-loxP-KanMX-loxP | This study |
| WYAJ | PGK1p-mleAJ(Oenococcus oeni)-PGK1t-loxP-KanMX-loxP | This study |
| WYm2 | PGK1p-MAE2(Schizosaccharomyces pombe)-PGK1t-loxP-KanMX-loxP | This study |
| WYPN | PGK1p-mlePN (Lactococcus lactis)-PGK1t-loxP-KanMX-loxP | This study |
| WYPZ | PGK1p-mlePZ(Lactobacillus plantarum)-PGK1t-loxP-KanMX-loxP | This study |
| WYPJ | PGK1p-mlePJ(Oenococcus oeni)-PGK1t-loxP-KanMX-loxP | This study |
| WYm1 | PGK1p-MAE1(Schizosaccharomyces pombe)-PGK1t-loxP-KanMX-loxP | This study |
| WYPE | PGK1p-mlePN(Lactococcus lactis)-PGK1t-PGK1p-MAE1-PGK1t-loxP -KanMX-loxP | This study |
| WYPSN | PGK1p-mlePN(Lactococcus lactis)-PGK1t-PGK1p-mleSN(Lactococcus lactis)-PGK1t-loxP- KanMX-loxP | This study |
| WYPSZ | PGK1p-mlePN(Lactococcus lactis)-PGK1t-PGK1p-mleSZ(Lactobacillus plantarum)-PGK1t-loxP- KanMX-loxP | This study |
| WYPAJ | PGK1p-mlePN(Lactococcus lactis)-PGK1t-PGK1p-mleAJ(Oenococcus oeni)-PGK1t-loxP- KanMX-loxP | This study |
| WYPm2 | PGK1p-mlePN(Lactococcus lactis)-PGK1t-PGK1p-MAE2(Schizosaccharomyces pombe)-PGK1t-loxP- KanMX-loxP | This study |
| WYm1E | PGK1p-MAE1(Schizosaccharomyces pombe)-PGK1t-PGK1p-MAE1-PGK1t-loxP -KanMX-loxP | This study |
| WYm1SN | PGK1p-MAE1(Schizosaccharomyces pombe)-PGK1t-PGK1p-mleSN(Lactococcus lactis)-PGK1t-loxP- KanMX-loxP | This study |
| WYm1SZ | PGK1p-MAE1(Schizosaccharomyces pombe)-PGK1t-PGK1p-mleSZ(Lactobacillus plantarum)-PGK1t-loxP- KanMX-loxP | This study |
| WYm1AJ | PGK1p-MAE1(Schizosaccharomyces pombe)-PGK1t-PGK1p-mleAJ(Oenococcus oeni)-PGK1t-loxP- KanMX-loxP | This study |
| WYm1m2 | PGK1p-MAE1(Schizosaccharomyces pombe)-PGK1t-PGK1p-MAE2(Schizosaccharomyces pombe)-PGK1t-loxP- KanMX-loxP | This study |
| Plasmids | ||
| Yep352 pUG6 | Apr, cloning vector E. coli/S. cerevisiae shuttle vector, containing Amp+ and loxP-KanMX-loxP cassette | This lab This study |
| pPGK1 | E. coli/S. cerevisiae shuttle vector, containing Amp+and PGK1p and PGK1t | This study |
| pSH-Zeocin | Zeor, Cre expression vector | This study |
| Primers | Sequence (5′→3′) |
|---|---|
| PGK-U | CGCGGATCCTCTAACTGATCTATCCAAAACTG |
| PGK-D | ACGCGTCGACTAACGAACGCAGAATTTTCGAG |
| MAE-U | GAATTCCAGATCTCCTCGAGATGCTTAGAACCAGACTATCCG |
| MAE-D | TCTATCGCAGATCCCTCGAGCTACAATTGGTTGGTGTGCAC |
| MAE2-U | GAATTCCAGATCTCCTCGAGGCACGTGGACCGTCTTACC |
| MAE2-D | TCTATCGCAGATCCCTCGAGAGTTGATGAATAACAATAGGAGAAA |
| mleSN-U | GAATTCCAGATCTCCTCGAGATGCGTGCACATGAAATTT |
| mleSN-D | TCTATCGCAGATCCCTCGAGTTAGTACTCTGGATACCATTTAAGA |
| K(ApaI)-U | CCGCTAACAATACCTGGGCCCCAGCTGAAGCTTCGTACGC |
| K(ApaI)-D | GCACACGGTGTGGTGGGCCCGCATAGGCCACTAGTGGATCTG |
| MAE1-U | GAATTCCAGATCTCCTCGAGTTCATTTTCTCTCTTGGCCAC |
| MAE1-D | TCTATCGCAGATCCCTCGAGCTTTTGTCATGAAATCCCTCTTA |
| mlePN-U | GAATTCCAGATCTCCTCGAGATGAAAAAACTTAAAGAAACGA |
| mlePN-D | TCTATCGCAGATCCCTCGAGTTAATAAAAGAATCGTATAAGAATT |
| PGK(SmaI)-U | GTACCCGGGTCTAACTGATCTATCCAAAACTGA |
| PGK(SmaI)-D | GATCCCCGGGTAACGAACGCAGAATTTTC |
| mleSZ-U | GAATTCCAGATCTCCTCGAGATGACAAAAACTGCAAGTGAAAT |
| mleSZ-D | TCTATCGCAGATCCCTCGAGCTATTTGCTGATGGCCCG |
| mleAJ-U | GAATTCCAGATCTCCTCGAGATGACAGATCCAGTAAGTATT |
| mleAJ-D | TCTATCGCAGATCCCTCGAGATTAGTATTTCGGATCCCACT |
| YmlePN-U | ATGAAAAAACTTAAAGAAACGA |
| YmlePN-D | TTAATAAAAGAATCGTATAAGAATT |
| YmleSZ-U | ATGACAAAAACTGCAAGTGAAAT |
| YmleSZ-D | CTATTTGCTGATGGCCCG |
| YmleAJ-U | ATGACAGATCCAGTAAGTATT |
| YmleAJ-D | ATTAGTATTTCGGATCCCACT |
| K-U | CAGCTGAAGCTTCGTACGC |
| K-D | GCATAGGCCACTAGTGGATCTG |
| ApaI-U | CCTGCTTCAAACCGCTAACAATA |
| ApaI-D | CGAATGCACACGGTGTGGT |
| SmaI-U | TTCGAGCTCGGTACCCG |
| SmaI-D | AGTTAGAGGATCCCCGGG |
| Strains | n-Propanol (mg/L) | Isobutyl Alcohol (mg/L) | Isoamyl Alcohol (mg/L) | Phenethyl Alcohol (mg/L) | Ethyl Acetate (mg/L) | Ethyl Lactate (mg/L) |
|---|---|---|---|---|---|---|
| WY1 | 51.83 ± 2.94 | 35.354 ± 1.54 | 201.53 ± 2.67 | 13.521 ± 0.98 | 23.301 ± 1.04 | ≈0 |
| WY0 | 52.342 ± 3.212 | 35.421 ± 1.89 | 204.321 ± 2.85 | 13.395 ± 1.13 | 23.721 ± 1.75 | ≈0 |
| WYE | 51.921 ± 1.97 | 35.015 ± 2.16 | 205.127 ± 3.57 | 13.832 ± 0.97 | 23.55 ± 2.53 | ≈0 |
| WYm2 | 52.044 ± 1.72 | 34.199 ± 1.05 | 212.105 ± 7.61 * | 14.145 ± 1.32 | 28.521 ± 1.93 * | ≈0 |
| WYSN | 50.697 ± 1.56 | 29.955 ± 3.52 * | 176.282 ± 4.05 * | 14.290 ± 1.53 | 21.716 ± 1.00 * | 11.115 ± 0.59 * |
| WYAJ | 51.017 ± 1.05 | 31.832 ± 1.35 * | 189.205 ± 2.13 * | 14.315 ± 1.05 | 21.573 ± 0.45 * | 5.065 ± 0.84 * |
| WYSZ | 50.538 ± 1.35 | 32.375 ± 2.30 * | 191.372 ± 1.85 * | 13.580 ± 1.38 | 21.063 ± 0.75 * | 6.130 ± 0.05 * |
| WYPN | 53.757 ± 2.45 | 36.335 ± 0.15 * | 217.858 ± 5.93 * | 14.033 ± 0.75 | 27.473 ± 1.89 * | ≈0 |
| WYm1 | 55.646 ± 0.89 | 45.244 ± 1.92 * | 223.662 ± 3.19 * | 14.257 ± 0.62 | 25.914 ± 2.33 * | ≈0 |
| WYPE | 52.216 ± 3.67 | 40.785 ± 0.98 * | 219.903 ± 4.97 * | 13.889 ± 1.05 | 26.530 ± 1.83 * | ≈0 |
| WYPSN | 52.385 ± 2.41 | 34.639 ± 1.53 | 210.775 ± 6.05 * | 14.496 ± 1.42 | 29.741 ± 2.04 * | 13.942 ± 1.42 * |
| WYPSZ | 52.353 ± 2.86 | 35.745 ± 2.05 | 205.62 ± 3.55 * | 13.753 ± 1.95 | 24.342 ± 1.05 * | 7.431 ± 1.55 * |
| WYPAJ | 51.964 ± 2.67 | 35.069 ± 1.80 | 204.175 ± 2.42 | 13.766 ± 1.65 | 20.156 ± 0.05 | 5.905 ± 1.80 * |
| WYPm2 | 54.791 ± 1.98 | 34.881 ± 2.94 | 211.031 ± 2.45 * | 14.468 ± 1.08 | 29.896 ± 1.52 * | ≈0 |
| WYm1E | 55.521 ± 4.05 * | 45.004 ± 5.08 * | 220.291 ± 3.12 * | 14.823 ± 0.67 | 26.393 ± 2.74 * | ≈0 |
| WYm1SN | 51.068 ± 1.54 | 28.184 ± 1.06 * | 171.756 ± 1.09 * | 14.630 ± 0.32 | 15.850 ± 1.08 * | 13.992 ± 1.08 * |
| WYm1SZ | 53.532 ± 2.05 | 34.977 ± 2.04 | 202.437 ± 4.96 | 12.966 ± 0.85 | 27.906 ± 2.48 * | 9.327 ± 0.76 * |
| WYm1AJ | 52.462 ± 1.77 | 35.728 ± 3.62 | 182.482 ± 4.96 * | 12.964 ± 1.05 | 14.874 ± 2.16 * | 5.594 ± 0.63 * |
| WYm1m2 | 53.705 ± 3.06 | 32.144 ± 2.95 * | 210.968 ± 3.25 * | 13.989 ± 1.93 | 32.705 ± 1.33 * | ≈0 |
| Strains | Weight Losses of CO2 (g) | Ethanol (%, v/v) | Residual Sugar (g/L) |
|---|---|---|---|
| WY1 | 14.95 ± 0.15 | 11.02 ± 0.12 | 2.65 ± 0.10 |
| WY0 | 14.83 ± 0.25 | 11.03 ± 0.15 | 2.78 ± 0.21 |
| WYE | 14.90 ± 0.14 | 11.04 ± 0.15 | 2.66 ± 0.10 |
| WYm2 | 14.79 ± 0.12 | 10.95 ± 0.20 | 2.70 ± 0.08 |
| WYSN | 14.82 ± 0.20 | 10.80 ± 0.14 | 2.60 ± 0.05 |
| WYAJ | 14.92 ± 0.22 | 11.16 ± 0.22 | 2.61 ± 0.12 |
| WYSZ | 14.93 ± 0.20 | 12.10 ± 0.25 | 2.78 ± 0.27 |
| WYPN | 14.72 ± 0.12 | 11.00 ± 0.15 | 2.67 ± 0.20 |
| WYm1 | 14.79 ± 0.12 | 10.95 ± 0.18 | 2.62 ± 0.14 |
| WYPE | 14.75 ± 0.14 | 11.05 ± 0.21 | 2.7 ± 0.15 |
| WYPSN | 14.82 ± 0.20 | 10.95 ± 0.10 | 2.65 ± 0.15 |
| WYPSZ | 14.85 ± 0.15 | 11.05 ± 0.05 | 2.70 ± 0.02 |
| WYPAJ | 14.80 ± 0.22 | 11.00 ± 0.13 | 2.72 ± 0.13 |
| WYPm2 | 14.79 ± 0.12 | 11.10 ± 0.15 | 2.66 ± 0.12 |
| WYm1E | 14.75 ± 0.14 | 11.00 ± 0.14 | 2.65 ± 0.15 |
| WYm1SN | 14.86 ± 0.24 | 11.05 ± 0.10 | 2.64 ± 0.14 |
| WYm1SZ | 14.83 ± 0.12 | 11.00 ± 0.06 | 2.70 ± 0.10 |
| WYm1AJ | 14.62 ± 0.15 | 10.83 ± 0.12 * | 2.97 ± 0.05 * |
| WYm1m2 | 14.82 ± 0.20 | 11.02 ± 0.15 | 2.62 ± 0.06 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, P.; Song, W.; Wang, Y.; Li, X.; Wu, S.; Li, B.; Zhang, C. Effects of Heterologous Expression of Genes Related L–Malic acid Metabolism in Saccharomyces uvarum on Flavor Substances Production in Wine. Foods 2024, 13, 2038. https://doi.org/10.3390/foods13132038
Li P, Song W, Wang Y, Li X, Wu S, Li B, Zhang C. Effects of Heterologous Expression of Genes Related L–Malic acid Metabolism in Saccharomyces uvarum on Flavor Substances Production in Wine. Foods. 2024; 13(13):2038. https://doi.org/10.3390/foods13132038
Chicago/Turabian StyleLi, Ping, Wenjun Song, Yumeng Wang, Xin Li, Shankai Wu, Bingjuan Li, and Cuiying Zhang. 2024. "Effects of Heterologous Expression of Genes Related L–Malic acid Metabolism in Saccharomyces uvarum on Flavor Substances Production in Wine" Foods 13, no. 13: 2038. https://doi.org/10.3390/foods13132038
APA StyleLi, P., Song, W., Wang, Y., Li, X., Wu, S., Li, B., & Zhang, C. (2024). Effects of Heterologous Expression of Genes Related L–Malic acid Metabolism in Saccharomyces uvarum on Flavor Substances Production in Wine. Foods, 13(13), 2038. https://doi.org/10.3390/foods13132038






