Low Gluten Beers Contain Variable Gluten and Immunogenic Epitope Content
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Sample Collection
2.3. In-Solution Enzymatic Digest
2.4. Discovery Proteomics
2.5. Targeted Proteomics
2.6. Statistical Analysis and Visualisation
2.7. Epitope Mapping
3. Results
3.1. Measuring Gluten Protein Content in Beer Using Competitive ELISA
3.2. Discovery Proteomics on Trypsin- and Chymotrypsin-Digested Beer Samples
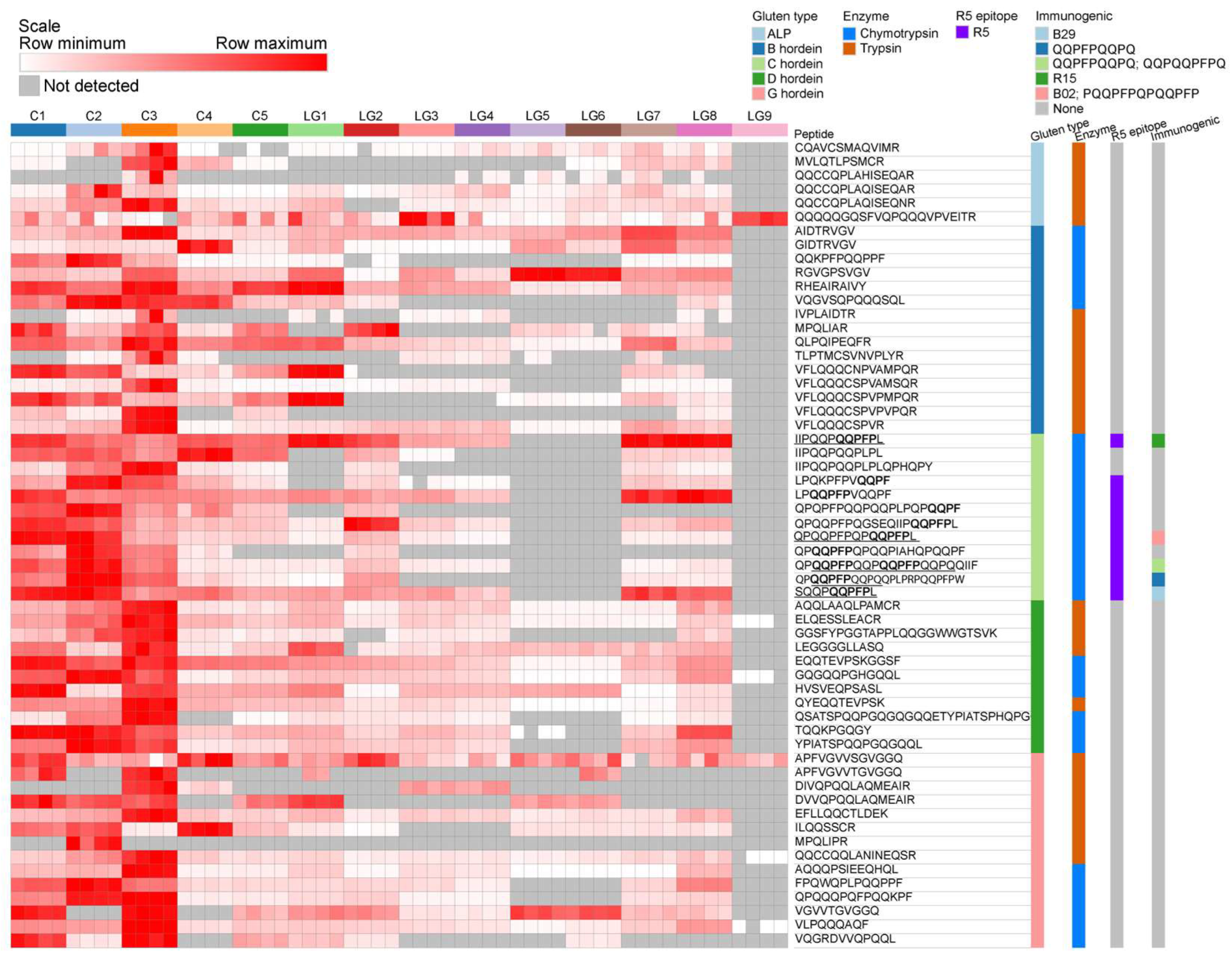
3.2.1. Identification of Hordein Proteins in LG Beer
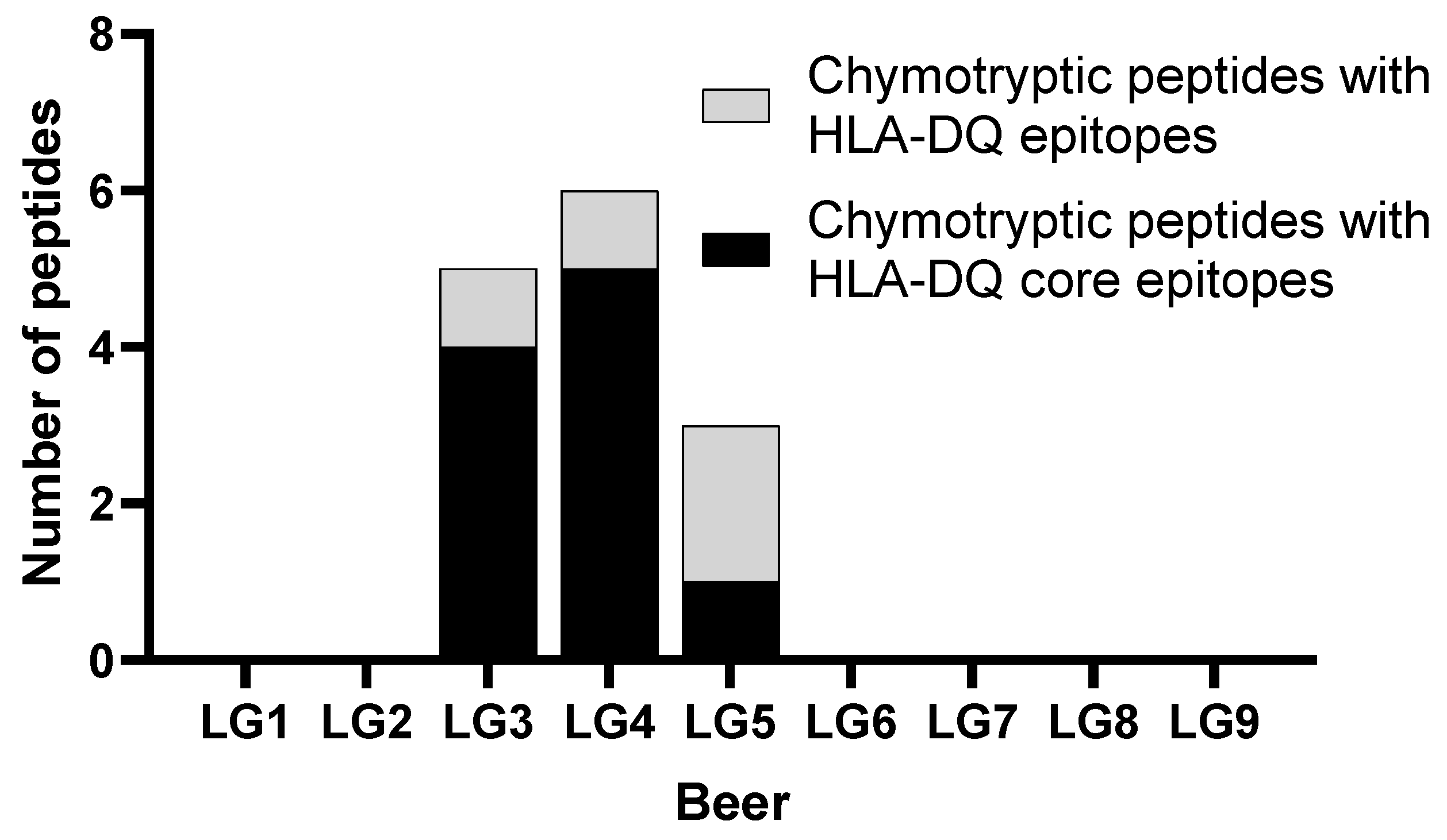
3.2.2. Identification of Hordein Peptides in LG Beer
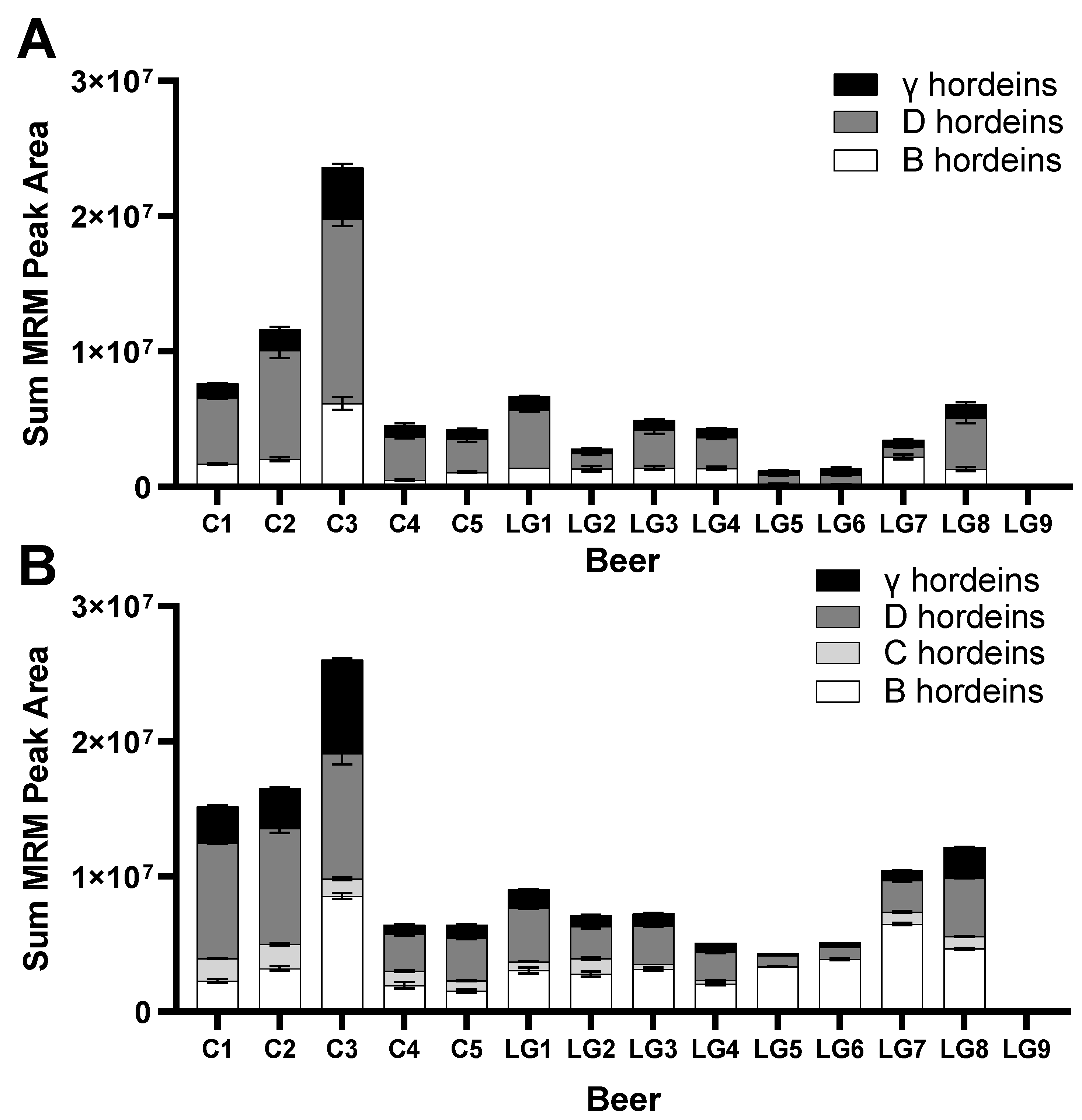
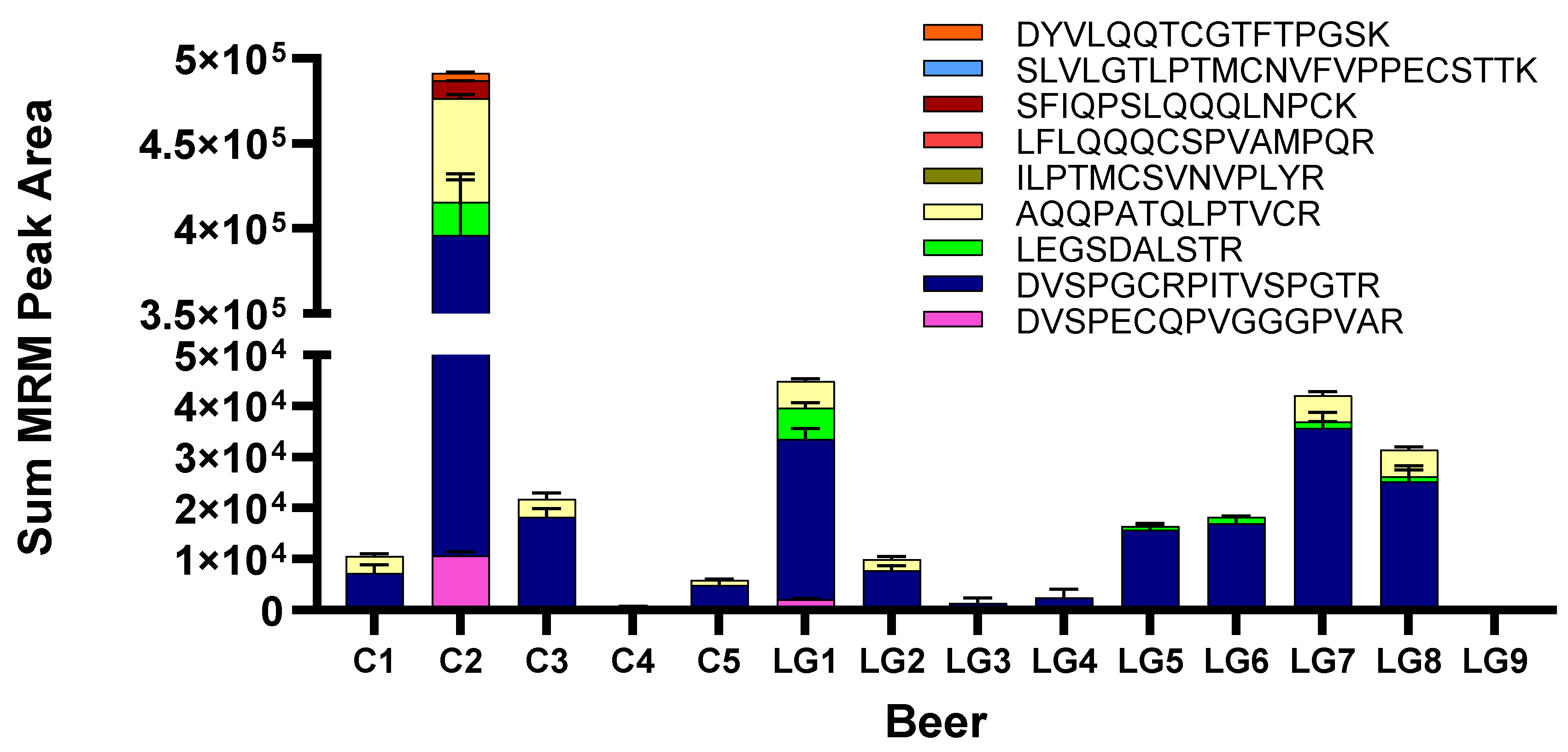
3.3. Targeted MRM-MS for Hordein Quantification
3.4. Measurement of Relative Abundance of Gluten Proteins
3.5. Wheat Detection
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Tye-Din, J.A.; Galipeau, H.J.; Agardh, D. Celiac disease: A review of current concepts in pathogenesis, prevention, and novel therapies. Front. Pediatr. 2018, 6, 350. [Google Scholar] [CrossRef] [PubMed]
- Juhász, A.; Belova, T.; Florides, C.G.; Maulis, C.; Fischer, I.; Gell, G.; Birinyi, Z.; Ong, J.; Keeble-Gagnère, G.; Maharajan, A. Genome mapping of seed-borne allergens and immunoresponsive proteins in wheat. Sci. Adv. 2018, 4, eaar8602. [Google Scholar] [CrossRef]
- Jabri, B.; Sollid, L.M. Tissue-mediated control of immunopathology in coeliac disease. Nat. Rev. Immunol. 2009, 9, 858–870. [Google Scholar] [CrossRef] [PubMed]
- Nurminen, S.; Kivelä, L.; Huhtala, H.; Kaukinen, K.; Kurppa, K. Extraintestinal manifestations were common in children with coeliac disease and were more prevalent in patients with more severe clinical and histological presentation. Acta Paediatr. 2019, 108, 681–687. [Google Scholar] [CrossRef] [PubMed]
- Catassi, C.; Fabiani, E.; Iacono, G.; D’Agate, C.; Francavilla, R.; Biagi, F.; Volta, U.; Accomando, S.; Picarelli, A.; De Vitis, I. A prospective, double-blind, placebo-controlled trial to establish a safe gluten threshold for patients with celiac disease. Am. J. Clin. Nutr. 2007, 85, 160–166. [Google Scholar] [CrossRef] [PubMed]
- Allred, L.K.; Lesko, K.; McKiernan, D.; Kupper, C.; Guandalini, S. The celiac patient antibody response to conventional and gluten-removed beer. J. AOAC Int. 2017, 100, 485–491. [Google Scholar] [CrossRef] [PubMed]
- CXS 118-1979; Food and Agriculture Organization/World Health Organization (FAO/WHO). Standard for Foods for Special Dietary Use for Persons Intolerant to Gluten; World Health Organization: Rome, Italy, 2015.
- Gluten Intolerance Group. GFCO Manual Rev. 2021.1; Gluten Free Certification Organization: Auburn, WA, USA, 2021. [Google Scholar]
- Food and Drug Administration. Food labeling: gluten-free labeling of foods. Final rule. Fed. Regist. 2013, 78, 47154–47179. [Google Scholar]
- Bugyi, Z.; Török, K.; Hajas, L.; Adonyi, Z.; Popping, B.; Tömösközi, S. Comparative study of commercially available gluten ELISA kits using an incurred reference material. Qual. Assur. Saf. Crops Foods 2013, 5, 79–87. [Google Scholar] [CrossRef]
- Lexhaller, B.; Tompos, C.; Scherf, K.A. Fundamental study on reactivities of gluten protein types from wheat, rye and barley with five sandwich ELISA test kits. Food Chem. 2017, 237, 320–330. [Google Scholar] [CrossRef]
- Mitea, C.; Kooy-Winkelaar, Y.; van Veelen, P.; de Ru, A.; Drijfhout, J.W.; Koning, F.; Dekking, L. Fine specificity of monoclonal antibodies against celiac disease–inducing peptides in the gluteome. Am. J. Clin. Nutr. 2008, 88, 1057–1066. [Google Scholar] [CrossRef]
- Morón, B.; Cebolla, A.; Manyani, H.; Alvarez-Maqueda, M.; Megìas, M.; del Carmen, T.M.; Lopez, M.C.; Sousa, C. Sensitive detection of cereal fractions that are toxic to celiac disease patients by using monoclonal antibodies to a main immunogenic wheat peptide. Am. J. Clin. Nutr. 2008, 87, 405–414. [Google Scholar] [CrossRef] [PubMed]
- Colgrave, M.L.; Goswami, H.; Howitt, C.A.; Tanner, G.J. What is in a Beer? Proteomic Characterization and Relative Quantification of Hordein (Gluten) in Beer. J. Proteome Res. 2012, 11, 386–396. [Google Scholar] [CrossRef] [PubMed]
- Osman, A.A.; Uhlig, H.H.; Valdes, I.; Amin, M.; Méndez, E.; Mothes, T. A monoclonal antibody that recognizes a potential coeliac-toxic repetitive pentapeptide epitope in gliadins. Eur. J. Gastroenterol. Hepatol. 2001, 13, 1189–1193. [Google Scholar] [CrossRef] [PubMed]
- Kahlenberg, F.; Sanchez, D.; Lachmann, I.; Tuckova, L.; Tlaskalova, H.; Méndez, E.; Mothes, T. Monoclonal antibody R5 for detection of putatively coeliac-toxic gliadin peptides. Eur. Food Res. Technol. 2006, 222, 78–82. [Google Scholar] [CrossRef]
- Valdés, I.; García, E.; Llorente, M.; Méndez, E. Innovative approach to low-level gluten determination in foods using a novel sandwich enzyme-linked immunosorbent assay protocol. Eur. J. Gastroenterol. Hepatol. 2003, 15, 465–747. [Google Scholar] [CrossRef] [PubMed]
- Skerritt, J.; Hill, A.; Andrews, J. Antigenicity of wheat prolamins: Detailed epitope analysis using a panel of monoclonal antibodies. J. Cereal Sci. 2000, 32, 259–279. [Google Scholar] [CrossRef]
- Skerritt, J.; Hill, A. Monoclonal antibody sandwich enzyme immunoassays for determination of gluten in foods. J. Agric. Food Chem. 1990, 38, 1771–1778. [Google Scholar] [CrossRef]
- Lexhaller, B.; Tompos, C.; Scherf, K.A. Comparative analysis of prolamin and glutelin fractions from wheat, rye, and barley with five sandwich ELISA test kits. Anal. Bioanal. Chem. 2016, 408, 6093–6104. [Google Scholar] [CrossRef]
- Rallabhandi, P.; Sharma, G.M.; Pereira, M.; Williams, K.M. Immunological characterization of the gluten fractions and their hydrolysates from wheat, rye and barley. J. Agric. Food Chem. 2015, 63, 1825–1832. [Google Scholar] [CrossRef]
- Tanner, G.J.; Blundell, M.J.; Colgrave, M.L.; Howitt, C.A. Quantification of hordeins by ELISA: The correct standard makes a magnitude of difference. PLoS ONE 2013, 8, e56456. [Google Scholar] [CrossRef]
- Tanabe, S. IgE-binding Abilities of Pentapeptides, QQPFP and PQQPF in Wheat Gliadin. J. Nutr. Sci. Vitaminol. 2004, 50, 367–370. [Google Scholar] [CrossRef] [PubMed]
- Bruins Slot, I.D.; Bremer, M.G.E.G.; Van Der Fels-Klerx, I.; Hamer, R.J. Evaluating the performance of gluten ELISA test kits: The numbers do not tell the tale. Cereal Chem. 2015, 92, 513–521. [Google Scholar] [CrossRef]
- Scherf, K.A.; Poms, R.E. Recent developments in analytical methods for tracing gluten. J. Cereal Sci. 2016, 67, 112–122. [Google Scholar] [CrossRef]
- Scherf, K.A. Gluten analysis of wheat starches with seven commercial ELISA test kits—Up to six different values. Food Anal. Methods 2017, 10, 234–246. [Google Scholar] [CrossRef]
- Diaz-Amigo, C.; Popping, B. Accuracy of ELISA detection methods for gluten and reference materials: A realistic assessment. J. Agric. Food Chem. 2013, 61, 5681–5688. [Google Scholar] [CrossRef]
- Thompson, T.; Méndez, E. Commercial Assays to Assess Gluten Content of Gluten-Free Foods: Why They Are Not Created Equal. J. Am. Diet. Assoc. 2008, 108, 1682–1687. [Google Scholar] [CrossRef] [PubMed]
- Haas-Lauterbach, S.; Immer, U.; Richter, M.; Koehler, P. Gluten fragment detection with a competitive ELISA. J. AOAC Int. 2012, 95, 377–381. [Google Scholar] [CrossRef]
- Dostálek, P.; Hochel, I.; Méndez, E.; Hernando, A.; Gabrovská, D. Immunochemical determination of gluten in malts and beers. Food Addit. Contam. 2006, 23, 1074–1078. [Google Scholar] [CrossRef]
- Colgrave, M.L.; Byrne, K.; Howitt, C.A. Liquid Chromatography–Mass Spectrometry Analysis Reveals Hydrolyzed Gluten in Beers Crafted to Remove Gluten. J. Agric. Food Chem. 2017, 65, 9715–9725. [Google Scholar] [CrossRef]
- Taylor, J.P.; Zannini, E.; Jacob, F.; Arendt, E.K. A study on malt modification, used as a tool to reduce levels of beer hordeins. J. Inst. Brew. 2018, 124, 143–147. [Google Scholar] [CrossRef]
- Leiper, K.A.; Stewart, G.G.; McKeown, I.P.; Nock, T.; Thompson, M.J. Optimising beer stabilisation by the selective removal of tannoids and sensitive proteins. J. Inst. Brew. 2005, 111, 118–127. [Google Scholar] [CrossRef]
- Taylor, J.; Jacob, F.; Arendt, E. Fundamental study on the impact of transglutaminase on hordein levels in beer. J. Am. Soc. Brew. Chem. 2015, 73, 253–260. [Google Scholar] [CrossRef]
- Wieser, H.; Koehler, P. Detoxification of gluten by means of enzymatic treatment. J. AOAC Int. 2012, 95, 356–363. [Google Scholar] [CrossRef] [PubMed]
- Edens, L.; Dekker, P.; Van Der Hoeven, R.; Deen, F.; de Roos, A.; Floris, R. Extracellular prolyl endoprotease from Aspergillus niger and its use in the debittering of protein hydrolysates. J. Agric. Food Chem. 2005, 53, 7950–7957. [Google Scholar] [CrossRef] [PubMed]
- Fiedler, K.L.; Cao, W.; Zhang, L.; Naziemiec, M.; Bedford, B.; Yin, L.; Smith, N.; Arbuckle, M.; Lopez-Hernandez, A.; Jackson, L.S. Detection of gluten in a pilot-scale barley-based beer produced with and without a prolyl endopeptidase enzyme. Food Addit. Contam. Part A 2019, 36, 1151–1162. [Google Scholar] [CrossRef] [PubMed]
- Ribeiro, M.; Nunes, F.M.; Guedes, S.; Domingues, P.; Silva, A.M.; Carrillo, J.M.; Rodriguez-Quijano, M.; Branlard, G.; Igrejas, G. Efficient chemo-enzymatic gluten detoxification: Reducing toxic epitopes for celiac patients improving functional properties. Sci. Rep. 2015, 5, 18041. [Google Scholar] [CrossRef] [PubMed]
- Tanner, G.J.; Blundell, M.J.; Colgrave, M.L.; Howitt, C.A. Creation of the first ultra-low gluten barley (Hordeum vulgare L.) for coeliac and gluten-intolerant populations. Plant Biotechnol. J. 2016, 14, 1139–1150. [Google Scholar] [CrossRef] [PubMed]
- Gumienna, M.; Górna, B. Gluten hypersensitivities and their impact on the production of gluten-free beer. Eur. Food Res. Technol. 2020, 246, 2147–2160. [Google Scholar] [CrossRef]
- Colgrave, M.L.; Goswami, H.; Blundell, M.; Howitt, C.A.; Tanner, G.J. Using mass spectrometry to detect hydrolysed gluten in beer that is responsible for false negatives by ELISA. J. Chromatogr. A 2014, 1370, 105–114. [Google Scholar] [CrossRef]
- Food and Drug Administration. Food Labeling; Gluten-Free Labeling of Fermented or Hydrolyzed Foods; Docket No. FDA-2014-N-1021; Food and Drug Administration: Rockville, MD, USA, 2020; pp. 49240–49261. [Google Scholar]
- Tang, W.H.; Shilov, I.V.; Seymour, S.L. Nonlinear fitting method for determining local false discovery rates from decoy database searches. J. Proteome Res. 2008, 7, 3661–3667. [Google Scholar] [CrossRef]
- MacLean, B.; Tomazela, D.M.; Shulman, N.; Chambers, M.; Finney, G.L.; Frewen, B.; Kern, R.; Tabb, D.L.; Liebler, D.C.; MacCoss, M.J. Skyline: An open source document editor for creating and analyzing targeted proteomics experiments. Bioinformatics 2010, 26, 966–968. [Google Scholar] [CrossRef]
- Colgrave, M.L.; Byrne, K.; Blundell, M.; Howitt, C.A. Identification of barley-specific peptide markers that persist in processed foods and are capable of detecting barley contamination by LC-MS/MS. J. Proteom. 2016, 147, 169–176. [Google Scholar] [CrossRef] [PubMed]
- Ouyang, S.; Buell, C.R. The TIGR Plant Repeat Databases: A collective resource for the identification of repetitive sequences in plants. Nucleic Acids Res. 2004, 32, D360–D363. [Google Scholar] [CrossRef] [PubMed]
- Sollid, L.M.; Qiao, S.-W.; Anderson, R.P.; Gianfrani, C.; Koning, F. Nomenclature and listing of celiac disease relevant gluten T-cell epitopes restricted by HLA-DQ molecules. Immunogenetics 2012, 64, 455–460. [Google Scholar] [CrossRef] [PubMed]
- Sollid, L.M.; Tye-Din, J.A.; Qiao, S.-W.; Anderson, R.P.; Gianfrani, C.; Koning, F. Update 2020: Nomenclature and listing of celiac disease–relevant gluten epitopes recognized by CD4+ T cells. Immunogenetics 2020, 72, 85–88. [Google Scholar] [CrossRef]
- Tye-Din, J.A.; Stewart, J.A.; Dromey, J.A.; Beissbarth, T.; van Heel, D.A.; Tatham, A.; Henderson, K.; Mannering, S.I.; Gianfrani, C.; Jewell, D.P.; et al. Comprehensive, quantitative mapping of T cell epitopes in gluten in celiac disease. Sci. Transl. Med. 2010, 2, 41ra51. [Google Scholar] [CrossRef] [PubMed]
- Shilov, I.V.; Seymour, S.L.; Patel, A.A.; Loboda, A.; Tang, W.H.; Keating, S.P.; Hunter, C.L.; Nuwaysir, L.M.; Schaeffer, D.A. The Paragon Algorithm, a Next Generation Search Engine That Uses Sequence Temperature Values and Feature Probabilities to Identify Peptides from Tandem Mass Spectra. Mol. Cell. Proteom. 2007, 6, 1638–1655. [Google Scholar] [CrossRef]
- Vita, R.; Mahajan, S.; Overton, J.A.; Dhanda, S.K.; Martini, S.; Cantrell, J.R.; Wheeler, D.K.; Sette, A.; Peters, B. The Immune Epitope Database (IEDB): 2018 update. Nucleic Acids Res. 2019, 47, D339–D343. [Google Scholar] [CrossRef]
- Shewry, P.; Bunce, N.; Kreis, M.; Forde, B. Polymorphism at the Hor 1 locus of barley (Hordeum vulgare L.). Biochem. Genet. 1985, 23, 391–404. [Google Scholar] [CrossRef]
- Gu, Y.Q.; Anderson, O.D.; Londeorë, C.F.; Kong, X.; Chibbar, R.N.; Lazo, G.R. Structural organization of the barley D-hordein locus in comparison with its orthologous regions of wheat genomes. Genome 2003, 46, 1084–1097. [Google Scholar] [CrossRef]
- Shewry, P.R.; Tatham, A.; Field, J.M.; Fords, B.G.; Clark, J.; Gallois, P.; Marris, C.; Halford, N.; Fords, J.; Kreis, M. The Structures of Barley and Wheat Prolamins and their Genes. Biochem. Und Physiol. Der Pflanz. 1988, 183, 117–127. [Google Scholar] [CrossRef]
- Shewry, P.; Kreis, M.; Parmar, S.; Lew, E.-L.; Kasarda, D. Identification of γ-type hordeins in barley. FEBS Lett. 1985, 190, 61–64. [Google Scholar] [CrossRef]
- Colgrave, M.L.; Goswami, H.; Byrne, K.; Blundell, M.; Howitt, C.A.; Tanner, G.J. Proteomic Profiling of 16 Cereal Grains and the Application of Targeted Proteomics To Detect Wheat Contamination. J. Proteome Res. 2015, 14, 2659–2668. [Google Scholar] [CrossRef] [PubMed]
- Pérot, M.; Lupi, R.; Guyot, S.; Delayre-Orthez, C.; Gadonna-Widehem, P.; Thébaudin, J.-Y.; Bodinier, M.; Larré, C. Polyphenol Interactions Mitigate the Immunogenicity and Allergenicity of Gliadins. J. Agric. Food Chem. 2017, 65, 6442–6451. [Google Scholar] [CrossRef] [PubMed]
- Plundrich, N.J.; Kulis, M.; White, B.L.; Grace, M.H.; Guo, R.; Burks, A.W.; Davis, J.P.; Lila, M.A. Novel strategy to create hypoallergenic peanut protein–polyphenol edible matrices for oral immunotherapy. J. Agric. Food Chem. 2014, 62, 7010–7021. [Google Scholar] [CrossRef] [PubMed]
- Chung, S.-Y.; Champagne, E.T. Reducing the allergenic capacity of peanut extracts and liquid peanut butter by phenolic compounds. Food Chem. 2009, 115, 1345–1349. [Google Scholar] [CrossRef]
- Siebert, K.J.; Troukhanova, N.V.; Lynn, P.Y. Nature of polyphenol− protein interactions. J. Agric. Food Chem. 1996, 44, 80–85. [Google Scholar] [CrossRef]
- Baell, J.; Walters, M.A. Chemistry: Chemical con artists foil drug discovery. Nature 2014, 513, 481–483. [Google Scholar] [CrossRef]
- R-Biopharm AG. Enzyme immunoassay for the quantitative determination of gliadins and corresponding proteins. In RIDASCREEN® Gliadin R7001; R-Biopharm AG: Darmstadt, Germany, 2012. [Google Scholar]
- R-Biopharm AG. Enzyme immunoassay for the quantitative determination of peptide fragments of gliadins and corresponding proteins. In RIDASCREEN® Gliadin Competitive R7021; R-Biopharm AG: Darmstadt, Germany, 2015. [Google Scholar]
- Allred, L.K.; Nye-Wood, M.G.; Colgrave, M.L. Analysis of gluten in dried yeast and yeast-containing products. Foods 2020, 9, 1790. [Google Scholar] [CrossRef]

| Beer Reference Number | Description | Nation of Origin | R5 Competitive ELISA Gluten Concentration (mg/kg) |
|---|---|---|---|
| C1 | Adjunct lager | Australia | 60.0 |
| C2 | Cream ale | Australia | 146.3 |
| C3 | Strong lager | Denmark | 55.9 |
| C4 | Adjunct lager | Mexico | 9.0 |
| C5 | Adjunct lager | Australia | 5.4 |
| LG1 | Pilsner | Germany | 4.7 |
| LG2 | Pale lager | Australia | 14.7 |
| LG3 | IPA | Scotland | 4.6 |
| LG4 | Pilsner | Scotland | 5.3 |
| LG5 | Pale lager (Oct 2018) | Finland | 4.2 |
| LG6 | Pale lager (May 2019) | Finland | 4.9 |
| LG7 | Pilsner | Finland | 7.5 |
| LG8 | Helles | Finland | 6.0 |
| LG9 | Lager | Finland | 2.0 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nye-Wood, M.G.; Byrne, K.; Stockwell, S.; Juhász, A.; Bose, U.; Colgrave, M.L. Low Gluten Beers Contain Variable Gluten and Immunogenic Epitope Content. Foods 2023, 12, 3252. https://doi.org/10.3390/foods12173252
Nye-Wood MG, Byrne K, Stockwell S, Juhász A, Bose U, Colgrave ML. Low Gluten Beers Contain Variable Gluten and Immunogenic Epitope Content. Foods. 2023; 12(17):3252. https://doi.org/10.3390/foods12173252
Chicago/Turabian StyleNye-Wood, Mitchell G., Keren Byrne, Sally Stockwell, Angéla Juhász, Utpal Bose, and Michelle L. Colgrave. 2023. "Low Gluten Beers Contain Variable Gluten and Immunogenic Epitope Content" Foods 12, no. 17: 3252. https://doi.org/10.3390/foods12173252
APA StyleNye-Wood, M. G., Byrne, K., Stockwell, S., Juhász, A., Bose, U., & Colgrave, M. L. (2023). Low Gluten Beers Contain Variable Gluten and Immunogenic Epitope Content. Foods, 12(17), 3252. https://doi.org/10.3390/foods12173252