ACE-Inhibitory Activity of Whey Proteins Fractions Derived of Fermentation by Lacticaseibacillus rhamnosus GG and Streptococcus thermophilus SY-102
Abstract
:1. Introduction
2. Materials and Methods
2.1. Culture Preparation
2.2. Fermentation
2.3. Proteolytic Profile Analysis
2.3.1. Analysis of Free Amino Groups
2.3.2. Peptide Separation by Tris-Tricine-SDS-PAGE
2.3.3. Separation of Peptides by SEC-HPLC
2.4. ACE Inhibitory Activity
2.5. Statistical Analysis
3. Results and Discussion
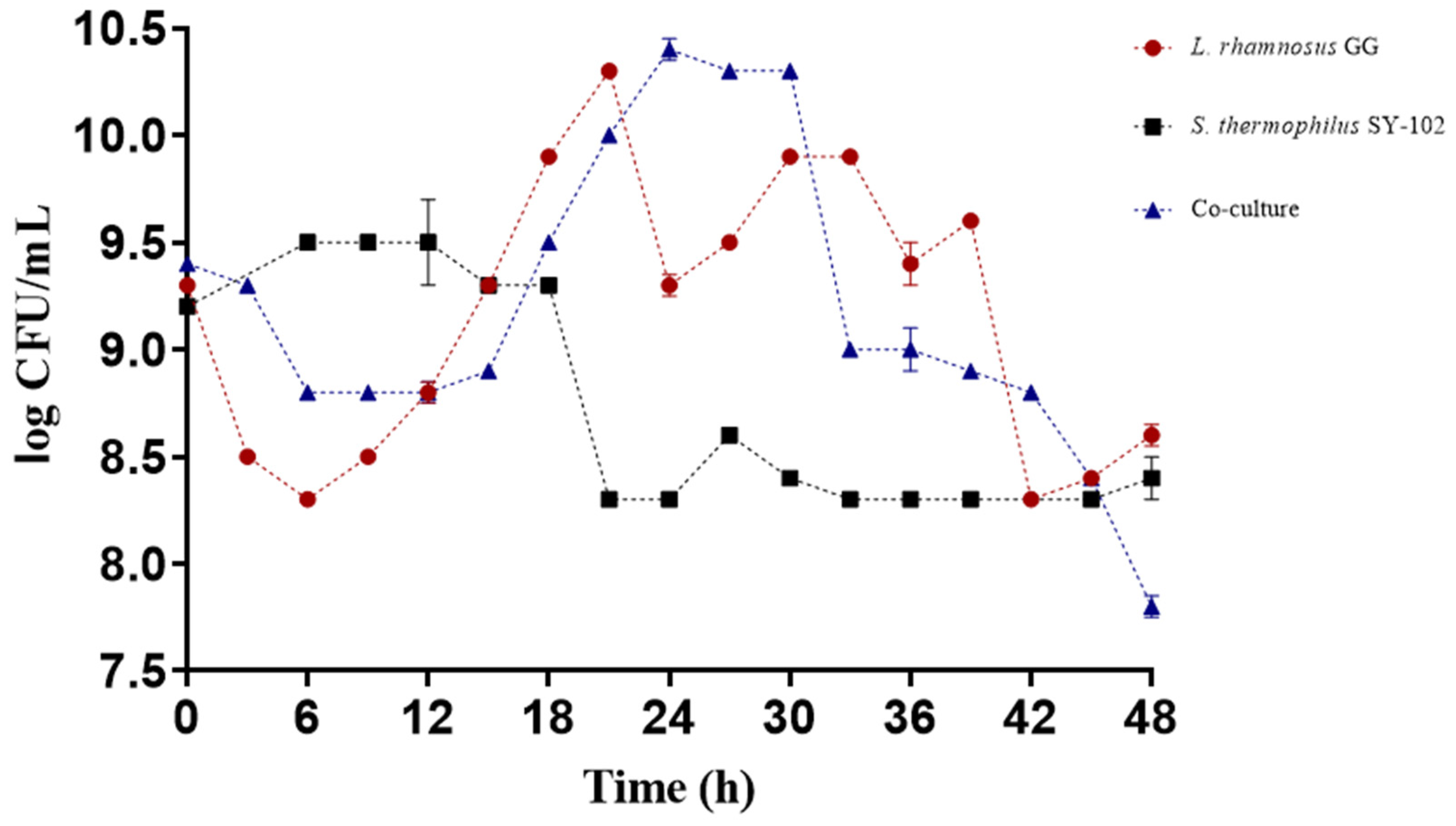
3.1. Growth during Fermentation
3.2. pH Changes of the Medium during the Time of Fermentation
3.3. Proteolysis
3.3.1. Determination of Free Amino Groups by Technique of TNBS
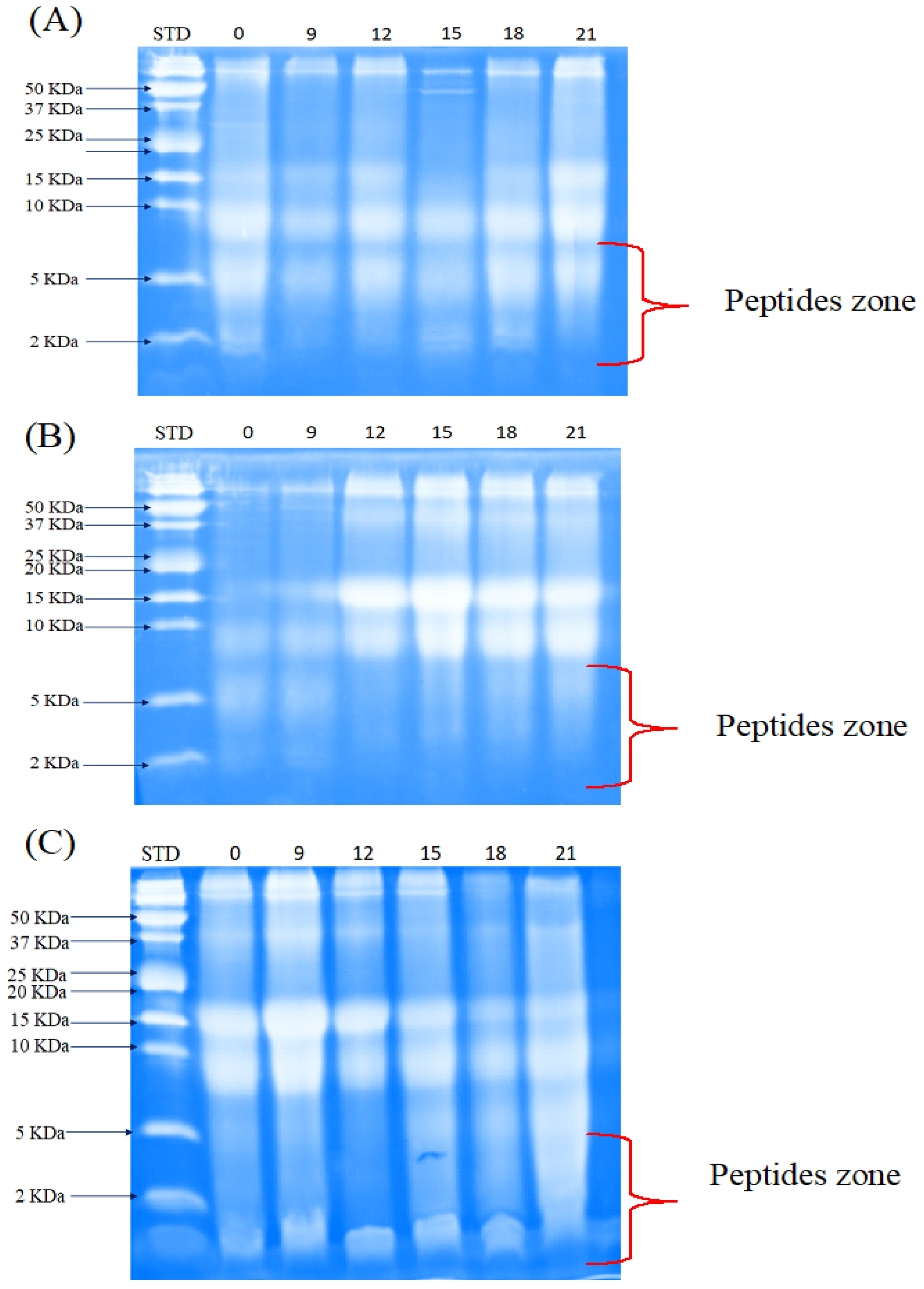
3.3.2. Peptide Separation by Tris-Tricine-SDS-PAGE
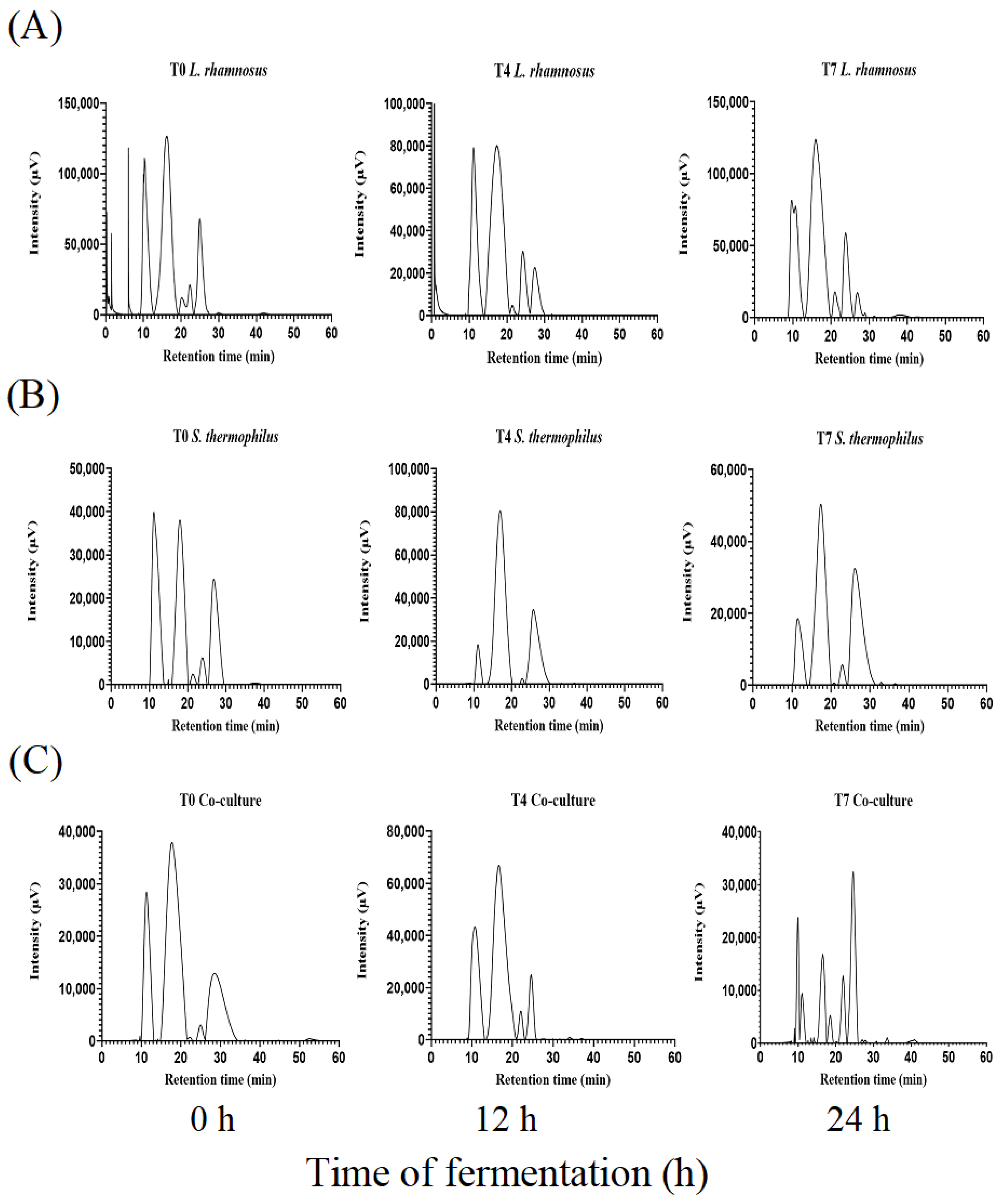
3.3.3. Separation of Peptides by SEC-HPLC
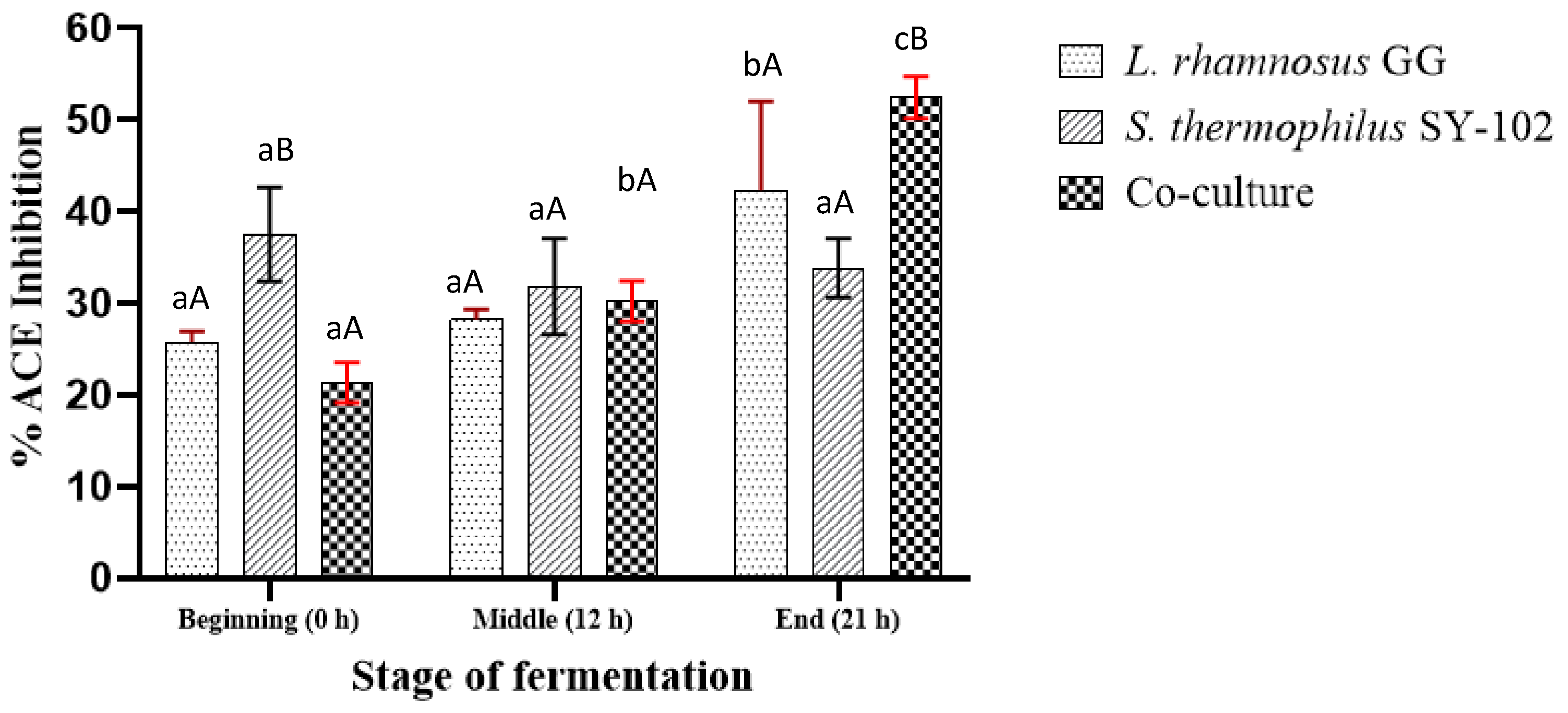
3.4. Determination of ACE Inhibition
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Mollea, C.; Marmo, L.; Bosco, F. Valorisation of Cheese Whey, a By-Product from the Dairy Industry. In Food Industry; Muzzalupo, I., Ed.; InTech: London, UK, 2013; ISBN 978-953-51-0911-2. [Google Scholar]
- Kareb, O.; Aïder, M. Whey and Its Derivatives for Probiotics, Prebiotics, Synbiotics, and Functional Foods: A Critical Review. Probiotics Antimicrob. Prot. 2019, 11, 348–369. [Google Scholar] [CrossRef]
- Hannibal, B.; Santillán, A.; Mercy, A.; Ramos, E.; Paola, V.; Rincon, A. Aprovechamiento Del Suero de Leche Como Bebida Energizante Para Minimizar El Impacto Ambiental. Eur. Sci. J. 2015, 11, 257–268. [Google Scholar]
- Oriach, C.S.; Robertson, R.C.; Stanton, C.; Cryan, J.F.; Dinan, T.G. Food for Thought: The Role of Nutrition in the Microbiota-Gut–Brain Axis. Clin. Nutr. Exp. 2016, 6, 25–38. [Google Scholar] [CrossRef] [Green Version]
- Olvera-Rosales, L.B.; Cruz-Guerrero, A.E.; García-Garibay, J.M.; Gómez-Ruíz, L.C.; Contreras-López, E.; Guzmán-Rodríguez, F.; González-Olivares, L.G. Bioactive Peptides of Whey: Obtaining, Activity, Mechanism of Action, and Further Applications. Crit. Rev. Food Sci. Nutr 2022, 1–31. [Google Scholar] [CrossRef]
- Dullius, A.; Goettert, M.I.; de Souza, C.F.V. Whey Protein Hydrolysates as a Source of Bioactive Peptides for Functional Foods—Biotechnological Facilitation of Industrial Scale-Up. J. Func. Foods 2018, 42, 58–74. [Google Scholar] [CrossRef]
- Daliri, E.B.-M.; Lee, B.H.; Park, B.-J.; Kim, S.-H.; Oh, D.-H. Antihypertensive Peptides from Whey Proteins Fermented by Lactic Acid Bacteria. Food Sci. Biotechnol. 2018, 27, 1781–1789. [Google Scholar] [CrossRef]
- Corrêa, A.P.F.; Daroit, D.J.; Fontoura, R.; Meira, S.M.M.; Segalin, J.; Brandelli, A. Hydrolysates of Sheep Cheese Whey as a Source of Bioactive Peptides with Antioxidant and Angiotensin-Converting Enzyme Inhibitory Activities. Peptides 2014, 61, 48–55. [Google Scholar] [CrossRef] [Green Version]
- Fu, Z.; Lin, J. An Overview of Bioinformatics Tools and Resources in Allergy. In Food Allergens; Lin, J., Alcocer, M., Eds.; Methods in Molecular Biology; Springer New York: New York, NY, USA, 2017; Volume 1592, pp. 223–245. ISBN 978-1-4939-6923-4. [Google Scholar]
- Xia, Y.; Yu, J.; Xu, W.; Shuang, Q. Purification and Characterization of Angiotensin-I-Converting Enzyme Inhibitory Peptides Isolated from Whey Proteins of Milk Fermented with Lactobacillus plantarum QS670. J. Dairy Sci. 2020, 103, 4919–4928. [Google Scholar] [CrossRef] [PubMed]
- Raveschot, C.; Cudennec, B.; Coutte, F.; Flahaut, C.; Fremont, M.; Drider, D.; Dhulster, P. Production of Bioactive Peptides by Lactobacillus Species: From Gene to Application. Front. Microbiol. 2018, 9, 2354. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, M.; Bayjanov, J.R.; Renckens, B.; Nauta, A.; Siezen, R.J. The Proteolytic System of Lactic Acid Bacteria Revisited: A Genomic Comparison. BMC Genom. 2010, 11, 36. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Martin, M.; Hagemann, D.; Henle, T.; Deussen, A. The Angiotensin Converting Enzyme-Inhibitory Effects of the Peptide Isoleucine-Tryptophan after Oral Intake via Whey Hydrolysate in Men. J. Hypertens. 2018, 36, e220. [Google Scholar] [CrossRef]
- Onuh, J.O.; Aluko, R.E. Metabolomics as a Tool to Study the Mechanism of Action of Bioactive Protein Hydrolysates and Peptides: A Review of Current Literature. Trends Food Sci. Technol. 2019, 91, 625–633. [Google Scholar] [CrossRef]
- Madureira, A.R.; Tavares, T.; Gomes, A.M.P.; Pintado, M.E.; Malcata, F.X. Invited Review: Physiological Properties of Bioactive Peptides Obtained from Whey Proteins. J. Dairy Sci. 2010, 93, 437–455. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tondo, A.R.; Caputo, L.; Mangiatordi, G.F.; Monaci, L.; Lentini, G.; Logrieco, A.F.; Montaruli, M.; Nicolotti, O.; Quintieri, L. Structure-Based Identification and Design of Angiotensin Converting Enzyme-Inhibitory Peptides from Whey Proteins. J. Agric. Food Chem. 2019, 68, 541–548. [Google Scholar] [CrossRef] [PubMed]
- de Oliveira, M.E.G.; Garcia, E.F.; de Oliveira, C.E.V.; Gomes, A.M.P.; Pintado, M.M.E.; Madureira, A.R.M.F.; da Conceição, M.L.; do EgyptoQueiroga, R.D.C.R.; de Souza, E.L. Addition of Probiotic Bacteria in a Semi-Hard Goat Cheese (Coalho): Survival to Simulated Gastrointestinal Conditions and Inhibitory Effect against Pathogenic Bacteria. Food Res. Int. 2014, 64, 241–247. [Google Scholar] [CrossRef]
- Schägger, H.; Von Jagow, G. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal. Biochem. 1987, 166, 368–379. [Google Scholar] [CrossRef]
- González-Olivares, L.G.; Añorve-Morga, J.; Castañeda-Ovando, A.; Contreras-López, E.; Jaimez-Ordaz, J. Peptide Separation of Commercial Fermented Milk during Refrigerated Storage. Food Sci. Technol. 2014, 34, 674–679. [Google Scholar] [CrossRef] [Green Version]
- Cushman, D.W.; Wang, F.L.; Fung, W.C.; Harvey, C.M.; DeForrest, J.M. Differentiation of Angiotensin–Converting Enzyme (ACE) Inhibitors by Their Selective Inhibition of ACE in Physiologically Important Target Organs. Am. J. Hypertens. 1989, 2, 294–306. [Google Scholar] [CrossRef]
- Liu, E.; Zheng, H.; Shi, T.; Ye, L.; Konno, T.; Oda, M.; Shen, H.; Ji, Z.-S. Relationship between Lactobacillus bulgaricus and Streptococcus thermophilus under Whey Conditions: Focus on Amino Acid Formation. Int. Dairy J. 2016, 56, 141–150. [Google Scholar] [CrossRef]
- Crittenden, R.G.; Martinez, N.R.; Playne, M.J. Synthesis and Utilisation of Folate by Yoghurt Starter Cultures and Probiotic Bacteria. Int. J. Food Microbiol. 2003, 80, 217–222. [Google Scholar] [CrossRef]
- Yu, Y.; Yu, W.; Jin, Y. Peptidomic Analysis of Milk Fermented by Lactobacillus delbrueckii subsp. bulgaricus and Streptococcus thermophilus. FHFH 2021, 1, 100033. [Google Scholar]
- Hols, P.; Hancy, F.; Fontaine, L.; Grossiord, B.; Prozzi, D.; Leblondbourget, N.; Decaris, B.; Bolotin, A.; Delorme, C.; Duskoehrlich, S. New Insights in the Molecular Biology and Physiology of Revealed by Comparative Genomics. FEMS Microbiol. Rev. 2005, 29, 435–463. [Google Scholar] [CrossRef]
- Sebastián-Nicolas, J.L.; Contreras-López, E.; Ramírez-Godínez, J.; Cruz-Guerrero, A.E.; Rodríguez-Serrano, G.M.; Añorve-Morga, J.; Jaimez-Ordaz, J.; Castañeda-Ovando, A.; Pérez-Escalante, E.; Ayala-Niño, A.; et al. Milk Fermentation by Lacticaseibacillus rhamnosus GG and Streptococcus thermophilus SY-102: Proteolytic Profile and ACE-Inhibitory Activity. Fermentation 2021, 7, 215. [Google Scholar] [CrossRef]
- Vinderola, C.G.; Mocchiutti, P.; Reinheimer, J.A. Interactions among Lactic Acid Starter and Probiotic Bacteria Used for Fermented Dairy Products. J. Dairy Sci 2002, 85, 721–729. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez-Serrano, G.M.; Garcia-Garibay, J.M.; Cruz-Guerrero, A.E.; del Carmen Gomez-Ruiz, L.; Ayala-Nino, A.; Castaneda-Ovando, A.; Gonzalez-Olivares, L.G. Proteolytic System of Streptococcus thermophilus. J. Microbiol. Biotechnol. 2018, 28, 1581–1588. [Google Scholar] [CrossRef]
- Hekmat, S.; Soltani, H.; Reid, G. Growth and Survival of Lactobacillus reuteri RC-14 and Lactobacillus rhamnosus GR-1 in Yogurt for Use as a Functional Food. Innov. Food Sci. Emerg. Technol. 2009, 10, 293–296. [Google Scholar] [CrossRef]
- Sun, J.; Chen, H.; Qiao, Y.; Liu, G.; Leng, C.; Zhang, Y.; Lv, X.; Feng, Z. The Nutrient Requirements of Lactobacillus rhamnosus GG and Their Application to Fermented Milk. J. Dairy Sci. 2019, 102, 5971–5978. [Google Scholar] [CrossRef]
- Courtin, P.; Monnet, V.; Rul, F. Cell-Wall Proteinases PrtS and PrtB Have a Different Role in Streptococcus thermophilus/Lactobacillus bulgaricus Mixed Cultures in Milk. Microbiology 2002, 148, 3413–3421. [Google Scholar] [CrossRef] [Green Version]
- Chang, O.K.; Roux, É.; Awussi, A.A.; Miclo, L.; Jardin, J.; Jameh, N.; Dary, A.; Humbert, G.; Perrin, C. Use of a Free Form of the Streptococcus thermophilus Cell Envelope Protease PrtS as a Tool to Produce Bioactive Peptides. Int. Dairy J. 2014, 38, 104–115. [Google Scholar] [CrossRef]
- Ali, E.; Nielsen, S.D.; Aal, A.-E.; El-Leboudy, A.; Saleh, E.; LaPointe, G. Use of Mass Spectrometry to Profile Peptides in Whey Protein Isolate Medium Fermented by Lactobacillus helveticus LH-2 and Lactobacillus acidophilus La-5. Front. Nutr. 2019, 6, 152. [Google Scholar] [CrossRef] [Green Version]
- Hafeez, Z.; Cakir-Kiefer, C.; Lecomte, X.; Miclo, L.; Dary-Mourot, A. The X-Prolyl Dipeptidyl-Peptidase PepX of Streptococcus thermophilus Initially Described as Intracellular Is Also Responsible for Peptidase Extracellular Activity. J. Dairy Sci. 2019, 102, 113–123. [Google Scholar] [CrossRef] [Green Version]
- Hu, T.; Cui, Y.; Qu, X. Analysis of the Proteolytic System of Streptococcus thermophilus Strains CS5, CS9, CS18 and CS20. Int. Dairy J. 2021, 118, 105025. [Google Scholar] [CrossRef]
- Innocente, N.; Biasutti, M.; Rita, F.; Brichese, R.; Comi, G.; Iacumin, L. Effect of Indigenous Lactobacillus rhamnosus Isolated from Bovine Milk on Microbiological Characteristics and Aromatic Profile of Traditional Yogurt. LWT 2016, 66, 158–164. [Google Scholar] [CrossRef]
- Quirós, A.; Ramos, M.; Muguerza, B.; Delgado, M.A.; Miguel, M.; Aleixandre, A.; Recio, I. Identification of Novel Antihypertensive Peptides in Milk Fermented with Enterococcus faecalis. Int. Dairy J. 2007, 17, 33–41. [Google Scholar] [CrossRef]
- Murray, B.A.; FitzGerald, R.J. Angiotensin Converting Enzyme Inhibitory Peptides Derived from Food Proteins: Biochemistry, Bioactivity and Production. Curr. Pharm. Des. 2007, 13, 773–791. [Google Scholar] [CrossRef]
- Yu, Y.; Jin, Y.; Wang, F.; Yan, J.; Qi, Y.; Ye, M. Protein Digestomic Analysis Reveals the Bioactivity of Deer Antler Velvet in Simulated Gastrointestinal Digestion. Food Res. Int. 2017, 96, 182–190. [Google Scholar] [CrossRef] [PubMed]
- Li, G.-H.; Le, G.-W.; Shi, Y.-H.; Shrestha, S. Angiotensin I–Converting Enzyme Inhibitory Peptides Derived from Food Proteins and Their Physiological and Pharmacological Effects. Nutr. Res. 2004, 24, 469–486. [Google Scholar] [CrossRef]

| Free Amino Groups μg/mL | |||
|---|---|---|---|
| Time of Fermentation (h) | L. rhamnosus GG | S. thermophilus SY-102 | L. rhamnosus GG + S. thermophilus SY-102 |
| 0 | 366.91 ± 0.02 aAB | 463.43 ± 0.13 aA | 226.59 ± 0.15 aAB |
| 9 | 277.67 ± 0.01 abA | 264.63 ± 0.02 bA | 285.28 ± 0.13 aA |
| 12 | 208.97 ± 0.02 abcA | 167.34 ± 0.05 bcA | 363.32 ± 0.02 aB |
| 15 | 239.85 ± 0.02 abcA | 255.06 ± 0.00 bA | 253.76 ± 0.20 aA |
| 18 | 277.23 ± 0.07 abA | 350.60 ± 0.03 abdA | 268.97 ± 0.17 aA |
| 21 | 293.54 ± 0.05 abA | 323.54 ± 0.04 abAC | 452.67 ± 0.06 aB |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Olvera-Rosales, L.B.; Pérez-Escalante, E.; Castañeda-Ovando, A.; Contreras-López, E.; Cruz-Guerrero, A.E.; Regal-López, P.; Cardelle-Cobas, A.; González-Olivares, L.G. ACE-Inhibitory Activity of Whey Proteins Fractions Derived of Fermentation by Lacticaseibacillus rhamnosus GG and Streptococcus thermophilus SY-102. Foods 2023, 12, 2416. https://doi.org/10.3390/foods12122416
Olvera-Rosales LB, Pérez-Escalante E, Castañeda-Ovando A, Contreras-López E, Cruz-Guerrero AE, Regal-López P, Cardelle-Cobas A, González-Olivares LG. ACE-Inhibitory Activity of Whey Proteins Fractions Derived of Fermentation by Lacticaseibacillus rhamnosus GG and Streptococcus thermophilus SY-102. Foods. 2023; 12(12):2416. https://doi.org/10.3390/foods12122416
Chicago/Turabian StyleOlvera-Rosales, Laura Berenice, Emmanuel Pérez-Escalante, Araceli Castañeda-Ovando, Elizabeth Contreras-López, Alma Elizabeth Cruz-Guerrero, Patricia Regal-López, Alejandra Cardelle-Cobas, and Luis Guillermo González-Olivares. 2023. "ACE-Inhibitory Activity of Whey Proteins Fractions Derived of Fermentation by Lacticaseibacillus rhamnosus GG and Streptococcus thermophilus SY-102" Foods 12, no. 12: 2416. https://doi.org/10.3390/foods12122416







