Potential of Near-Infrared Spectroscopy (NIRS) for Efficient Classification Based on Postharvest Storage Time, Cultivar and Maturity in Coconut Water
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Treatments
2.2. Measurement of Biochemical Properties
2.3. NIRS Measurement
2.4. Chemometrics Analysis and Data Analysis
3. Results
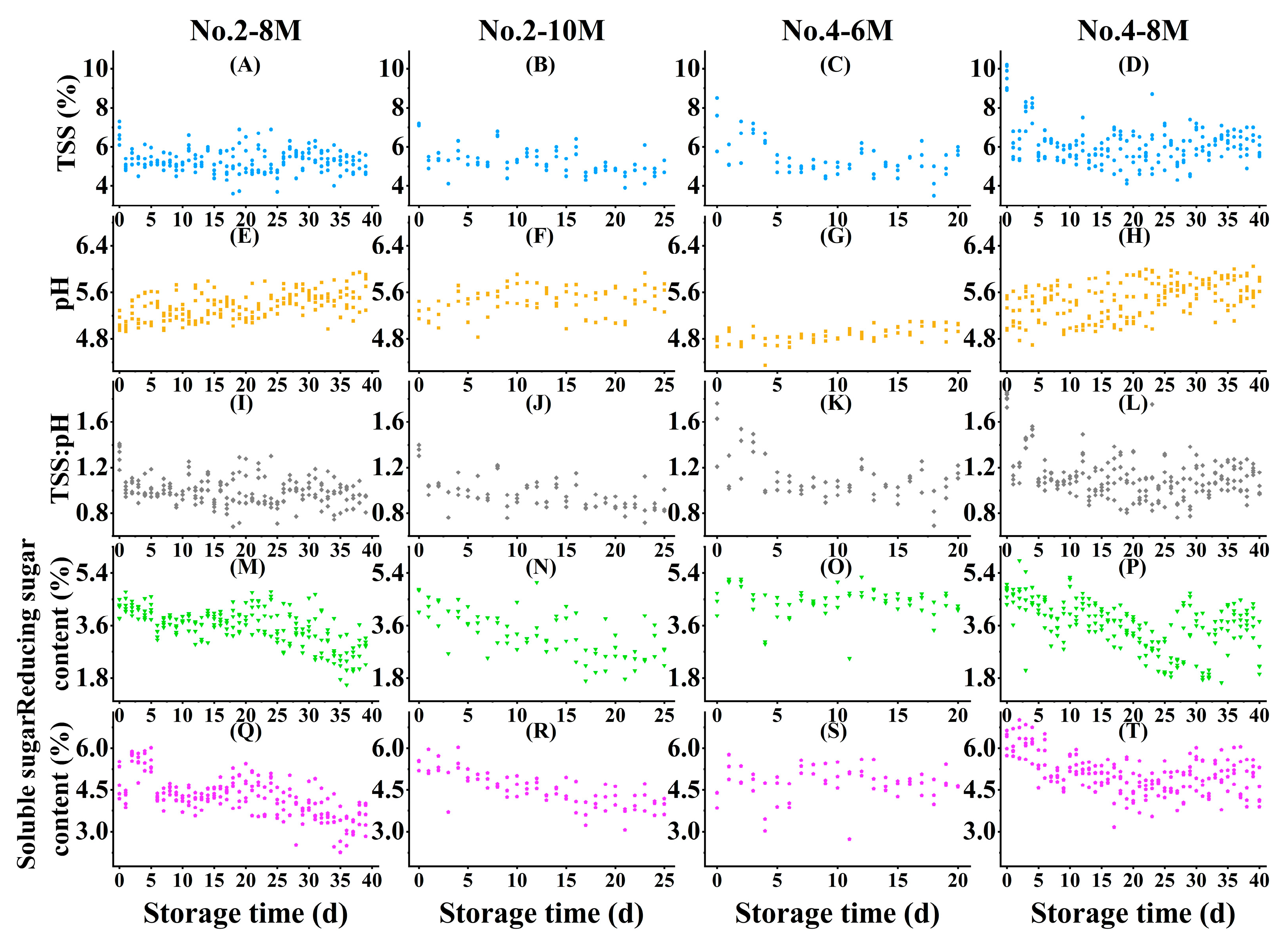
3.1. Distribution and Quantification of Reference Data
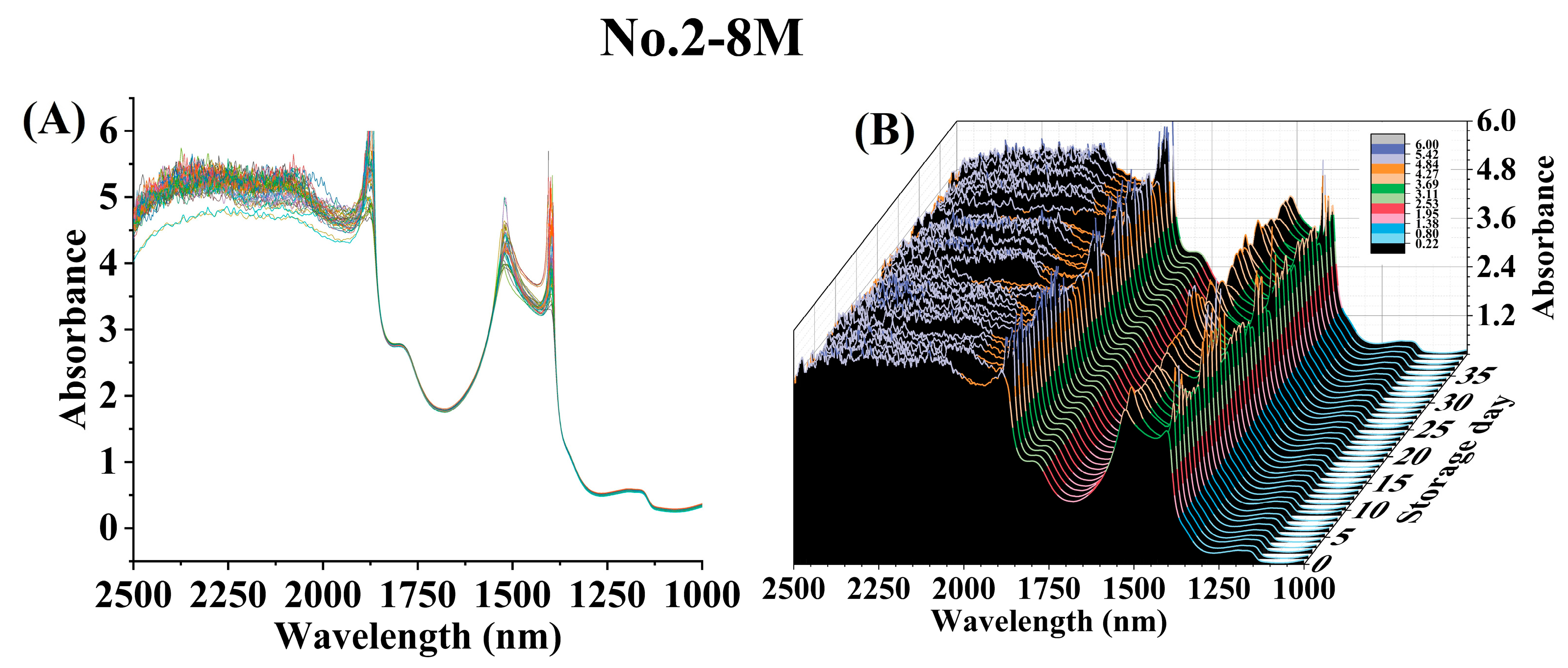
3.2. Spectral Characteristics
3.3. Model Development for Quantitative Analysis
3.4. Classification of CW Samples Based on Spectral Information
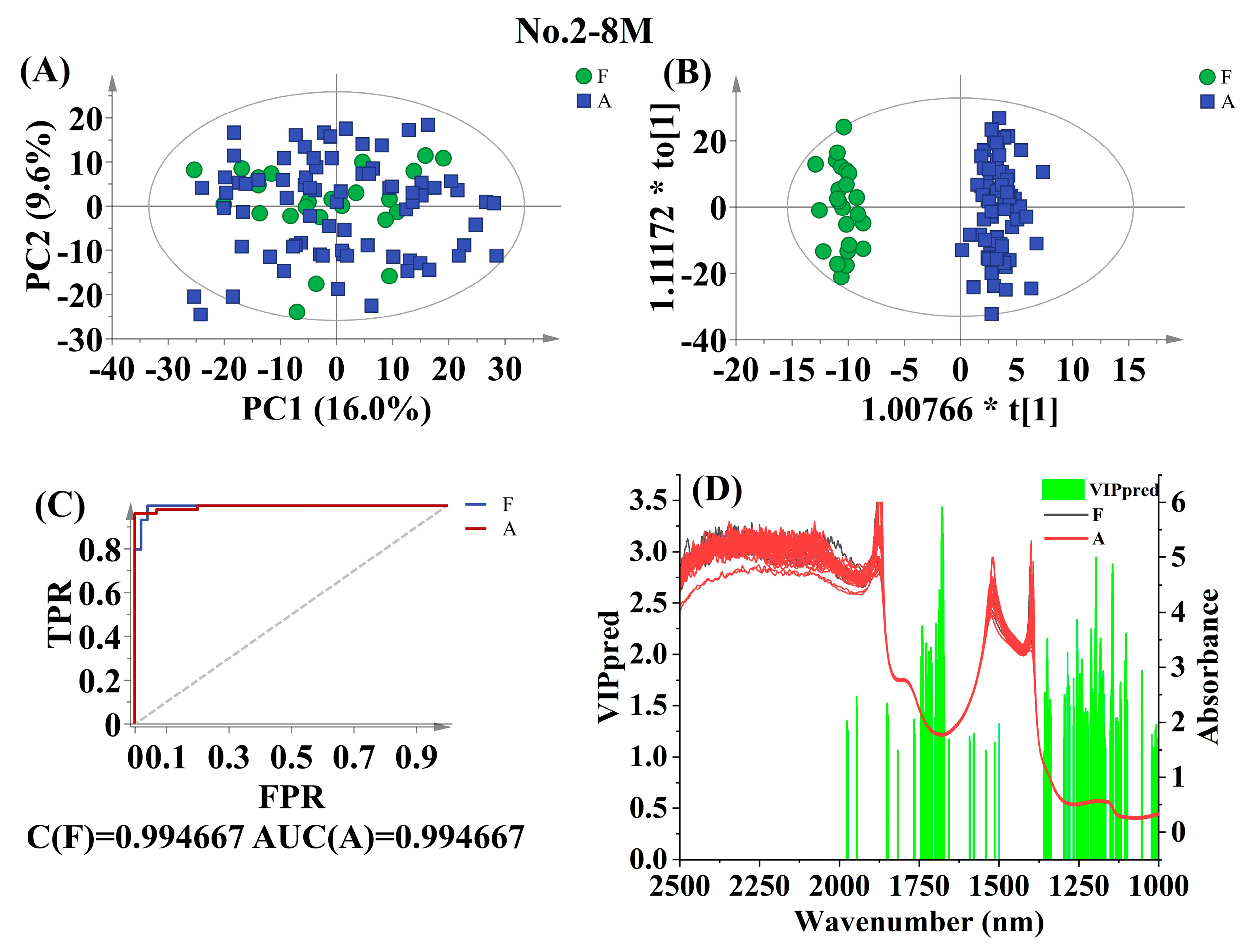
3.4.1. Discrimination using Postharvest Storage Time
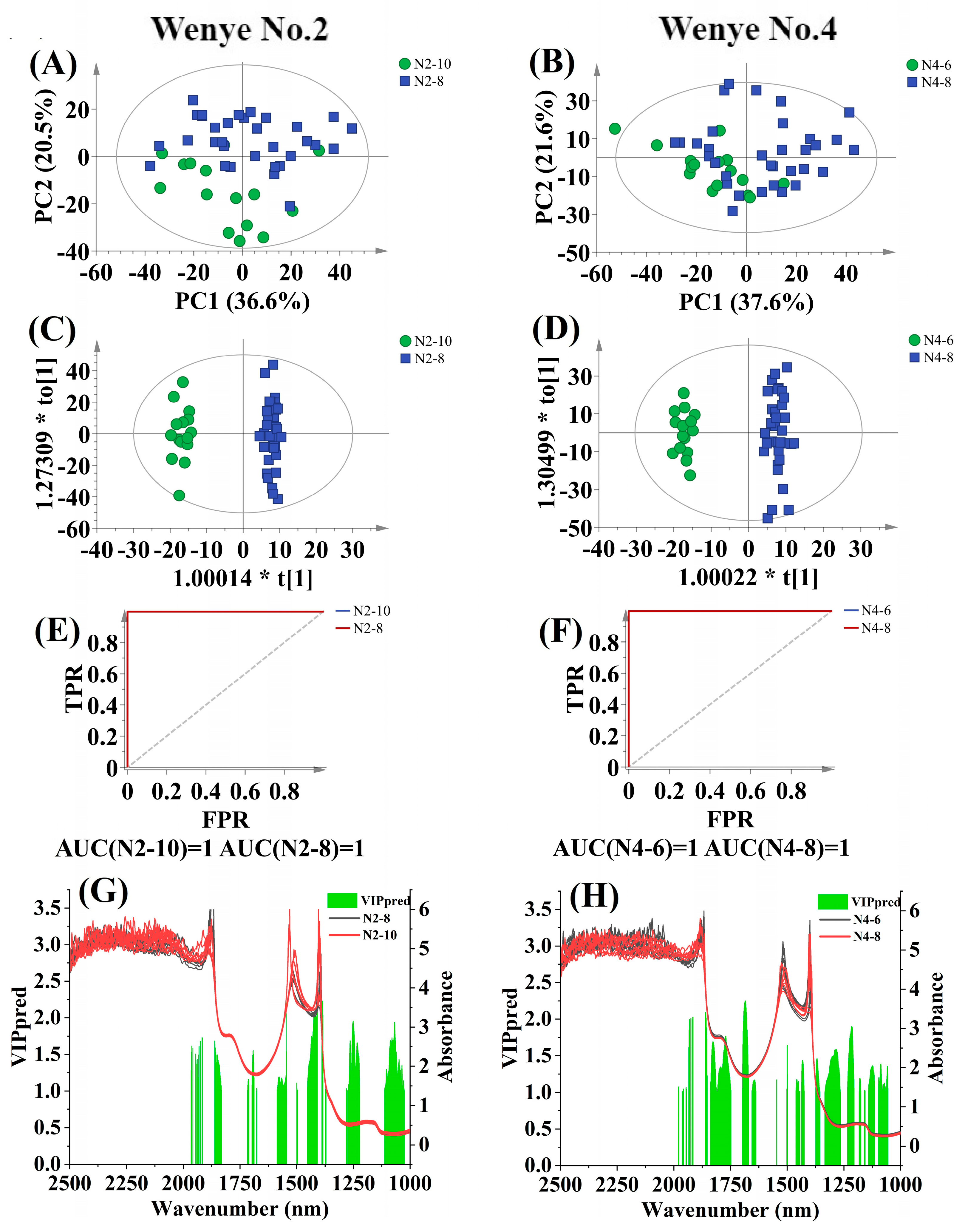
3.4.2. Discrimination by Coconut Cultivar
3.4.3. Discrimination by Maturity
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Pinto, R.O.; do Nascimento, R.B.; Jermolovicius, L.A.; Jurkiewicz, C.; Gut, J.A.; Pinto, U.M.; Landgraf, M. Microbiological feasibility of microwave processing of coconut water. LWT 2021, 145, 111344. [Google Scholar] [CrossRef]
- Lakshmanan, J.; Zhang, B.; Wright, K.; Motameni, A.T.; Jaganathan, V.; Schultz, D.J.; Klinge, C.M.; Harbrecht, B.G. Tender coconut water suppresses hepatic inflammation by activating AKT and JNK signaling pathways in an in vitro model of sepsis. J. Funct. Foods 2020, 64, 103637. [Google Scholar] [CrossRef]
- Mahnot, N.K.; Kalita, D.; Mahanta, C.L.; Chaudhuri, M.K. Effect of additives on the quality of tender coconut water processed by nonthermal two stage microfiltration technique. LWT 2014, 59, 1191–1195. [Google Scholar] [CrossRef]
- Kumar, M.; Saini, S.S.; Agrawal, P.K.; Roy, P.; Sircar, D. Nutritional and metabolomics characterization of the coconut water at different nut developmental stages. J. Food Compost. Anal. 2021, 96, 103738. [Google Scholar] [CrossRef]
- Shen, X.; Wang, Y.; Ran, L.; Liu, R.; Sun, X.; Hu, L.; Xiao, Y.; Chen, F. Flavor deterioration of liquid endosperm in postharvest tender coconut revealed by LC-MS-based metabolomics, GC-IMS and E-tongue. Postharvest Biol. Technol. 2022, 187, 111866. [Google Scholar] [CrossRef]
- Burns, D.T.; Johnston, E.L.; Walker, M.J. Authenticity and the potability of coconut water—A critical review. J. AOAC Int. 2020, 103, 800–806. [Google Scholar] [CrossRef]
- Perera, L.; Baudouin, L.; Mackay, I. SSR markers indicate a common origin of self-pollinating dwarf coconut in South-East Asia under domestication. Sci. Hortic. 2016, 211, 255–262. [Google Scholar] [CrossRef]
- Jirapong, C.; Changprasert, S.; Kanlayanarat, S.; Bodhipadma, K.; Noichinda, S.; Wongs-Aree, C. Characterization of the liquid endosperm attributes in young coconut fruit during storage. Int. Food Res. J. 2018, 25, 2650–2656. [Google Scholar]
- Meethaworn, K.; Luckanatinwong, V.; Zhang, B.; Chen, K.; Siriphanich, J. Off-flavor caused by cold storage is related to induced activity of LOX and HPL in young coconut fruit. LWT 2019, 114, 108329. [Google Scholar] [CrossRef]
- Cheevitsopon, E.; Sirisomboon, P. Rapid evaluation of fat content in curry soup containing coconut milk by using near infrared spectroscopy. J. Near Infrared Spectrosc. 2017, 26, 16–25. [Google Scholar] [CrossRef]
- Sirisomboon, P.; Nawayon, J. Evaluation of soluble solids of curry soup containing coconut milk by near infrared spectroscopy. J. Near Infrared Spectrosc. 2017, 25, 203–210. [Google Scholar] [CrossRef]
- Cheevitsopon, E.; Sirisomboon, P. Evaluation of salt content of curry soup containing coconut milk by near infrared spectroscopy. J. Near Infrared Spectrosc. 2018, 26, 149–158. [Google Scholar] [CrossRef]
- Thitibunjan, N.; Sirisomboon, P. Evaluation of pH of curry soup containing coconut milk by near infrared spectroscopy. IOP Conf. Ser. Earth Environ. Sci. 2019, 301, 012061. [Google Scholar] [CrossRef]
- Davrieux, F.; Prades, A.; Mialet-Serra, I.; Dupuis, S.; Assa, R.R. Physico-chemical changes in the fruits of two coconut (Cocos nucifera L.) hybrids during ripening: A NIRS-boosted study. In Proceedings of the FRUTIC 05, 7th Fruit, Nut and Vegetable Production Engineering Symposium: Information and Technology for Sustainable Fruit and Vegetable Production, Montpellier, France, 12–16 September 2005. [Google Scholar]
- Chen, W.; Zhang, G.; Chen, W.; Zhong, Q.; Chen, H. Metabolomic profiling of matured coconut water during post-harvest storage revealed discrimination and distinct changes in metabolites. RSC Adv. 2018, 8, 31396–31405. [Google Scholar] [CrossRef] [PubMed]
- Yun, Y.H.; Wang, W.T.; Tan, M.L.; Liang, Y.Z.; Li, H.D.; Cao, D.S.; Lu, H.M.; Xu, Q.S. A strategy that iteratively retains informative variables for selecting optimal variable subset in multivariate calibration. Anal. Chim. Acta 2014, 807, 36–43. [Google Scholar] [CrossRef] [PubMed]
- Yun, Y.H.; Bin, J.; Liu, D.L.; Xu, L.; Yan, T.L.; Cao, D.S.; Xu, Q.S. A hybrid variable selection strategy based on continuous shrinkage of variable space in multivariate calibration. Anal. Chim. Acta 2019, 1058, 58–69. [Google Scholar] [CrossRef] [PubMed]
- Song, X.; Huang, Y.; Yan, H.; Xiong, Y.; Min, S. A novel algorithm for spectral interval combination optimization. Anal. Chim. Acta 2016, 948, 19–29. [Google Scholar] [CrossRef]
- Chang, C.-W.; Laird, D.A.; Mausbach, M.J.; Hurburgh, C.R. Near-infrared reflectance spectroscopy–principal components regression analyses of soil properties. Soil Sci. Soc. Am. J. 2001, 65, 480–490. [Google Scholar] [CrossRef]
- Saeys, W.; Mouazen, A.M.; Ramon, H. Potential for onsite and online analysis of pig manure using visible and near infrared reflectance spectroscopy. Biosys. Eng. 2005, 91, 393–402. [Google Scholar] [CrossRef]
- Ghidini, S.; Varra, M.O.; Dall’Asta, C.; Badiani, A.; Ianieri, A.; Zanardi, E. Rapid authentication of European sea bass (Dicentrarchus labrax L.) according to production method, farming system, and geographical origin by near infrared spectroscopy coupled with chemometrics. Food Chem. 2019, 280, 321–327. [Google Scholar] [CrossRef]
- Workman, J.; Weyer, L. Practical Guide and Spectral Atlas for Interpretive Near-Infrared Spectroscopy, 2nd ed.; CRC Press: Boca Raton, FL, USA, 2012. [Google Scholar]
- Turker-Kaya, S.; Huck, C.W. A Review of Mid-Infrared and Near-Infrared Imaging: Principles, Concepts and Applications in Plant Tissue Analysis. Molecules 2017, 22, 168. [Google Scholar] [CrossRef]
- Bittante, G.; Patel, N.; Cecchinato, A.; Berzaghi, P. Invited review: A comprehensive review of visible and near-infrared spectroscopy for predicting the chemical composition of cheese. J. Dairy Sci. 2022, 105, 1817–1836. [Google Scholar] [CrossRef]
- Barker, M.; Rayens, W. Partial least squares for discrimination. J. Chemom. 2003, 17, 166–173. [Google Scholar] [CrossRef]
- Hoorfar, J. Global Safety of Fresh Produce: A Handbook of Best Practice, Innovative Commercial Solutions and Case Studies; Woodhead Publishing: Sawston, UK, 2014. [Google Scholar]
- Mahayothee, B.; Koomyart, I.; Khuwijitjaru, P.; Siriwongwilaichat, P.; Nagle, M.; Müller, J. Phenolic compounds, antioxidant activity, and medium chain fatty acids profiles of coconut water and meat at different maturity stages. Int. J. Food Prop. 2016, 19, 2041–2051. [Google Scholar] [CrossRef]
- Saradhuldhat, P.; Paull, R.E. Pineapple organic acid metabolism and accumulation during fruit development. Sci. Hortic. 2007, 112, 297–303. [Google Scholar] [CrossRef]
- Zhang, Y.; Chen, W.; Chen, H.; Zhong, Q.; Yun, Y.; Chen, W. Metabolomics analysis of the deterioration mechanism and storage time limit of tender coconut water during storage. Foods 2020, 9, 46. [Google Scholar] [CrossRef]
- Prades, A.; Dornier, M.; Diop, N.; Pain, J.-P. Coconut water uses, composition and properties: A review. Fruits 2012, 67, 87–107. [Google Scholar] [CrossRef]
- Cao, X.; Ding, H.; Yang, L.; Huang, J.; Zeng, L.; Tong, H.; Su, L.; Ji, X.; Wu, M.; Yang, Y. Near-infrared spectroscopy as a tool to assist Sargassum fusiforme quality grading: Harvest time discrimination and polyphenol prediction. Postharvest Biol. Technol. 2022, 192, 112030. [Google Scholar] [CrossRef]
- Gordon, A.; Jackson, J. 7—Case study: Application of appropriate technologies to improve the quality and safety of coconut water. In Food Safety and Quality Systems in Developing Countries; Gordon, A., Ed.; Academic Press: San Diego, CA, USA, 2017; pp. 185–216. [Google Scholar]
- Haseena, M.; Kasturi Bai, K.V.; Padmanabhan, S. Post-harvest quality and shelf-life of tender coconut. J. Food Sci. Technol. 2010, 47, 686–689. [Google Scholar] [CrossRef] [PubMed]
- Saensuk, C.; Wanchana, S.; Choowongkomon, K.; Wongpornchai, S.; Kraithong, T.; Imsabai, W.; Chaichoompu, E.; Ruanjaichon, V.; Toojinda, T.; Vanavichit, A.; et al. De novo transcriptome assembly and identification of the gene conferring a “pandan-like” aroma in coconut (Cocos nucifera L.). Plant Sci. 2016, 252, 324–334. [Google Scholar] [CrossRef] [PubMed]

| Attributes | Number of Collected Samples | Number of Outliers | Variable Selection Methods | Number of Latent Variables | Number of Variables | Training Set | External Validation Set | |||
|---|---|---|---|---|---|---|---|---|---|---|
| RMSEF | RMSEP | RPD | ||||||||
| TSS | 544 | 37 | Full-spectrum | 6 | 1131 | 0.1516 | 0.7041 | −0.0462 | 0.6578 | 0.9777 |
| CARS | 6 | 21 | 0.1608 | 0.7003 | −0.0473 | 0.6582 | 0.9633 | |||
| VCPA | 8 | 11 | 0.4232 | 0.5806 | 0.2316 | 0.5637 | 1.1077 | |||
| ICO | 5 | 128 | 0.4213 | 0.5815 | 0.2395 | 0.5609 | 1.1467 | |||
| pH | 544 | 15 | Full-spectrum | 1 | 1131 | 0.0260 | 0.3048 | 0.0265 | 0.3137 | 1.0135 |
| CARS | 2 | 7 | 0.0610 | 0.2993 | 0.0191 | 0.3149 | 1.0073 | |||
| VCPA | 12 | 12 | 0.6354 | 0.1865 | 0.4776 | 0.2298 | 1.2636 | |||
| ICO | 14 | 157 | 0.6570 | 0.1809 | 0.4609 | 0.2334 | 1.3619 | |||
| TSS:pH | 544 | 31 | Full-spectrum | 11 | 1131 | 0.2540 | 0.1225 | −0.0876 | 0.1299 | 0.9589 |
| CARS | 2 | 14 | 0.0894 | 0.1354 | −0.0995 | 0.1306 | 0.9280 | |||
| VCPA | 9 | 10 | 0.4584 | 0.1044 | 0.1687 | 0.1136 | 1.0785 | |||
| ICO | 14 | 68 | 0.3656 | 0.1130 | 0.1811 | 0.1127 | 1.1051 | |||
| Reducing sugar content | 544 | 19 | Full-spectrum | 20 | 1131 | 0.4972 | 0.5436 | 0.2713 | 0.6322 | 1.1715 |
| CARS | 7 | 31 | 0.7359 | 0.3940 | 0.7207 | 0.3913 | 1.8235 | |||
| VCPA | 10 | 11 | 0.7682 | 0.3691 | 0.7081 | 0.4001 | 1.7267 | |||
| ICO | 6 | 93 | 0.7552 | 0.3793 | 0.6999 | 0.4057 | 1.8255 | |||
| Soluble sugar content | 544 | 25 | Full-spectrum | 20 | 1131 | 0.5037 | 0.5366 | 0.2635 | 0.5818 | 1.1652 |
| CARS | 6 | 107 | 0.6978 | 0.4188 | 0.5962 | 0.4307 | 1.5417 | |||
| VCPA | 8 | 11 | 0.7046 | 0.4140 | 0.6197 | 0.4181 | 1.5680 | |||
| ICO | 17 | 120 | 0.7209 | 0.4024 | 0.6047 | 0.4262 | 1.5906 | |||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shen, X.; Wang, T.; Wei, J.; Li, X.; Deng, F.; Niu, X.; Wang, Y.; Kan, J.; Zhang, W.; Yun, Y.-H.; et al. Potential of Near-Infrared Spectroscopy (NIRS) for Efficient Classification Based on Postharvest Storage Time, Cultivar and Maturity in Coconut Water. Foods 2023, 12, 2415. https://doi.org/10.3390/foods12122415
Shen X, Wang T, Wei J, Li X, Deng F, Niu X, Wang Y, Kan J, Zhang W, Yun Y-H, et al. Potential of Near-Infrared Spectroscopy (NIRS) for Efficient Classification Based on Postharvest Storage Time, Cultivar and Maturity in Coconut Water. Foods. 2023; 12(12):2415. https://doi.org/10.3390/foods12122415
Chicago/Turabian StyleShen, Xiaojun, Tao Wang, Jingyi Wei, Xin Li, Fuming Deng, Xiaoqing Niu, Yuanyuan Wang, Jintao Kan, Weimin Zhang, Yong-Huan Yun, and et al. 2023. "Potential of Near-Infrared Spectroscopy (NIRS) for Efficient Classification Based on Postharvest Storage Time, Cultivar and Maturity in Coconut Water" Foods 12, no. 12: 2415. https://doi.org/10.3390/foods12122415
APA StyleShen, X., Wang, T., Wei, J., Li, X., Deng, F., Niu, X., Wang, Y., Kan, J., Zhang, W., Yun, Y.-H., & Chen, F. (2023). Potential of Near-Infrared Spectroscopy (NIRS) for Efficient Classification Based on Postharvest Storage Time, Cultivar and Maturity in Coconut Water. Foods, 12(12), 2415. https://doi.org/10.3390/foods12122415









