Biochemical and Genomic Characterization of Two New Strains of Lacticaseibacillus paracasei Isolated from the Traditional Corn-Based Beverage of South Africa, Mahewu, and Their Comparison with Strains Isolated from Kefir Grains
Abstract
1. Introduction
2. Materials and Methods
2.1. Isolation, Identification and Profile of Enzymatic Activities
2.2. Inhibition of Pathogens and Antagonistic Interactions
2.3. Fermentation of Milk
2.4. Genome Sequencing, Assembly and Annotation
2.5. Statistical Data Manipulations
3. Results and Discussion
3.1. Isolation and Taxonomic Assignment of the Mahewu-Derived Strains
3.2. Comparative Functional Characterization of the Mahewu- and Kefir-Derived Strains
3.2.1. Biochemical Characterization: Ability to Utilize Different Substrates and Profile of Enzymatic Activities
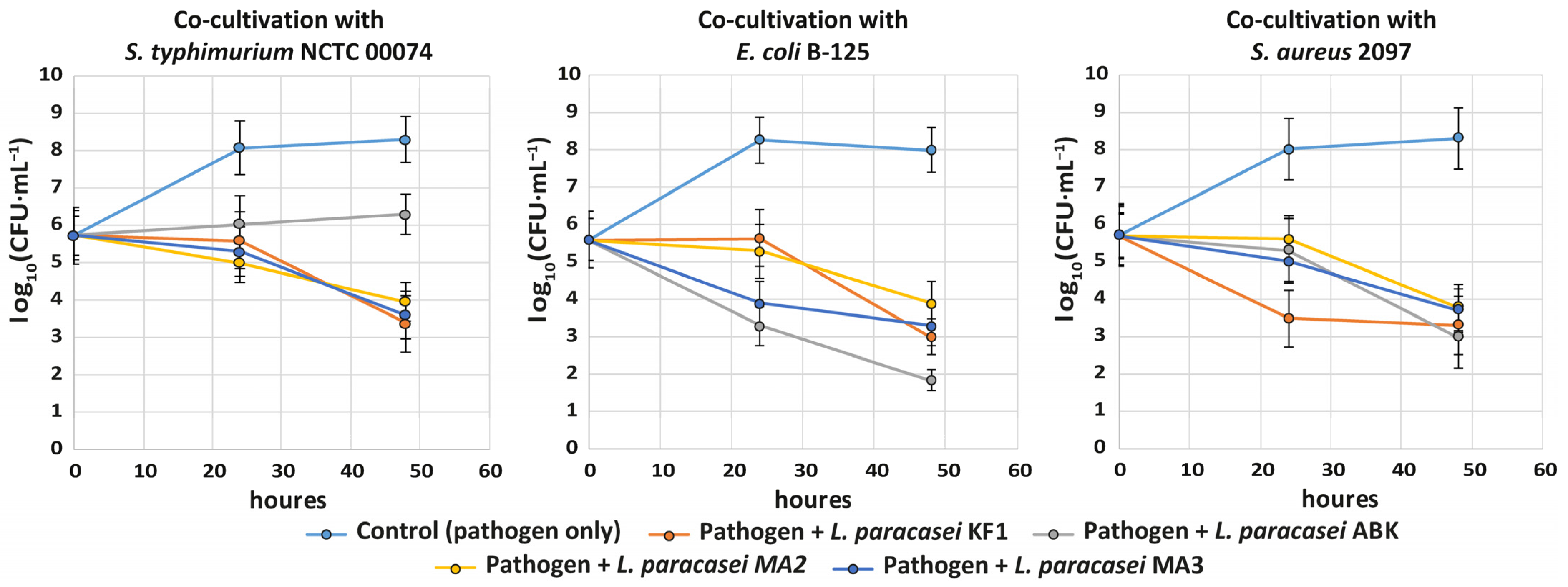
3.2.2. Inhibition of Pathogens and Antagonistic Interactions
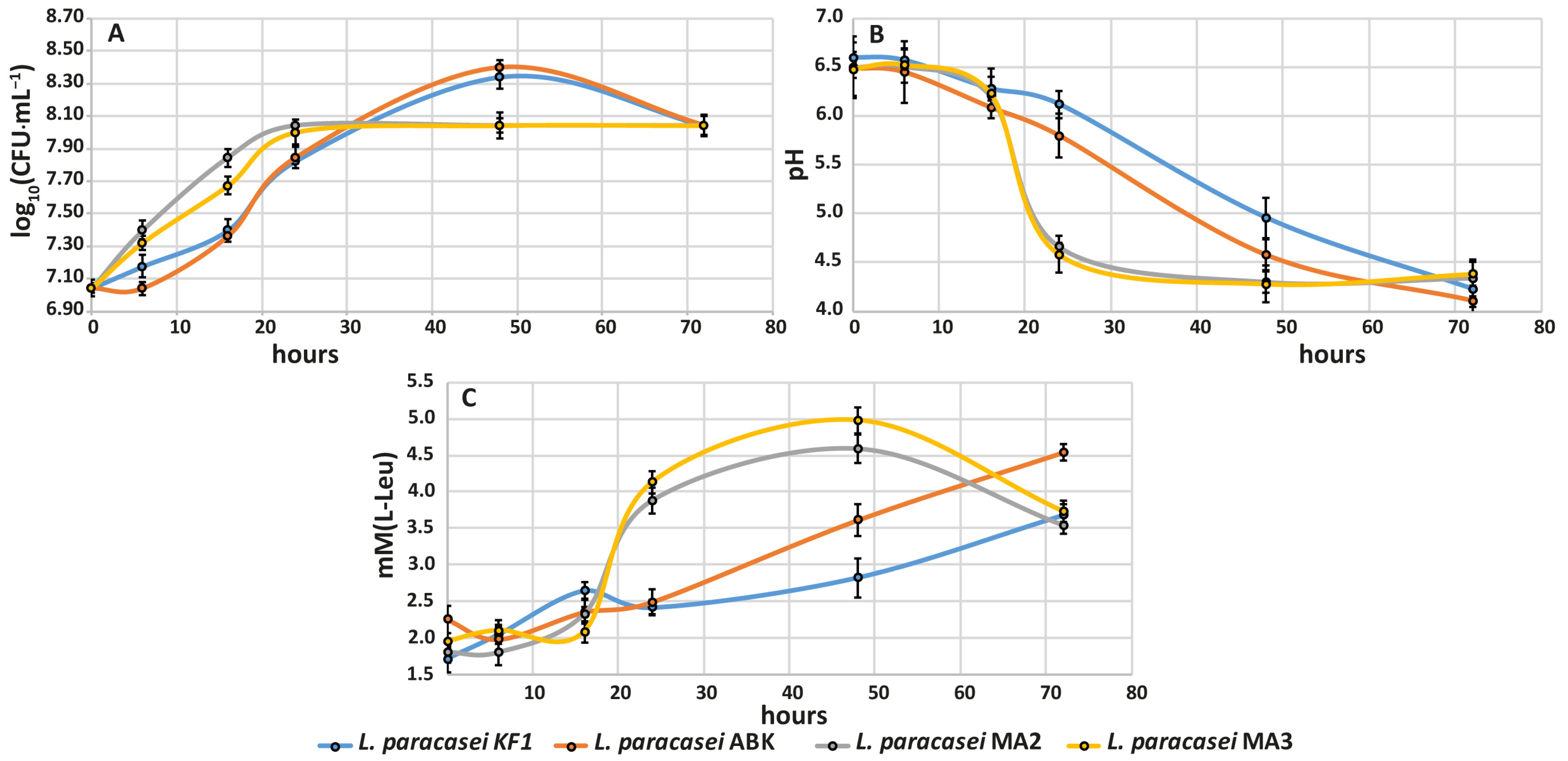
3.2.3. Growth Ability, Acidification Capability and Proteolytic Activity during Milk Fermentation
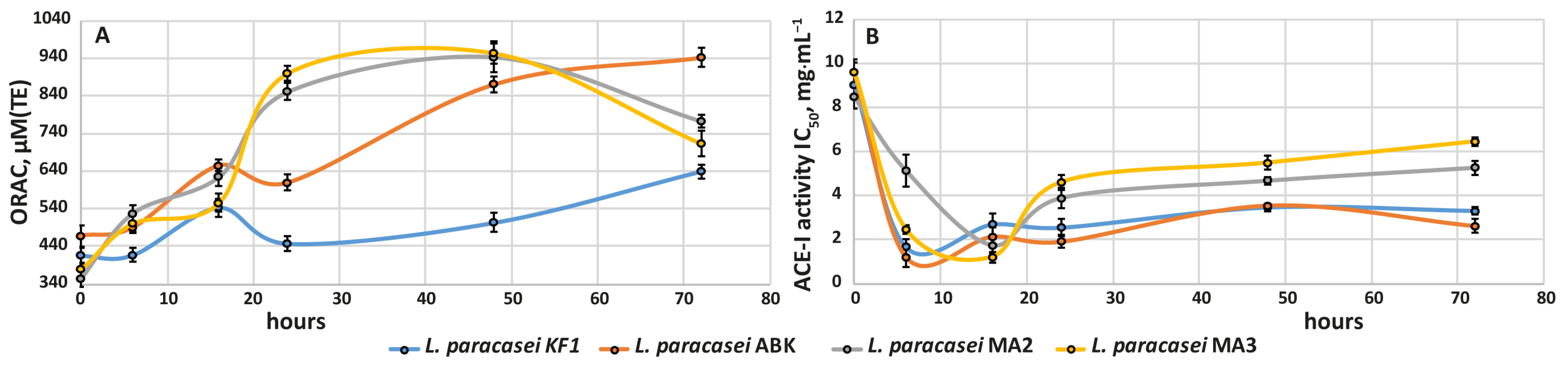
3.2.4. Development of Antioxidant and Antihypertensive Properties during Milk Fermentation
3.3. Comparative Genomic Characterization of the Mahewu- and Kefir-Derived Strains
3.3.1. Genome Sequencing, Assembly and Annotation
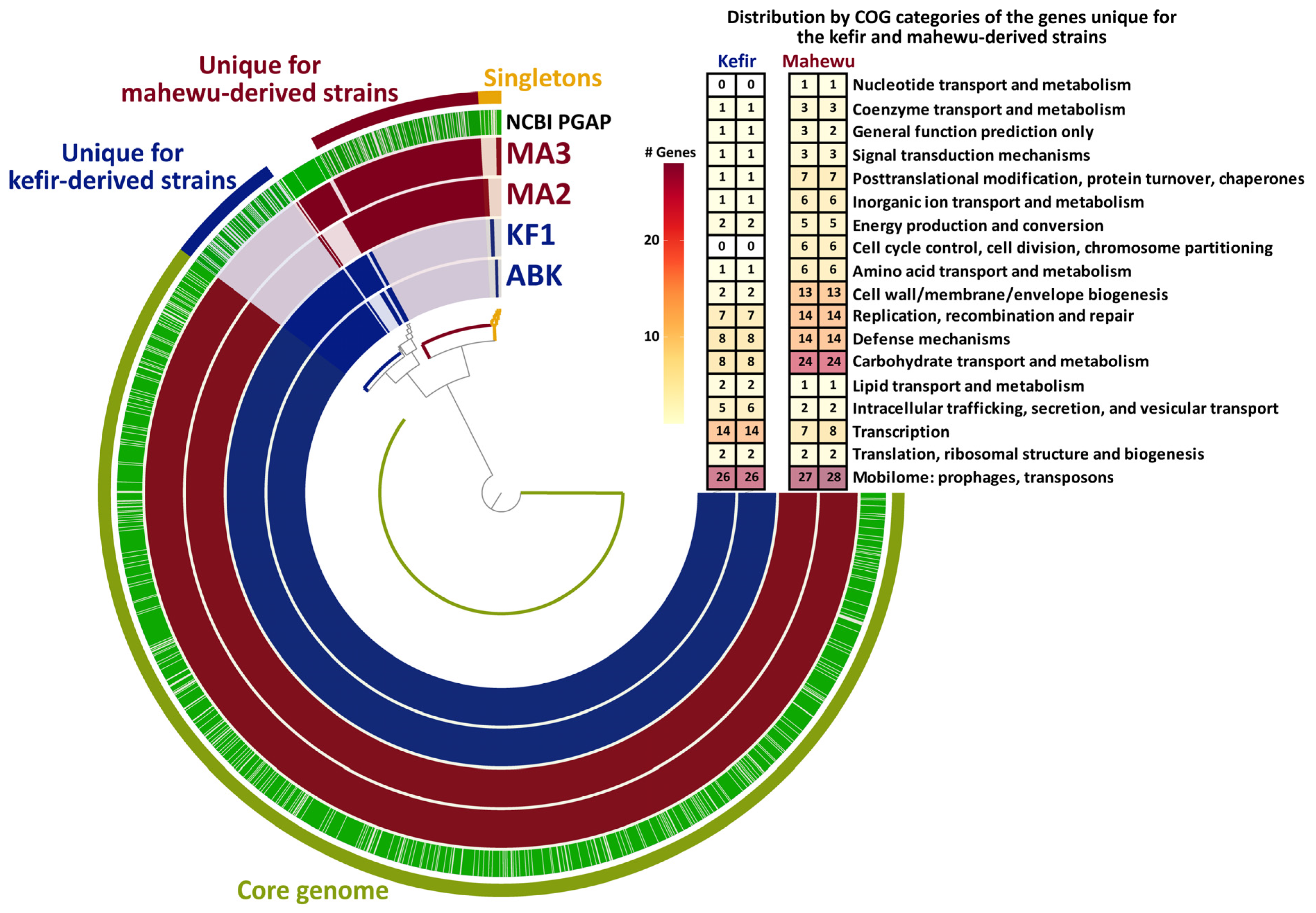
3.3.2. Functional Annotation and Pan-Genomic Analysis
3.3.3. Genome Stability
3.3.4. Bacteriocin Genome Content
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Bintsis, T. Lactic acid bacteria: Their applications in foods. J. Bacteriol. Mycol. Open Access 2018, 6, 89–94. [Google Scholar] [CrossRef]
- Sharma, R.; Garg, P.; Kumar, P.; Bhatia, S.K.; Kulshrestha, S. Microbial Fermentation and Its Role in Quality Improvement of Fermented Foods. Fermentation 2020, 6, 106. [Google Scholar] [CrossRef]
- Leis, R.; de Castro, M.-J.; de Lamas, C.; Picáns, R.; Couce, M.L. Effects of Prebiotic and Probiotic Supplementation on Lactase Deficiency and Lactose Intolerance: A Systematic Review of Controlled Trials. Nutrients 2020, 12, 1487. [Google Scholar] [CrossRef]
- Nourmohammadi, E.; Mahoonak, A.S. Health Implications of Bioactive Peptides: A Review. Int. J. Vitam. Nutr. Res. 2018, 88, 319–343. [Google Scholar] [CrossRef]
- Ooi, L.-G.; Liong, M.-T. Cholesterol-Lowering Effects of Probiotics and Prebiotics: A Review of in Vivo and in Vitro Findings. Int. J. Mol. Sci. 2010, 11, 2499–2522. [Google Scholar] [CrossRef]
- Adebo, O.A.; Gabriela Medina-Meza, I. Impact of Fermentation on the Phenolic Compounds and Antioxidant Activity of Whole Cereal Grains: A Mini Review. Molecules 2020, 25, 927. [Google Scholar] [CrossRef]
- García-Burgos, M.; Moreno-Fernández, J.; Alférez, M.J.M.; Díaz-Castro, J.; López-Aliaga, I. New perspectives in fermented dairy products and their health relevance. J. Funct. Foods 2020, 72, 104059. [Google Scholar] [CrossRef]
- Natividad, J.M.M.; Verdu, E.F. Modulation of intestinal barrier by intestinal microbiota: Pathological and therapeutic implications. Pharmacol. Res. 2013, 69, 42–51. [Google Scholar] [CrossRef]
- Hiippala, K.; Jouhten, H.; Ronkainen, A.; Hartikainen, A.; Kainulainen, V.; Jalanka, J.; Satokari, R. The Potential of Gut Commensals in Reinforcing Intestinal Barrier Function and Alleviating Inflammation. Nutrients 2018, 10, 988. [Google Scholar] [CrossRef]
- Boricha, A.A.; Shekh, S.L.; Pithva, S.P.; Ambalam, P.S.; Manuel Vyas, B.R. In vitro evaluation of probiotic properties of Lactobacillus species of food and human origin. LWT 2019, 106, 201–208. [Google Scholar] [CrossRef]
- Oleskin, A.V.; Shenderov, B.A. Probiotics and Psychobiotics: The Role of Microbial Neurochemicals. Probiotics Antimicrob. Proteins 2019, 11, 1071–1085. [Google Scholar] [CrossRef]
- Servin, A.L. Antagonistic activities of lactobacilli and bifidobacteria against microbial pathogens. FEMS Microbiol. Rev. 2004, 28, 405–440. [Google Scholar] [CrossRef]
- Maldonado Galdeano, C.; Cazorla, S.I.; Lemme Dumit, J.M.; Vélez, E.; Perdigón, G. Beneficial effects of probiotic consumption on the immune system. Ann. Nutr. Metab. 2019, 74, 115–124. [Google Scholar] [CrossRef]
- Zheng, J.; Wittouck, S.; Salvetti, E.; Franz, C.M.A.P.; Harris, H.M.B.; Mattarelli, P.; O’Toole, P.W.; Pot, B.; Vandamme, P.; Walter, J.; et al. A taxonomic note on the genus Lactobacillus: Description of 23 novel genera, emended description of the genus Lactobacillus Beijerinck 1901, and union of Lactobacillaceae and Leuconostocaceae. Int. J. Syst. Evol. Microbiol. 2020, 70, 2782–2858. [Google Scholar] [CrossRef]
- Agarwala, R.; Barrett, T.; Beck, J.; Benson, D.A.; Bollin, C.; Bolton, E.; Bourexis, D.; Brister, J.R.; Bryant, S.H.; Canese, K.; et al. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2018, 46, D8–D13. [Google Scholar] [CrossRef]
- Sanders, M.E.; Benson, A.; Lebeer, S.; Merenstein, D.J.; Klaenhammer, T.R. Shared mechanisms among probiotic taxa: Implications for general probiotic claims. Curr. Opin. Biotechnol. 2018, 49, 207–216. [Google Scholar] [CrossRef]
- MARSHALL, V.M. Starter cultures for milk fermentation and their characteristics. Int. J. Dairy Technol. 1993, 46, 49–56. [Google Scholar] [CrossRef]
- Bintsis, T. Lactic acid bacteria as starter cultures: An update in their metabolism and genetics. AIMS Microbiol. 2018, 4, 665–684. [Google Scholar] [CrossRef]
- Marco, M.L.; Sanders, M.E.; Gänzle, M.; Arrieta, M.C.; Cotter, P.D.; De Vuyst, L.; Hill, C.; Holzapfel, W.; Lebeer, S.; Merenstein, D.; et al. The International Scientific Association for Probiotics and Prebiotics (ISAPP) consensus statement on fermented foods. Nat. Rev. Gastroenterol. Hepatol. 2021, 18, 196–208. [Google Scholar] [CrossRef]
- Jones, R.M. The Use of Lactobacillus casei and Lactobacillus paracasei in Clinical Trials for the Improvement of Human Health. In The Microbiota in Gastrointestinal Pathophysiology; Elsevier: Amsterdam, The Netherlands, 2017; pp. 99–108. [Google Scholar]
- Hill, D.; Sugrue, I.; Tobin, C.; Hill, C.; Stanton, C.; Ross, R.P. The Lactobacillus casei Group: History and Health Related Applications. Front. Microbiol. 2018, 9, 2107. [Google Scholar] [CrossRef]
- Huang, C.-H.; Li, S.-W.; Huang, L.; Watanabe, K. Identification and Classification for the Lactobacillus casei Group. Front. Microbiol. 2018, 9, 1974. [Google Scholar] [CrossRef]
- Duar, R.M.; Lin, X.B.; Zheng, J.; Martino, M.E.; Grenier, T.; Pérez-Muñoz, M.E.; Leulier, F.; Gänzle, M.; Walter, J. Lifestyles in transition: Evolution and natural history of the genus Lactobacillus. FEMS Microbiol. Rev. 2017, 41, S27–S48. [Google Scholar] [CrossRef]
- Martino, M.E.; Bayjanov, J.R.; Caffrey, B.E.; Wels, M.; Joncour, P.; Hughes, S.; Gillet, B.; Kleerebezem, M.; van Hijum, S.A.F.T.; Leulier, F. Nomadic lifestyle of Lactobacillus plantarum revealed by comparative genomics of 54 strains isolated from different habitats. Environ. Microbiol. 2016, 18, 4974–4989. [Google Scholar] [CrossRef]
- Kiousi, D.E.; Efstathiou, C.; Tegopoulos, K.; Mantzourani, I.; Alexopoulos, A.; Plessas, S.; Kolovos, P.; Koffa, M.; Galanis, A. Genomic Insight Into Lacticaseibacillus paracasei SP5, Reveals Genes and Gene Clusters of Probiotic Interest and Biotechnological Potential. Front. Microbiol. 2022, 13, 922689. [Google Scholar] [CrossRef]
- Smokvina, T.; Wels, M.; Polka, J.; Chervaux, C.; Brisse, S.; Boekhorst, J.; van Hylckama Vlieg, J.E.T.; Siezen, R.J. Lactobacillus paracasei Comparative Genomics: Towards Species Pan-Genome Definition and Exploitation of Diversity. PLoS ONE 2013, 8, e68731. [Google Scholar] [CrossRef]
- Zhang, Z.-G.; Ye, Z.-Q.; Yu, L.; Shi, P. Phylogenomic reconstruction of lactic acid bacteria: An update. BMC Evol. Biol. 2011, 11, 1. [Google Scholar] [CrossRef]
- Fidanza, M.; Panigrahi, P.; Kollmann, T.R. Lactiplantibacillus plantarum–Nomad and Ideal Probiotic. Front. Microbiol. 2021, 12, 2911. [Google Scholar] [CrossRef]
- Torres-Miranda, A.; Melis-Arcos, F.; Garrido, D. Characterization and Identification of Probiotic Features in Lacticaseibacillus Paracasei Using a Comparative Genomic Analysis Approach. Probiotics Antimicrob. Proteins 2022, 14, 1211–1224. [Google Scholar] [CrossRef]
- Golicz, A.A.; Bayer, P.E.; Bhalla, P.L.; Batley, J.; Edwards, D. Pangenomics Comes of Age: From Bacteria to Plant and Animal Applications. Trends Genet. 2020, 36, 132–145. [Google Scholar] [CrossRef]
- Begunova, A.V.; Savinova, O.S.; Moiseenko, K.V.; Glazunova, O.A.; Rozhkova, I.V.; Fedorova, T.V. Characterization and Functional Properties of Lactobacilli Isolated from Kefir Grains. Appl. Biochem. Microbiol. 2021, 57, 458–467. [Google Scholar] [CrossRef]
- Moiseenko, K.V.; Glazunova, O.A.; Savinova, O.S.; Ajibade, B.O.; Ijabadeniyi, O.A.; Fedorova, T.V. Analytical characterization of the widely consumed commercialized fermented beverages from russia (Kefir and ryazhenka) and south africa (amasi and mahewu): Potential functional properties and profiles of volatile organic compounds. Foods 2021, 10, 3082. [Google Scholar] [CrossRef]
- Dasen, G.; Smutny, J.; Teuber, M.; Meile, L. Classification and Identification of Propionibacteria Based on Ribosomal RNA Genes and PCR. Syst. Appl. Microbiol. 1998, 21, 251–259. [Google Scholar] [CrossRef]
- Stamatakis, A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 2014, 30, 1312–1313. [Google Scholar] [CrossRef]
- Miller, M.A.; Pfeiffer, W.; Schwartz, T. Creating the CIPRES Science Gateway for inference of large phylogenetic trees. In Proceedings of the 2010 Gateway Computing Environments Workshop (GCE), New Orleans, LA, USA, 14 November 2010; IEEE: Piscataway, NJ, USA, 2010; pp. 1–8. [Google Scholar]
- Begunova, A.V.; Rozhkova, I.V.; Glazunova, O.A.; Moiseenko, K.V.; Savinova, O.S.; Fedorova, T.V. Fermentation Profile and Probiotic-Related Characteristics of Bifidobacterium longum MC-42. Fermentation 2021, 7, 101. [Google Scholar] [CrossRef]
- Savinova, O.S.; Glazunova, O.A.; Moiseenko, K.V.; Begunova, A.V.; Rozhkova, I.V.; Fedorova, T.V. Exoproteome Analysis of Antagonistic Interactions between the Probiotic Bacteria Limosilactobacillus reuteri LR1 and Lacticaseibacillus rhamnosus F and Multidrug Resistant Strain of Klebsiella pneumonia. Int. J. Mol. Sci. 2021, 22, 10999. [Google Scholar] [CrossRef]
- Adler-Nissen, J. Determination of the degree of hydrolysis of food protein hydrolysates by trinitrobenzenesulfonic acid. J. Agric. Food Chem. 1979, 27, 1256–1262. [Google Scholar] [CrossRef]
- Torkova, A.A.; Ryazantseva, K.A.; Agarkova, E.Y.; Kruchinin, A.G.; Tsentalovich, M.Y.; Fedorova, T.V. Rational design of enzyme compositions for the production of functional hydrolysates of cow milk whey proteins. Appl. Biochem. Microbiol. 2017, 53, 669–679. [Google Scholar] [CrossRef]
- Tatusova, T.; DiCuccio, M.; Badretdin, A.; Chetvernin, V.; Nawrocki, E.P.; Zaslavsky, L.; Lomsadze, A.; Pruitt, K.D.; Borodovsky, M.; Ostell, J. NCBI prokaryotic genome annotation pipeline. Nucleic Acids Res. 2016, 44, 6614–6624. [Google Scholar] [CrossRef]
- Siguier, P. ISfinder: The reference centre for bacterial insertion sequences. Nucleic Acids Res. 2006, 34, D32–D36. [Google Scholar] [CrossRef]
- Arndt, D.; Grant, J.R.; Marcu, A.; Sajed, T.; Pon, A.; Liang, Y.; Wishart, D.S. PHASTER: A better, faster version of the PHAST phage search tool. Nucleic Acids Res. 2016, 44, W16–W21. [Google Scholar] [CrossRef] [PubMed]
- Carattoli, A.; Zankari, E.; García-Fernández, A.; Voldby Larsen, M.; Lund, O.; Villa, L.; Møller Aarestrup, F.; Hasman, H. In Silico Detection and Typing of Plasmids using PlasmidFinder and Plasmid Multilocus Sequence Typing. Antimicrob. Agents Chemother. 2014, 58, 3895–3903. [Google Scholar] [CrossRef] [PubMed]
- van Heel, A.J.; de Jong, A.; Song, C.; Viel, J.H.; Kok, J.; Kuipers, O.P. BAGEL4: A user-friendly web server to thoroughly mine RiPPs and bacteriocins. Nucleic Acids Res. 2018, 46, W278–W281. [Google Scholar] [CrossRef] [PubMed]
- Eren, A.M.; Kiefl, E.; Shaiber, A.; Veseli, I.; Miller, S.E.; Schechter, M.S.; Fink, I.; Pan, J.N.; Yousef, M.; Fogarty, E.C.; et al. Community-led, integrated, reproducible multi-omics with anvi’o. Nat. Microbiol. 2020, 6, 3–6. [Google Scholar] [CrossRef] [PubMed]
- Delmont, T.O.; Eren, A.M. Linking pangenomes and metagenomes: The Prochlorococcus metapangenome. PeerJ 2018, 6, e4320. [Google Scholar] [CrossRef] [PubMed]
- Raak, N.; Rohm, H.; Jaros, D. Enzymatic Cross-Linking of Casein Facilitates Gel Structure Weakening Induced by Overacidification. Food Biophys. 2017, 12, 261–268. [Google Scholar] [CrossRef]
- Jung, S.H.; Hong, D.K.; Bang, S.-J.; Heo, K.; Sim, J.-J.; Lee, J.-L. The Functional Properties of Lactobacillus casei HY2782 Are Affected by the Fermentation Time. Appl. Sci. 2021, 11, 2481. [Google Scholar] [CrossRef]
- Begunova, A.V.; Savinova, O.S.; Glazunova, O.A.; Moiseenko, K.V.; Rozhkova, I.V.; Fedorova, T.V. Development of Antioxidant and Antihypertensive Properties during Growth of Lactobacillus helveticus, Lactobacillus rhamnosus and Lactobacillus reuteri on Cow’s Milk: Fermentation and Peptidomics Study. Foods 2020, 10, 17. [Google Scholar] [CrossRef]
- Sarmadi, B.H.; Ismail, A. Antioxidative peptides from food proteins: A review. Peptides 2010, 31, 1949–1956. [Google Scholar] [CrossRef]
- Pihlanto, A. Antioxidative peptides derived from milk proteins. Int. Dairy J. 2006, 16, 1306–1314. [Google Scholar] [CrossRef]
- Udenigwe, C.C.; Aluko, R.E. Food Protein-Derived Bioactive Peptides: Production, Processing, and Potential Health Benefits. J. Food Sci. 2012, 77, R11–R24. [Google Scholar] [CrossRef]
- Donkor, O.N.; Henriksson, A.; Vasiljevic, T.; Shah, N.P. Proteolytic activity of dairy lactic acid bacteria and probiotics as determinant of growth and in vitro angiotensin-converting enzyme inhibitory activity in fermented milk. Lait 2007, 87, 21–38. [Google Scholar] [CrossRef]
- Beltrán-Barrientos, L.M.; Hernández-Mendoza, A.; Torres-Llanez, M.J.; González-Córdova, A.F.; Vallejo-Córdoba, B. Invited review: Fermented milk as antihypertensive functional food. J. Dairy Sci. 2016, 99, 4099–4110. [Google Scholar] [CrossRef] [PubMed]
- Goodhead, I.; Darby, A.C. Taking the pseudo out of pseudogenes. Curr. Opin. Microbiol. 2015, 23, 102–109. [Google Scholar] [CrossRef]
- Mira, A.; Pushker, R. The Silencing of Pseudogenes. Mol. Biol. Evol. 2005, 22, 2135–2138. [Google Scholar] [CrossRef][Green Version]
- Brockhurst, M.A.; Harrison, E.; Hall, J.P.J.; Richards, T.; McNally, A.; MacLean, C. The Ecology and Evolution of Pangenomes. Curr. Biol. 2019, 29, R1094–R1103. [Google Scholar] [CrossRef]
- Song, Y.; He, Q.; Zhang, J.; Qiao, J.; Xu, H.; Zhong, Z.; Zhang, W.; Sun, Z.; Yang, R.; Cui, Y.; et al. Genomic Variations in Probiotic Lactobacillus plantarum P-8 in the Human and Rat Gut. Front. Microbiol. 2018, 9, 893. [Google Scholar] [CrossRef] [PubMed]
- Sybesma, W.; Molenaar, D.; van IJcken, W.; Venema, K.; Kort, R. Genome Instability in Lactobacillus rhamnosus GG. Appl. Environ. Microbiol. 2013, 79, 2233–2239. [Google Scholar] [CrossRef] [PubMed]
- Feng, C.; Zhang, F.; Wang, B.; Zhang, L.; Dong, Y.; Shao, Y. Genome-wide analysis of fermentation and probiotic trait stability in Lactobacillus plantarum during continuous culture. J. Dairy Sci. 2020, 103, 117–127. [Google Scholar] [CrossRef] [PubMed]
- Darmon, E.; Leach, D.R.F. Bacterial Genome Instability. Microbiol. Mol. Biol. Rev. 2014, 78, 1–39. [Google Scholar] [CrossRef] [PubMed]
- Idowu, O.; Fadahunsi, I.F.; Onabiyi, O.A. Production and Nutritional Evaluation of Mahewu: A Non-Alcoholic Fermented Beaverage of South Africa. Int. J. Res. Pharm. Biosci. 2016, 3, 27. [Google Scholar]
- Fadahunsi, I.; Soremekun, O. Production, Nutritional and Microbiological Evaluation of Mahewu a South African Traditional Fermented Porridge. J. Adv. Biol. Biotechnol. 2017, 14, 1–10. [Google Scholar] [CrossRef]
- Holzapfel, W.; Leonie Taljaard, J. Industrialization of Mageu Fermentation in South Africa. In Industrialization of Indigenous Fermented Foods; Steinkraus, K., Ed.; CRC Press: Boca Raton, FL, USA, 2004. [Google Scholar]
- Odunfa, S.A.; Oyewole, O.B. African fermented foods. In Microbiology of Fermented Foods; Springer: Boston, MA, USA, 1998; pp. 713–752. [Google Scholar]
- Simango, C. Lactic acid fermentation of sour porridge and mahewu, a non-alcoholic fermented cereal beverage. JASSA J. Appl. Sci. S. Afr. 2005, 8, 89–98. [Google Scholar] [CrossRef]
- Simatende, P.; Siwela, M.; Gadaga, T.H. Identification of lactic acid bacteria and determination of selected biochemical properties in emasi and emahewu. S. Afr. J. Sci. 2019, 115, 2–8. [Google Scholar] [CrossRef] [PubMed]
- Vandecraen, J.; Chandler, M.; Aertsen, A.; Van Houdt, R. The impact of insertion sequences on bacterial genome plasticity and adaptability. Crit. Rev. Microbiol. 2017, 43, 709–730. [Google Scholar] [CrossRef] [PubMed]
- Gänzle, M.G.; Loponen, J.; Gobbetti, M. Proteolysis in sourdough fermentations: Mechanisms and potential for improved bread quality. Trends Food Sci. Technol. 2008, 19, 513–521. [Google Scholar] [CrossRef]
- Papadimitriou, K.; Zoumpopoulou, G.; Georgalaki, M.; Alexandraki, V.; Kazou, M.; Anastasiou, R.; Tsakalidou, E. Sourdough Bread. In Innovations in Traditional Foods; Elsevier: Amsterdam, The Netherlands, 2019; pp. 127–158. [Google Scholar]
- Sauer, M.; Russmayer, H.; Grabherr, R.; Peterbauer, C.K.; Marx, H. The Efficient Clade: Lactic Acid Bacteria for Industrial Chemical Production. Trends Biotechnol. 2017, 35, 756–769. [Google Scholar] [CrossRef]
- Ricke, S. Perspectives on the use of organic acids and short chain fatty acids as antimicrobials. Poult. Sci. 2003, 82, 632–639. [Google Scholar] [CrossRef]
- Carr, F.J.; Chill, D.; Maida, N. The Lactic Acid Bacteria: A Literature Survey. Crit. Rev. Microbiol. 2002, 28, 281–370. [Google Scholar] [CrossRef]
- Almeida-Santos, A.C.; Novais, C.; Peixe, L.; Freitas, A.R. Enterococcus spp. as a Producer and Target of Bacteriocins: A Double-Edged Sword in the Antimicrobial Resistance Crisis Context. Antibiotics 2021, 10, 1215. [Google Scholar] [CrossRef]
- Browne, K.; Chakraborty, S.; Chen, R.; Willcox, M.D.; Black, D.S.; Walsh, W.R.; Kumar, N. A New Era of Antibiotics: The Clinical Potential of Antimicrobial Peptides. Int. J. Mol. Sci. 2020, 21, 7047. [Google Scholar] [CrossRef]
- Todorov, S.D.; Popov, I.; Weeks, R.; Chikindas, M.L. Use of Bacteriocins and Bacteriocinogenic Beneficial Organisms in Food Products: Benefits, Challenges, Concerns. Foods 2022, 11, 3145. [Google Scholar] [CrossRef] [PubMed]
- Zimina, M.; Babich, O.; Prosekov, A.; Sukhikh, S.; Ivanova, S.; Shevchenko, M.; Noskova, S. Overview of Global Trends in Classification, Methods of Preparation and Application of Bacteriocins. Antibiotics 2020, 9, 553. [Google Scholar] [CrossRef] [PubMed]
- Perez, R.H.; Perez, M.T.M.; Elegado, F.B. Bacteriocins from Lactic Acid Bacteria: A Review of Biosynthesis, Mode of Action, Fermentative Production, Uses, and Prospects. Int. J. Philipp. Sci. Technol. 2015, 8, 61–67. [Google Scholar] [CrossRef]
- Veskovic-Moracanin, S.; Djukic, D.; Memisi, N. Bacteriocins produced by lactic acid bacteria: A review. Acta Period. Technol. 2014, 283, 271–283. [Google Scholar] [CrossRef]
- Cotter, P.D.; Ross, R.P.; Hill, C. Bacteriocins—A viable alternative to antibiotics ? Nat. Rev. Microbiol. 2012, 11, 95–105. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, S.; Sarangi, A.N.; Mukherjee, M.; Bhowmick, S.; Tripathy, S. Reanalysis of Lactobacillus paracasei Lbs2 Strain and Large-Scale Comparative Genomics Places Many Strains into Their Correct Taxonomic Position. Microorganisms 2019, 7, 487. [Google Scholar] [CrossRef]

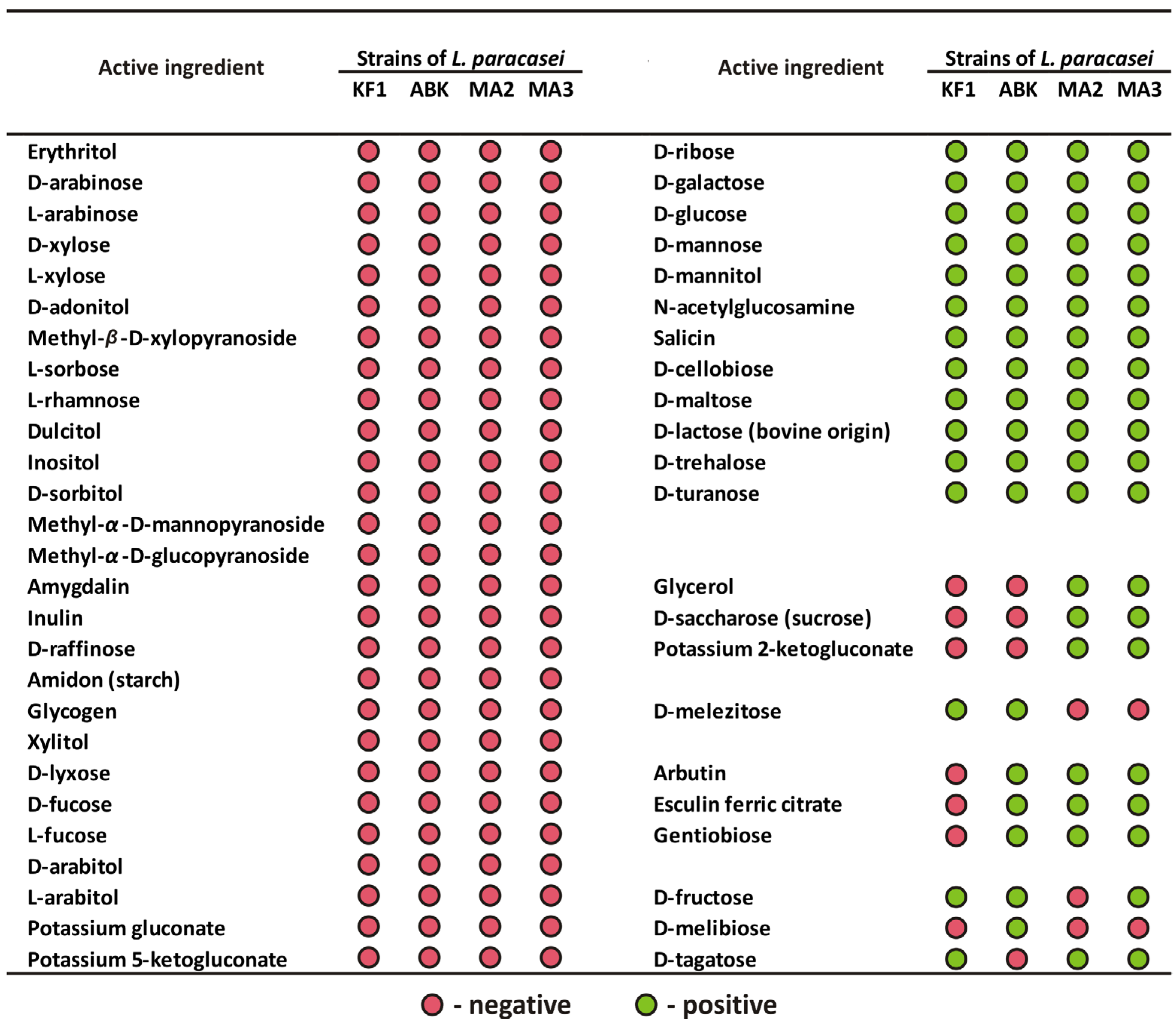
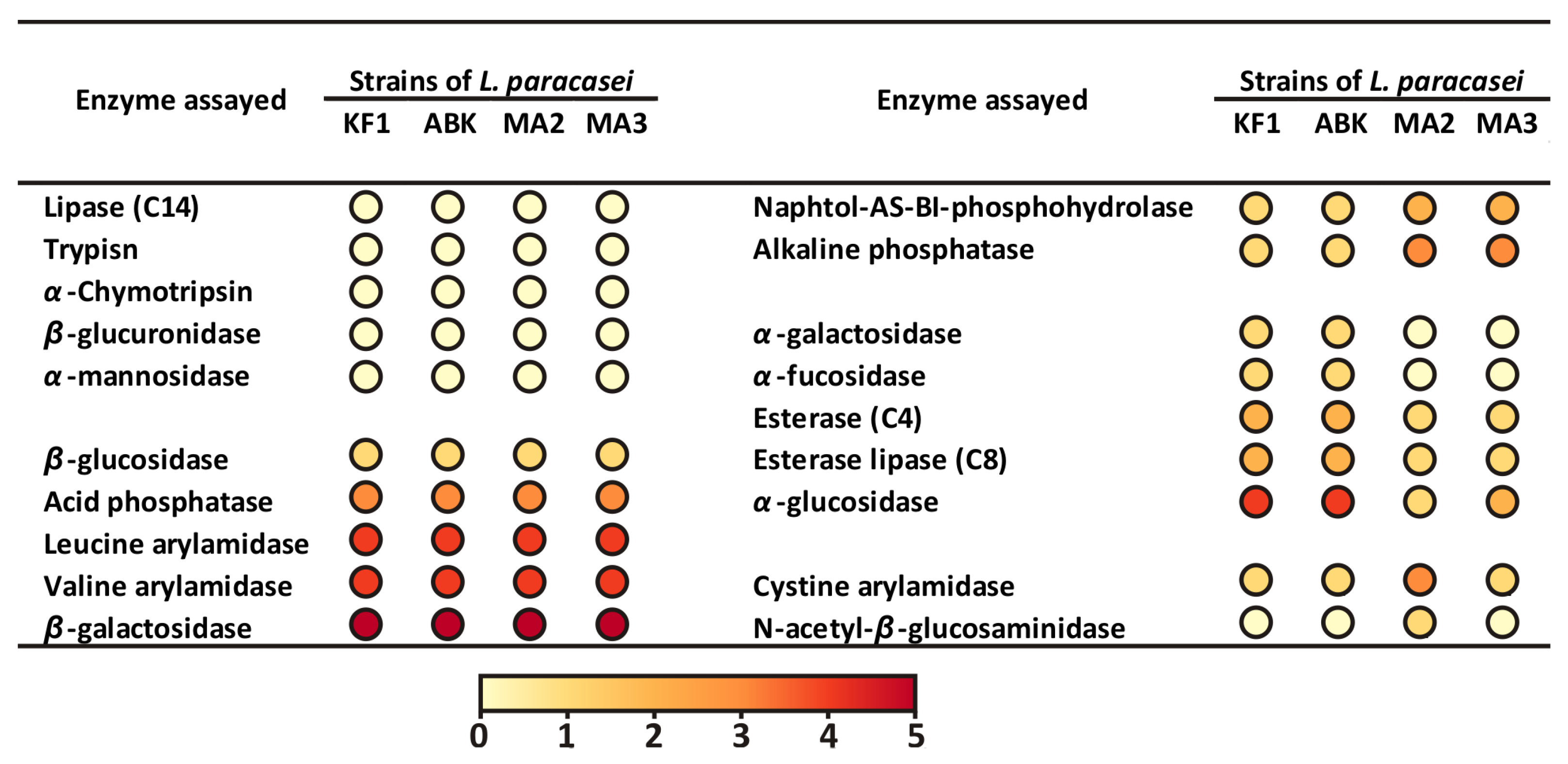
| Strains of L. paracasei | |||
|---|---|---|---|
| KF1 | ABK | MA2 | |
| ABK | - | ||
| MA2 | + | + | |
| MA3 | + | + | - |
| Kefir-Derived Strains | |||||||
| Lacticaseibacillus paracasei KF1 (GB Accession: GCA_023470645.1) | Lacticaseibacillus paracasei ABK (GB Accession: GCA_018967025.1) | ||||||
| Sequencing | Sequencing | ||||||
| Sequencing technology | Ion Torrent | Number of reads | 3,432,445 | Sequencing technology | Ion Torrent | Number of reads | 3,635,019 |
| Mean read size | 208 bp | Mean read size | 208 bp | ||||
| Assembly | Structural annotation | Assembly | Structural annotation | ||||
| Assembly size, bp | 2,697,398 | Genes (total): | 2791 | Assembly size, Mb | 2,698,106 | Genes (total): | 2796 |
| Overall coverage | 100× | - Protein coding | 2517 | Overall coverage | 100× | - Protein coding | 2524 |
| Number of contigs | 248 | - RNA coding | 78 | Number of contigs | 246 | - RNA coding | 78 |
| Longest contig, bp | 212,858 | - Pseudogenes | 196 | Longest contig, bp | 212,860 | - Pseudogenes | 194 |
| N50 contig size, bp | 36,612 | CRISPR arrays | 0 | N50 contig size, bp | 36,610 | CRISPR arrays | 0 |
| Mean contig size, bp | 10,572 | Mean contig size, bp | 10,773 | ||||
| Mahewu-Derived Strains | |||||||
| Lacticaseibacillus paracasei MA2 (GB Accession: GCA_018966985.1) | Lacticaseibacillus paracasei MA3 (GB Accession: GCA_023470655.1) | ||||||
| Sequencing | Sequencing | ||||||
| Sequencing technology | Ion Torrent | Number of reads | 2,972,024 | Sequencing technology | Ion Torrent | Number of reads | 3,314,216 |
| Mean read size, bp | 209 bp | Mean read size, bp | 212 bp | ||||
| Assembly | Structural annotation | Assembly | Structural annotation | ||||
| Assembly size, bp | 2,878,977 | Genes (total): | 2977 | Assembly size, Mb | 2,870,266 | Genes (total): | 2965 |
| Overall coverage | 100× | - Protein coding | 2651 | Overall coverage | 100× | - Protein coding | 2650 |
| Number of contigs | 363 | - RNA coding | 79 | Number of contigs | 350 | - RNA coding | 79 |
| Longest contig, bp | 170,621 | - Pseudogenes | 247 | Longest contig, bp | 170,654 | - Pseudogenes | 236 |
| N50 contig size, bp | 37,017 | CRISPR arrays | 0 | N50 contig size, bp | 37,018 | CRISPR arrays | 0 |
| Mean contig size, bp | 7622 | Mean contig size, bp | 8022 | ||||
| Strains of L. paracasei | |||||
|---|---|---|---|---|---|
| KF1 | ABK | MA2 | MA3 | ||
| Insertion sequences | |||||
| IS Family | Origin | BLAST hit | |||
| IS5 | Lactobacillus rhamnosus | ISLrh2 | ISLrh2 | ISLrh2 | ISLrh2 |
| IS5 | Lactobacillus rhamnosus | ISLrh3 | ISLrh3 | ISLrh3 | ISLrh3 |
| IS5 | Lactobacillus casei | ISLca2 | ISLca2 | ISLca2 | ISLca2 |
| IS3 | Lactobacillus casei | ISL1 | ISL1 | ISL1 | ISL1 |
| IS30 | Lactobacillus plantarum | ISLpl1 | ISLpl1 | ISLpl1 | ISLpl1 |
| IS3 | Lactobacillus sanfranciscensis | IS153 | IS153 | IS153 | IS153 |
| ISL3 | Leuconostoc mesenteroides | IS1165 | IS1165 | IS1165 | IS1165 |
| IS30 | Pediococcus pentosaceus | ISPp1 | ISPp1 | ISPp1 | ISPp1 |
| IS256 | Enterococcus hirae | IS1310 | IS1310 | IS1310 | IS1310 |
| IS6 | Leuconostoc mesenteroides | - | - | IS1297 | IS1297 |
| IS6 | Lactococcus lactis | - | - | ISS1N | ISS1N |
| IS6 | Lactococcus lactis | - | - | ISS1E | ISS1E |
| IS6 | Lactococcus lactis | - | - | ISS1M | ISS1M |
| IS6 | Lactococcus lactis | - | - | ISS1D | ISS1D |
| IS6 | Lactococcus lactis | - | - | ISS1CH | ISS1CH |
| IS6 | Lactococcus lactis | - | - | ISS1A | ISS1A |
| IS6 | Lactococcus lactis | - | - | IS946V | IS946V |
| IS6 | Lactococcus lactis | - | - | ISS1T | ISS1T |
| IS6 | Lactococcus lactis | - | - | ISS1S | ISS1S |
| IS6 | Lactococcus lactis | - | - | ISS1RS | ISS1RS |
| IS6 | Lactococcus lactis | - | - | ISS1B | ISS1B |
| IS6 | Lactococcus lactis | - | - | ISS1X | ISS1X |
| IS6 | Lactococcus lactis | - | - | ISS1Z | ISS1Z |
| IS6 | Lactococcus garvieae | - | - | ISLgar4 | ISLgar4 |
| IS5 | Streptococcus thermophilus | - | - | IS1194 | - |
| IS1182 | Streptococcus agalactiae | - | - | ISSag8 | - |
| IS1182 | Streptococcus agalactiae | - | - | IS1563 | - |
| Prophages | |||||
| Most common phage name | Completeness | Number of Total Proteins | |||
| PHAGE_Lactob_phijl1_NC_006936 | Intact | 57 | 59 | - | - |
| PHAGE_Lactob_BH1_NC_048737 | Questionable | 29 | 28 | - | - |
| PHAGE_Lactob_iLp84_NC_028783 | Incomplete | 18 | 18 | - | - |
| PHAGE_Staphy_phiPV83_NC_002486 | Incomplete | 9 | 10 | - | - |
| PHAGE_Staphy_SPbeta_like_NC_029119 | Incomplete | 22 | 19 | 23 | 19 |
| PHAGE_Lactob_iLp1308_NC_028911 | Incomplete | 29 | 26 | 29 | 29 |
| PHAGE_Lister_LP_101_NC_024387 | Intact | - | - | 19 | 19 |
| PHAGE_Lactob_iA2_NC_028830 | Intact | - | - | 48 | 48 |
| PHAGE_Lactob_Lc_Nu_NC_007501 | Incomplete | - | - | 16 | 16 |
| Plasmids | |||||
| Best BLAST hit | Origin | Presence | |||
| pLDW-11 | Companilactobacillus alimentarius DSM 20249 | No | No | Yes | Yes |
| Bacteriocin-Containing Cluster | Strains of L. paracasei | |||
|---|---|---|---|---|
| KF1 | ABK | MA2 | MA3 | |
| Butyrivibriocin AR10 | Yes | Yes | Yes | Yes |
| ComC/Lactococcin/LSEI_2386 | Yes | Yes | Yes | Yes |
| Carnocin CP52 | Yes | Yes | Yes | Yes |
| LSEI 2163 | Yes | Yes | Yes | Yes |
| ComC/Acidocin 8912/Acidocin A | No | No | Yes | Yes |
| Enterolysin A | Yes | Yes | No | No |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Moiseenko, K.V.; Begunova, A.V.; Savinova, O.S.; Glazunova, O.A.; Rozhkova, I.V.; Fedorova, T.V. Biochemical and Genomic Characterization of Two New Strains of Lacticaseibacillus paracasei Isolated from the Traditional Corn-Based Beverage of South Africa, Mahewu, and Their Comparison with Strains Isolated from Kefir Grains. Foods 2023, 12, 223. https://doi.org/10.3390/foods12010223
Moiseenko KV, Begunova AV, Savinova OS, Glazunova OA, Rozhkova IV, Fedorova TV. Biochemical and Genomic Characterization of Two New Strains of Lacticaseibacillus paracasei Isolated from the Traditional Corn-Based Beverage of South Africa, Mahewu, and Their Comparison with Strains Isolated from Kefir Grains. Foods. 2023; 12(1):223. https://doi.org/10.3390/foods12010223
Chicago/Turabian StyleMoiseenko, Konstantin V., Anna V. Begunova, Olga S. Savinova, Olga A. Glazunova, Irina V. Rozhkova, and Tatyana V. Fedorova. 2023. "Biochemical and Genomic Characterization of Two New Strains of Lacticaseibacillus paracasei Isolated from the Traditional Corn-Based Beverage of South Africa, Mahewu, and Their Comparison with Strains Isolated from Kefir Grains" Foods 12, no. 1: 223. https://doi.org/10.3390/foods12010223
APA StyleMoiseenko, K. V., Begunova, A. V., Savinova, O. S., Glazunova, O. A., Rozhkova, I. V., & Fedorova, T. V. (2023). Biochemical and Genomic Characterization of Two New Strains of Lacticaseibacillus paracasei Isolated from the Traditional Corn-Based Beverage of South Africa, Mahewu, and Their Comparison with Strains Isolated from Kefir Grains. Foods, 12(1), 223. https://doi.org/10.3390/foods12010223






