Abstract
Droplet digital polymerase chain reaction (ddPCR) is one of the newest and most promising tools providing absolute quantification of target DNA molecules. Despite its emerging applications in microorganisms, few studies reported its use for detecting lactic acid bacteria. This study evaluated the applicability of a ddPCR assay targeting molecular genes obtained from in silico analysis for detecting Lactiplantibacillus plantarum subsp. plantarum, a bacterium mainly used as a starter or responsible for fermentation in food. The performance characteristics of a ddPCR were compared to those of a quantitative real-time PCR (qPCR). To compare the linearity and sensitivity of a qPCR and ddPCR, the calibration curve for a qPCR and the regression curve for a ddPCR were obtained using genomic DNA [102–108 colony-forming units (CFU)/mL] extracted from a pure culture and spiked food sample. Both the qPCR and ddPCR assays exhibited good linearity with a high coefficient of determination in the pure culture and spiked food sample (R2 ≥ 0.996). The ddPCR showed a 10-fold lower limit of detection, suggesting that a ddPCR is more sensitive than a qPCR. However, a ddPCR has limitations in the absolute quantitation of high bacterial concentrations (>106 CFU/mL). In conclusion, a ddPCR can be a reliable method for detecting and quantifying lactic acid bacteria in food.
1. Introduction
Lactic acid bacteria are involved in the spontaneous fermentations of foods such as meat, milk, fish, and vegetables [1]. They are commensal inhabitants of the human gastrointestinal tract and contribute to human health. As probiotics, lactic acid bacteria have shown that they may present beneficial effects, such as preventing diarrhea and inflammatory bowel disease [1,2,3]. The quantification of lactic acid bacteria is important for epidemiologic studies and their roles in various niche markets.
Lactiplantibacillus plantarum is a versatile species encountered in various niches, including meat, dairy, fish products, vegetables, and the human gastrointestinal tract [4,5]. L. plantarum is considered a safe probiotic, so it is widely used as an ingredient in fermented food and feed, such as cheese, milk, sauerkraut, and kimchi [6,7]. Between the two Lactiplantibacillus plantarum subspecies, L. plantarum subsp. plantarum provides a beneficial effect for the immune system, such as treating inflammatory diseases and mitigating pathogenic infections [8,9]. In a previous study, L. plantarum subsp. plantarum and Lactiplantibacillus plantarum subsp. argentoratensis affected the fermentation stage of vegetables, such as kimchi. L. plantarum subsp. plantarum was isolated only in fermented kimchi at a low temperature (4 °C), whereas L. plantarum subsp. argentoratensis was found only at a relatively high temperature (15 °C or 25 °C) [10]. Unlike L. plantarum subsp. plantarum, L. plantarum subsp. argentoratensis could not metabolize either methyl α-d-mannoside or melezitose [11].
A rapid method for the detection and quantification of L. plantarum subsp. plantarum in the food matrix is an essential tool for the food industry. However, prokaryotic systematics currently rely on labor- and time-consuming taxonomic approaches, including phenotypic characterization, variation analysis of 16S rRNA sequences, and DNA–DNA hybridization. Distinguishing closely related subspecies using these tools is difficult and often results in the misidentification of microorganisms [10]. Moreover, the molecular methods available for monitoring species or subspecies of L. plantarum are insufficient, as previous methods detect nontarget species of Lactiplantibacillus species or subspecies, including L. plantarum subsp. argentoratensis, Lactiplantibacillus paraplantarum, and Lactiplantibacillus pentosus [12,13]. Due to the limitations of these previous studies, there is an increasing demand for improving the current methods in studying prokaryotic systems [10].
The molecular-based detection and quantification of microorganisms have been successfully explored by real-time quantitative polymerase chain reaction (qPCR) in various food matrices [12,14]. A major advantage of a qPCR is that an amplification curve can be confirmed in hours instead of days, unlike conventional detection methods [15]. In addition, a qPCR is most commonly used as an efficient tool due to its high specificity and sensitivity [16]. A qPCR is a reliable and sensitive molecular tool applied and adopted in many different fields for the detection and quantification of genetically modified organisms (GMOs) [17,18], as well as for mutations and single nucleotide polymorphisms (SNPs) genotyping in the control of animal disease [19,20]. Recently, novel PCR-based methods for detecting and quantifying the molecular target have been introduced [21]. The droplet digital PCR (ddPCR) is a third-generation PCR tool [15]. A ddPCR mixture is divided into several partitions, each containing zero or at least one copy of the genomic DNA [15]. After amplification, partitions are counted as positive (presence of target gene) or negative (absence of target gene). The absolute quantification of the number of copies is performed using binomial Poisson statistics [15,22]. This allows a ddPCR to perform absolute quantification without using the calibration curve [15]. A ddPCR has been applied previously in various fields to quantify and detect genomic DNA targets, such as GMOs, viruses, pathogenic bacteria, antibiotic resistance genes, and vertebrate [23,24,25,26,27,28,29].
This study applied a ddPCR assay to detect and quantify L. plantarum subsp. plantarum in a food sample and compared the specificity and sensitivity of a qPCR targeting the ydiC gene. Moreover, the applicability of a ddPCR to detect lactic acid bacteria was discussed.
2. Materials and Methods
2.1. Bacterial Strains
L. plantarum subsp. plantarum and nontarget reference strains applied in this study were obtained from the Korean Collection for Type Cultures (KCTC, Daejeon, Korea), the Korean Culture Center of Microorganisms (KCCM, Seoul, Korea), the Korean Agricultural Culture Collection (KACC, Jeonju, Korea), the NITE Biological Resource Center (NBRC, Chiba, Japan), the Microorganism and Gene Bank (MGB, Gwangju, Korea), and the National Culture Collection for Pathogens (NCCP, Cheongju, Korea) (Table 1). All tested strains were grown in lactobacilli MRS (Difco Laboratories, Sparks, MD, USA) broth at 37 °C for 48 h. Genomic DNA of reference strains was extracted using DNeasy Blood & Tissue kit (Qiagen, Hilden, Germany). The genomic DNA concentration and quality were measured using a Maestrogen Nano spectrophotometer (Maestrogen, Las Vegas, NV, USA). Genomic DNA was stored at −20 °C before use as a template for qPCR and ddPCR [30].

Table 1.
List of 105 reference strains.
2.2. Primer and Probe Design
The ydiC gene (accession no. EFK30629.1) discovered by a pangenome analysis [12] was used as a target for detecting L. plantarum subsp. plantarum. The primer/probe set for detecting the ydiC gene was designed using the Primer Designer program (Scientific and Education Software, Durham, NC, USA). The primer and probe were designed considering the guanine-cytosine (GC) content and length to ensure high amplification conditions. In silico specificity was performed using in silico PCR amplification software (http://insilico.ehu.es/PCR/ accessed on 8 March 2022) [31] with genome sequences obtained from the GenBank sequence database. The sequences of the primer and probe used for qPCR and ddPCR assays and the amplicon size of the target gene are listed in Table 2.

Table 2.
Information for primer and probe for detecting L. plantarum subsp. plantarum.
2.3. qPCR Assay
The qPCR mixture consisted of 20 ng DNA template, 500 nM of each primer, 250 nM probe, 10 µL TaqMan™ Fast Universal PCR Master Mix (Applied Biosystems, Foster City, CA, USA), and purified water to a final volume of 20 µL. The amplification reaction was performed with the 7500 Fast Real-time PCR System (Applied Biosystems). The reaction was run at 50, hold for 2 min, followed by 95 °C for 10 min, then 40 cycles consisting of 95 °C for 15 s and 60 °C for 1 min per cycle. The output data were analyzed using ABI 7500 Fast Software (Applied Biosystems, Foster City, CA, USA). The specificity of primer and probe set was confirmed using 105 reference strains (Table 1). A calibration curve was constructed using genomic DNA from L. plantarum subsp. plantarum KACC 11451 with different concentrations [102–108 colony-forming units (CFU)/mL], as reported previously [32]. The viable cell was counted by the plate count method. Briefly, serial dilutions of cultured strain were grown on MRS agar and counted after incubation at 37 °C for 48 h. Amplification was performed thrice to construct the calibration curve. The calibration curves were constructed by plotting the Ct value and the number of cells.
2.4. ddPCR Assay
ddPCR was performed using 10 µL of 2 × ddPCR Supermix for probe (Bio-Rad, Pleasanton, CA, USA), 500 nM of each primer, 250 nM probe, 20 ng DNA template, and distilled water to a total volume of 20 µL. ddPCR was used to make the droplet mixture using the QX200 droplet generator (Bio-Rad). The droplet mixture was transferred to a PCR reaction plate and amplified with the following conditions: denaturation of 95 °C for 10 min, followed by 40 cycles of a two-step thermal profile consisting of 95 °C for 15 s and 60 °C for 60 s. The PCR product was incubated at 98 °C for 10 min and cooled to 4 °C until the droplets were read. The PCR reaction plate was transferred to the QX200 droplet reader (Bio-Rad). The number of positive (high level of fluorescence) and negative (low and constant level of fluorescence) droplets obtained were analyzed using QuantaSoft software (Bio-Rad, Pleasanton, CA, USA) [33].
2.5. Artificially Contaminated Milk Sample
To compare the performance characteristics of qPCR and ddPCR, L. plantarum subsp. plantarum was used to artificially contaminate food samples. The food sample was obtained from a market and confirmed absent of L. plantarum subsp. plantarum by qPCR. To prepare the spiked food sample, the milk sample was spiked with a pure culture of L. plantarum subsp. plantarum at a concentration of 108 CFU/mL [34,35] and mixed for 2 min using a homogenizer (Stomacher Circulator 400; Seward Ltd., London, UK). The number of bacteria was determined by the plate counting method according to a previous study [32]. Briefly, 0.1 mL of an appropriate dilution of bacteria was spread on MRS agar and incubation at 37 °C for 48 h for bacterial cell counting. An aliquot of 1 mL spiked food sample was transferred to a sterilized tube, used for genomic DNA extraction, and subjected to qPCR and ddPCR assays.
3. Results and Discussion
3.1. Specificity of Primer by In Silico PCR
Before performing a qPCR and ddPCR, the inclusivity and exclusivity of the new primer and probe set designed from the ydiC gene were tested by an in silico PCR assay. The ydiC gene is a novel genetic marker for detecting L. plantarum subsp. plantarum obtained from a pangenome analysis [12]. The sequence identity with the primer, in silico PCR result, and amplicon size obtained from the ydiC gene of 56 L. plantarum subsp. plantarum and 140 other species are represented in Table S1. The primer and probe generated a positive reaction with all L. plantarum subsp. plantarum strains, whereas the remaining nontarget species or subspecies produced a negative reaction. The amplicon size of all L. plantarum subsp. plantarum was 150 bp.
3.2. Evaluation of the Specificity and Sensitivity by qPCR
The accuracy of a qPCR and ddPCR depends on the specificity of the sequence or primer used in the experiment [36]. Many studies have reported detecting L. plantarum using the 16S rRNA sequence or housekeeping genes (atpD, recA, and dnaK) as qPCR marker genes [36,37,38]. However, these genes reported to date have high sequence homologies among other species or subspecies, such as L. plantarum subsp. argentoratensis and L. paraplantarum, and require an additional procedure for identification that is time-consuming and costly. Therefore, this study designed the primer and probe using the ydiC gene discovered by a pangenome analysis [12].
A qPCR was performed using L. plantarum subsp. plantarum and 104 other reference strains to determine the amplification efficiency and specificity of the primer targeting the ydiC gene. The target DNA of L. plantarum subsp. plantarum was successfully amplified (Figure 1A), whereas no amplification was observed for the 104 other reference strains tested, indicating that the primer was specific for L. plantarum subsp. plantarum detection, consistent with a previous result using the same target gene [12]. The calibration curve was constructed using the genomic DNA of L. plantarum subsp. plantarum ranging from 108 to 103 CFU/mL. When the calibration curves of a qPCR have R2 ≥ 0.98 and a slope from −3.1 to −3.6, it can be regarded as a high-efficiency primer [39]. A calibration curve had an amplification efficiency of 88.364% (Figure 1B), suggesting that the primer showed a high efficiency in detecting L. plantarum subsp. plantarum.
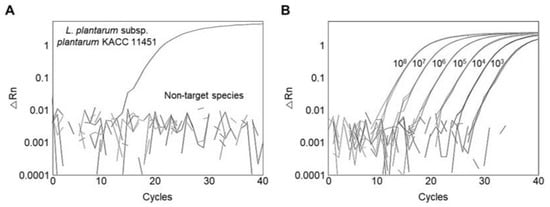
Figure 1.
Assessment of primer specificity and sensitivity using qPCR. (A) Amplification plot of qPCR; (B) amplification plot of serial dilution of genomic DNA of L. plantarum subsp. plantarum KACC 11451.
Propidium monoazide (PMA) combined with a PCR appears to be a potential method for distinguishing between living and dead cells [40,41,42]. Several studies have reported the application of PMA treatments to quantify viable bacterial cells in foods by a qPCR and ddPCR [40,43]. In this study, because genomic DNA was quantified without a PMA treatment, there is a disadvantage that viable, but non-cultivable (VBNC) cells cannot be quantified.
3.3. Evaluation of Specificity and Sensitivity by ddPCR
A ddPCR has the potential to be a robust method for the quantification and detection of microorganisms in food [44,45]. This study investigated the potential of a ddPCR for detecting L. plantarum subsp. plantarum. Similar to the qPCR assay, the high specificity of a primer was observed in the ddPCR method. The genomic DNA from the 105 strains was detected by a ddPCR, and no droplets were observed for the 104 nontarget reference strains, including Lactiplantibacillus species closely related to L. plantarum subsp. plantarum (Figure 2A). Moreover, the ddPCR assay was sensitive with good linearity (slope = 0.9197, R2 = 0.996 for detecting the ydiC gene) ranging from 108 to 102 CFU/mL (Figure 2B). The limit of detection in the genomic DNA obtained from the pure culture was 102 CFU/mL, with a quantification value of 0.4 ± 0.11 copies (mean ± standard deviation). However, the ddPCR analysis failed to quantify the DNA when the L. plantarum subsp. plantarum population was >106 CFU/mL (Table 3). In the ddPCR, reaction saturation was reached with more than 20,000 positive droplets, making it impossible to quantify this concentration [35].
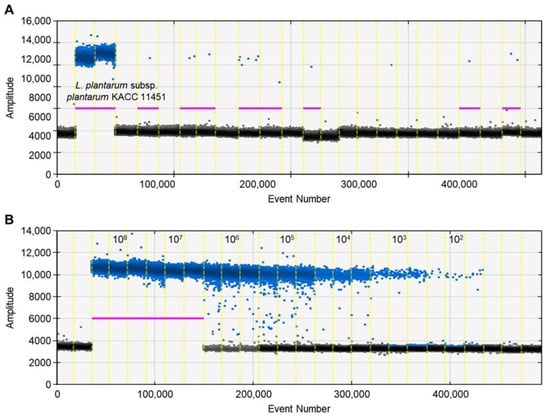
Figure 2.
Assessment of primer specificity and sensitivity using ddPCR. The positive and negative droplets classified as the thresholds of individual wells are indicated in blue and black, respectively. The threshold is determined by the droplet reader and is shown as a horizontal line. (A) Specificity of primer by ddPCR; (B) quantification of genomic DNA of L. plantarum subsp. plantarum KACC 11451 by ddPCR.

Table 3.
Quantification of genomic DNA extracted from pure culture and spiked milk sample.
3.4. Comparison of Sensitivity and Linearity of qPCR and ddPCR Assays
To compare the sensitivity and reliability, serial dilutions (108–102 CFU/mL) of the genomic DNA extracted from the pure culture and spiked food sample were used to determine the limit of detection and the limit of quantification. The limit of detection and the limit of quantification were calculated according to the previous studies [39,46]. The limit of detection of the qPCR was determined as 103 CFU/mL in both the pure culture and spiked food sample (Table 3). On the other hand, the limit of quantification was determined as 104 CFU/mL (Table 3). The qPCR showed a good linearity range of 108–103 CFU/mL with a 0.999 coefficient of determination (R2) in the genomic DNA of the pure culture and spiked food sample (Figure 3A,B). In contrast to the qPCR, the quantitative detection range of the ddPCR was from 106 to 102 CFU/mL (Table 3). The ddPCR assay showed the genomic DNA of the pure culture and spiked food sample with a good linearity (R2 = 0.996 and 0.998; Figure 3C,D). The ddPCR assay exhibited the lowest limit of detection value (102 CFU/mL) and limit of quantification value (103 CFU/mL) compared to the qPCR, showing that the ddPCR sensitivity was ten times higher than the qPCR detection (Table 3), consistent with previous studies in which the ddPCR is 10-fold more sensitive than the qPCR [15,44]. However, ddPCR droplets are positively saturated at >106 CFU/mL bacterial concentrations, making the Poisson distribution estimation invalid and resulting in a narrower dynamic range than the qPCR. The limitation of the ddPCR reported in previous studies was the quantification of the target DNA when bacterial concentrations exceeded 106 CFU/mL [15,22,44]. In this study, samples with a high bacterial abundance (>106 CFU/mL) were not quantified. To accurately quantify these samples, they must be diluted and run again on a ddPCR, thus increasing the time and cost required for the analysis [15]. Therefore, it is necessary to confirm that the amount of the target DNA is within the measurement range before performing the experiment.
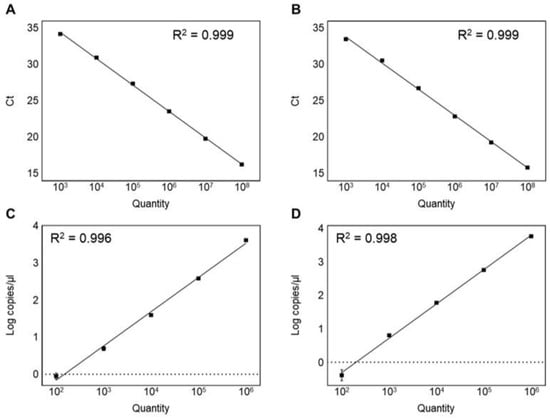
Figure 3.
Linearity and sensitivity of two assays. (A) qPCR in analyzing genomic DNA of L. plantarum subsp. plantarum KACC 11451; (B) qPCR in analyzing spiked milk sample; (C) ddPCR in analyzing genomic DNA of L. plantarum subsp. plantarum KACC 11451; (D) ddPCR in analyzing spiked milk sample.
This study evaluated the applicability of a ddPCR to detect L. plantarum subsp. plantarum. Unlike a qPCR, a ddPCR does not require a calibration curve. A ddPCR allowed quantifying the absolute number of the target DNA added to the mixture by partitioning the PCR reagents and using the Poisson algorithm [21,47]. This may reduce the bias introduced in the qPCR assay, as the calculated bacterial concentrations had to be constructed for the calibration curve. Moreover, the higher sensitivity of a ddPCR than of a qPCR has been reported previously [15,22,44]. This would be an advantage in food samples having PCR inhibitors or containing low copies of target molecules [27,28]. The difference in sensitivity between the assays was attributed to the higher resistance of the ddPCR to inhibitors occurring in food matrices [25,35]. In a ddPCR, the target DNA is distributed over thousands of droplets that constitute separate reaction compartments, leading to a higher tolerance to inhibitors [35,48]. Although more expensive and time-consuming than a qPCR, a ddPCR is currently a more reliable tool for detecting microorganisms, viruses, and GMOs [24,40,43,49]. Previous studies have demonstrated that a ddPCR is a suitable analytical tool for detecting foodborne pathogens in the food matrix [44,45,50]. Similar to foodborne pathogens, a ddPCR is a useful tool for detecting low-level L. plantarum subsp. plantarum. Therefore, a ddPCR can be a suitable analytical tool for the quantitative detection of lactic acid bacteria, which is important in probiotic products, dairy products, or fermented food.
4. Conclusions
To the best of our knowledge, this is the first report that demonstrated the ddPCR assay to quantify L. plantarum subsp. plantarum. The ddPCR observed a lower limit of detection than the qPCR, which could be advantageous in foods with a low number of target species. The ddPCR represents an innovation in the molecular world and is very useful, sensitive, and reliable for overcoming different limits for L. plantarum subsp. plantarum quantification. At the same time, it is not an instrument that is accessible and easy to use in any laboratories and industries, both for costs and for the type of analysis. In conclusion, this study can be used as preliminary data for a future robust assay optimization and validation. This method, which enables the quantification of L. plantarum subsp. plantarum in the food samples, can be a useful tool in the food industry to evaluate the quality of fermented food products.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/foods11091331/s1, Table S1: In silico PCR for specificity of primer and probe.
Author Contributions
Conceptualization, E.K. and H.-Y.K.; methodology, E.K. and S.-M.Y.; investigation, C.-H.C., S.-M.Y. and D.-S.K.; validation, S.-M.S. and G.-Y.L.; writing—original draft preparation, C.-H.C. and E.K.; writing—review and editing, H.-Y.K.; visualization, E.K.; project administration, H.-Y.K.; funding acquisition, H.-Y.K. All authors have read and agreed to the published version of the manuscript.
Funding
This work was carried out with the support of the “Cooperative Research Program for Agriculture Science & Technology Development (Project No. PJ01662001)” Rural Development Administration, Republic of Korea.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Data is contained within the article or supplementary material.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Furet, J.P.; Quénée, P.; Tailliez, P. Molecular quantification of lactic acid bacteria in fermented milk products using real-time quantitative PCR. Int. J. Food Microbiol. 2004, 97, 197–207. [Google Scholar] [CrossRef] [PubMed]
- Guarino, A.; Guandalini, S.; Vecchio, A.L. Probiotics for prevention and treatment of diarrhea. J. Clin. Gastroenterol. 2015, 49, S37–S45. [Google Scholar] [CrossRef] [PubMed]
- Peña, J.A.; Rogers, A.B.; Ge, Z.; Ng, V.; Li, S.Y.; Fox, J.G.; Versalovic, J. Probiotic Lactobacillus spp. diminish Helicobacter hepaticus-induced inflammatory bowel disease in interleukin-10-deficient mice. Infect. Immun. 2005, 73, 912–920. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Kim, E.; Chang, H.C.; Kim, H.Y. Complete genome sequence of Lactobacillus plantarum EM, a putative probiotic strain with the cholesterol-lowering effect and antimicrobial activity. Curr. Microbiol. 2020, 77, 1871–1882. [Google Scholar] [CrossRef]
- Martino, M.E.; Bayjanov, J.R.; Caffrey, B.E.; Wels, M.; Joncour, P.; Hughes, S.; Gillet, B.; Kleerebezem, M.; van Hijum, S.A.F.T.; Leulier, F. Nomadic lifestyle of Lactobacillus plantarum revealed by comparative genomics of 54 strains isolated from different habitats. Environ. Microbiol. 2016, 18, 4974–4989. [Google Scholar] [CrossRef]
- Jeong, C.H.; Sohn, H.; Hwang, H.; Lee, H.J.; Kim, T.W.; Kim, D.S.; Kim, C.S.; Han, S.G.; Hong, S.W. Comparison of the probiotic potential between Lactiplantibacillus plantarum isolated from kimchi and standard probiotic strains isolated from different sources. Foods 2021, 10, 2125. [Google Scholar] [CrossRef]
- Fernandes, P.; Loureiro, D.; Monteiro, V.; Ramos, C.; Nero, L.A.; Todorov, S.D.; Guerreiro, J.S. Lactobacillus plantarum isolated from cheese: Production and partial characterization of bacteriocin B391. Ann. Microbiol. 2017, 67, 433–442. [Google Scholar] [CrossRef]
- Wang, W.; Ma, H.; Yu, H.; Qin, G.; Tan, Z.; Wang, Y.; Pang, H. Screening of Lactobacillus plantarum subsp. plantarum with potential probiotic activities for inhibiting ETEC K88 in weaned piglets. Molecules 2020, 25, 4481. [Google Scholar] [CrossRef]
- Giri, S.S.; Kim, H.J.; Kim, S.G.; Kim, S.W.; Kwon, J.; Lee, S.B.; Woo, K.J.; Jung, W.J.; Kim, M.J.; Sukumaran, V.; et al. Effects of dietary Lactiplantibacillus plantarum subsp. plantarum L7, alone or in combination with Limosilactobacillus reuteri P16, on growth, mucosal immune responses, and disease resistance of Cyprinus carpio. Probiotics Antimicrob. Proteins 2021, 13, 1747–1758. [Google Scholar] [CrossRef]
- Jin, Y.J.; Park, Y.K.; Cho, M.S.; Lee, E.S.; Park, D.S. New insight and metrics to understand the ontogeny and succession of Lactobacillus plantarum subsp. plantarum and Lactobacillus plantarum subsp. argentoratensis. Sci. Rep. 2018, 8, 6029. [Google Scholar] [CrossRef]
- Bringel, F.; Castioni, A.; Olukoya, D.K.; Felis, G.E.; Torriani, S.; Dellaglio, F. Lactobacillus plantarum subsp. argentoratensis subsp. nov., isolated from vegetable matrices. Int. J. Syst. Evol. Microbiol. 2005, 55, 1629–1634. [Google Scholar] [CrossRef] [PubMed]
- Kim, E.; Kim, H.B.; Yang, S.M.; Kim, D.; Kim, H.Y. Real-time PCR assay for detecting Lactobacillus plantarum group using species/subspecies-specific genes identified by comparative genomics. LWT 2021, 138, 110789. [Google Scholar] [CrossRef]
- Huang, C.H.; Chen, C.C.; Lin, Y.C.; Chen, C.H.; Lee, A.Y.; Liou, J.S.; Gu, C.T.; Huang, L. The mutL gene as a genome-wide taxonomic marker for high resolution discrimination of Lactiplantibacillus plantarum and its closely related taxa. Microorganisms 2021, 9, 1570. [Google Scholar] [CrossRef] [PubMed]
- Wei, S.; Daliri, E.B.M.; Chelliah, R.; Park, B.J.; Lim, J.S.; Baek, M.A.; Nam, Y.S.; Seo, K.H.; Jin, Y.G.; Oh, D.H. Development of a multiplex real-time PCR for simultaneous detection of Bacillus cereus, Listeria monocytogenes, and Staphylococcus aureus in food samples. J. Food Saf. 2019, 39, e12558. [Google Scholar] [CrossRef]
- Porcellato, D.; Narvhus, J.; Skeie, S.B. Detection and quantification of Bacillus cereus group in milk by droplet digital PCR. J. Microbiol. Methods 2016, 127, 1–6. [Google Scholar] [CrossRef]
- Liu, Y.; Cao, Y.; Wang, T.; Dong, Q.; Li, J.; Niu, C. Detection of 12 common food-borne bacterial pathogens by taq man real-time PCR using a single set of reaction conditions. Front. Microbiol. 2019, 10, 222. [Google Scholar] [CrossRef]
- Pierboni, E.; Curcio, L.; Tovo, G.R.; Torricelli, M.; Rondini, C. Evaluation of systems for nopaline synthase terminator in fast and standard real-time PCR to screen genetically modified organisms. Food Anal. Methods 2016, 9, 1009–1019. [Google Scholar] [CrossRef]
- Fraiture, M.A.; Gobbo, A.; Marchesi, U.; Verginelli, D.; Papazova, N.; Roosens, N.H.C. Development of a real-time PCR marker targeting a new unauthorized genetically modified microorganism producing protease identified by DNA walking. Int. J. Food Microbiol. 2021, 354, 109330. [Google Scholar] [CrossRef]
- Lefever, S.; Rihani, A.; Van der Meulen, J.; Pattyn, F.; Van Maerken, T.; Van Dorpe, J.; Hellemans, J.; Vandesompele, J. Cost-effective and robust genotyping using double-mismatch allele-specific quantitative PCR. Sci. Rep. 2019, 9, 2150. [Google Scholar] [CrossRef]
- Torricelli, M.; Sebastiani, C.; Ciullo, M.; Ceccobelli, S.; Chiappini, B.; Vaccari, G.; Capocefalo, A.; Conte, M.; Giovannini, S.; Lasagna, E.; et al. PRNP polymorphisms in eight local goat populations/breeds from central and southern Italy. Animals 2021, 11, 333. [Google Scholar] [CrossRef]
- Hindson, B.J.; Ness, K.D.; Masquelier, D.A.; Belgrader, P.; Heredia, N.J.; Makarewicz, A.J.; Bright, I.J.; Lucero, M.Y.; Hiddessen, A.L.; Legler, T.C.; et al. High-throughput droplet digital PCR system for absolute quantitation of DNA copy number. Anal. Chem. 2011, 83, 8604–8610. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Xia, Q.; Yin, Y.; Wang, Z. Comparison of droplet digital PCR and quantitative PCR assays for quantitative detection of Xanthomonas citri subsp. citri. PLoS ONE 2016, 11, e0159004. [Google Scholar] [CrossRef]
- Ricchi, M.; Bertasio, C.; Boniotti, M.B.; Vicari, N.; Russo, S.; Tilola, M.; Bellotti, M.A.; Bertasi, B. Comparison among the quantification of bacterial pathogens by qPCR, dPCR, and cultural methods. Front. Microbiol. 2017, 8, 1174. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Zhai, S.; Gao, H.; Xiao, F.; Li, Y.; Wu, G.; Wu, Y. Development and assessment of a duplex droplet digital PCR method for quantification of GM rice Kemingdao. Anal. Bioanal. Chem. 2021, 413, 4341–4351. [Google Scholar] [CrossRef]
- Persson, S.; Eriksson, R.; Lowther, J.; Ellström, P.; Simonsson, M. Comparison between RT droplet digital PCR and RT real-time PCR for quantification of noroviruses in oysters. Int. J. Food Microbiol. 2018, 284, 73–83. [Google Scholar] [CrossRef] [PubMed]
- Rački, N.; Dreo, T.; Gutierrez-Aguirre, I.; Blejec, A.; Ravnikar, M. Reverse transcriptase droplet digital PCR shows high resilience to PCR inhibitors from plant, soil and water samples. Plant Methods 2014, 10, 42. [Google Scholar] [CrossRef]
- Xu, W.; Shen, P.; Li, R.; Liu, B.; Yang, L. Development of an event-specific droplet digital PCR assay for quantification and evaluation of the transgene DNAs in trace samples of GM PRNP-knockout goat. Foods 2022, 11, 868. [Google Scholar] [CrossRef] [PubMed]
- Park, S.; Rana, A.; Sung, W.; Munir, M. Competitiveness of quantitative polymerase chain reaction (qPCR) and droplet digital polymerase chain reaction (ddPCR) technologies, with a particular focus on detection of antibiotic resistance genes (ARGs). Appl. Microbiol. 2021, 1, 426–444. [Google Scholar] [CrossRef]
- Rice, L.M.; Robb, L.L.; Hartman, D.A.; Anderson, J.R.; Kading, R.C. Application of the droplet digital polymerase chain reaction (ddPCR) platform for detection and quantification of vertebrate host DNA in engorged mosquitoes. J. Med. Entomol. 2019, 56, 1150–1153. [Google Scholar] [CrossRef]
- Kim, E.; Kim, D.; Yang, S.-M.; Kim, H.-Y. Validation of probiotic species or subspecies identity in commercial probiotic products using high-resolution PCR method based on large-scale genomic analysis. Food Res. Int. 2022, 154, 111011. [Google Scholar] [CrossRef]
- In silico PCR amplification. Available online: http://insilico.ehu.es/PCR/ (accessed on 8 March 2022).
- Gómez-Rojo, E.M.; Romero-Santacreu, L.; Jaime, I.; Rovira, J. A novel real-time PCR assay for the specific identification and quantification of Weissella viridescens in blood sausages. Int. J. Food Microbiol. 2015, 215, 16–24. [Google Scholar] [CrossRef] [PubMed]
- Dupas, E.; Legendre, B.; Olivier, V.; Poliakoff, F.; Manceau, C.; Cunty, A. Comparison of real-time PCR and droplet digital PCR for the detection of Xylella fastidiosa in plants. J. Microbiol. Methods 2019, 162, 86–95. [Google Scholar] [CrossRef] [PubMed]
- Lei, S.; Gu, X.; Zhong, Q.; Duan, L.; Zhou, A. Absolute quantification of Vibrio parahaemolyticus by multiplex droplet digital PCR for simultaneous detection of tlh, tdh and ureR based on single intact cell. Food Control 2020, 114, 107207. [Google Scholar] [CrossRef]
- Cristiano, D.; Peruzy, M.F.; Aponte, M.; Mancusi, A.; Proroga, Y.T.R.; Capuano, F.; Murru, N. Comparison of droplet digital PCR vs real-time PCR for Yersinia enterocolitica detection in vegetables. Int. J. Food Microbiol. 2021, 354, 109321. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.; Huang, L.; Wu, C. Molecular discrimination of Lactobacillus plantarum group using comparative sequence analysis of the dnaJ gene and as a target for developing novel species-specific PCR primers. J. Chin. Soc. Anim. Sci. 2016, 45, 45–55. [Google Scholar]
- Galanis, A.; Kourkoutas, Y.; Tassou, C.C.; Chorianopoulos, N. Detection and identification of probiotic Lactobacillus plantarum strains by multiplex PCR using RAPD-derived primers. Int. J. Mol. Sci. 2015, 16, 25141–25153. [Google Scholar] [CrossRef] [PubMed]
- Schwendimann, L.; Kauf, P.; Fieseler, L.; Gantenbein-Demarchi, C.; Miescher Schwenninger, S. Development of a quantitative PCR assay for rapid detection of Lactobacillus plantarum and Lactobacillus fermentum in cocoa bean fermentation. J. Microbiol. Methods 2015, 115, 94–99. [Google Scholar] [CrossRef]
- Broeders, S.; Huber, I.; Grohmann, L.; Berben, G.; Taverniers, I.; Mazzara, M.; Roosens, N.; Morisset, D. Guidelines for validation of qualitative real-time PCR methods. Trends Food Sci. Technol. 2014, 37, 115–126. [Google Scholar] [CrossRef]
- Gobert, G.; Cotillard, A.; Fourmestraux, C.; Pruvost, L.; Miguet, J.; Boyer, M. Droplet digital PCR improves absolute quantification of viable lactic acid bacteria in faecal samples. J. Microbiol. Methods 2018, 148, 64–73. [Google Scholar] [CrossRef]
- Hansen, S.J.Z.; Morovic, W.; DeMeules, M.; Stahl, B.; Sindelar, C.W. Absolute enumeration of probiotic strains Lactobacillus acidophilus NCFM® and Bifidobacterium animalis subsp. lactis Bl-04® via chip-based digital PCR. Front. Microbiol. 2018, 9, 704. [Google Scholar] [CrossRef]
- Matsuki, T.; Watanabe, K.; Fujimoto, J.; Kado, Y.; Takada, T.; Matsumoto, K.; Tanaka, R. Quantitative PCR with 16S rRNA-gene-targeted species-specific primers for analysis of human intestinal bifidobacteria. Appl. Environ. Microbiol. 2004, 70, 167–173. [Google Scholar] [CrossRef] [PubMed]
- Pan, H.; Dong, K.; Rao, L.; Zhao, L.; Wu, X.; Wang, Y.; Liao, X. Quantitative detection of viable but nonculturable state Escherichia coli O157:H7 by ddPCR combined with propidium monoazide. Food Control 2020, 112, 107140. [Google Scholar] [CrossRef]
- Wang, M.; Yang, J.; Gai, Z.; Huo, S.; Zhu, J.; Li, J.; Wang, R.; Xing, S.; Shi, G.; Shi, F.; et al. Comparison between digital PCR and real-time PCR in detection of Salmonella Typhimurium in milk. Int. J. Food Microbiol. 2018, 266, 251–256. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Zhang, N.; Lv, J.; Zhu, P.; Pan, X.; Hu, J.; Wu, W.; Li, S.; Li, H. Comparing the performance of conventional PCR, RTQ-PCR, and droplet digital PCR assays in detection of Shigella. Mol. Cell. Probes 2020, 51, 101531. [Google Scholar] [CrossRef] [PubMed]
- Hougs, L.; Gatto, F.; Goerlich, O.; Grohmann, L.; Lieske, K.; Mazzara, M.; Narendja, F.; Ovesna, J.; Papazova, N.; Scholtens, I.M.J.; et al. Verification of analytical methods for GMO testing when implementing interlaboratory validated methods. In Testing and Analysis of GMO-Containing Foods and Feed; CRC Press: Boca Raton, FL, USA, 2017; pp. 245–266. [Google Scholar]
- Pinheiro, L.B.; Coleman, V.A.; Hindson, C.M.; Herrmann, J.; Hindson, B.J.; Bhat, S.; Emslie, K.R. Evaluation of a droplet digital polymerase chain reaction format for DNA copy number quantification. Anal. Chem. 2012, 84, 1003–1011. [Google Scholar] [CrossRef] [PubMed]
- Villamil, C.; Calderon, M.N.; Arias, M.M.; Leguizamon, J.E. Validation of droplet digital polymerase chain reaction for Salmonella spp. quantification. Front. Microbiol. 2020, 11, 1512. [Google Scholar] [CrossRef]
- Mairiang, D.; Songjaeng, A.; Hansuealueang, P.; Malila, Y.; Lertsethtakarn, P.; Silapong, S.; Poolpanichupatam, Y.; Klungthong, C.; Chin-Inmanu, K.; Thiemmeca, S.; et al. Application of one-step reverse transcription droplet digital PCR for dengue virus detection and quantification in clinical specimens. Diagnostics 2021, 11, 639. [Google Scholar] [CrossRef]
- Luo, J.; Li, J.; Yang, H.; Yu, J.; Wei, H. Accurate detection of methicillin-resistant Staphylococcus aureus in mixtures by use of single-bacterium duplex droplet digital PCR. J. Clin. Microbiol. 2017, 55, 2946–2955. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).