Comparative Transcriptomic Analysis of Staphylococcus aureus Reveals the Genes Involved in Survival at Low Temperature
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterial Strains and Culture Conditions
2.2. Preparation of Bacterial Suspensions and Cold Treatment
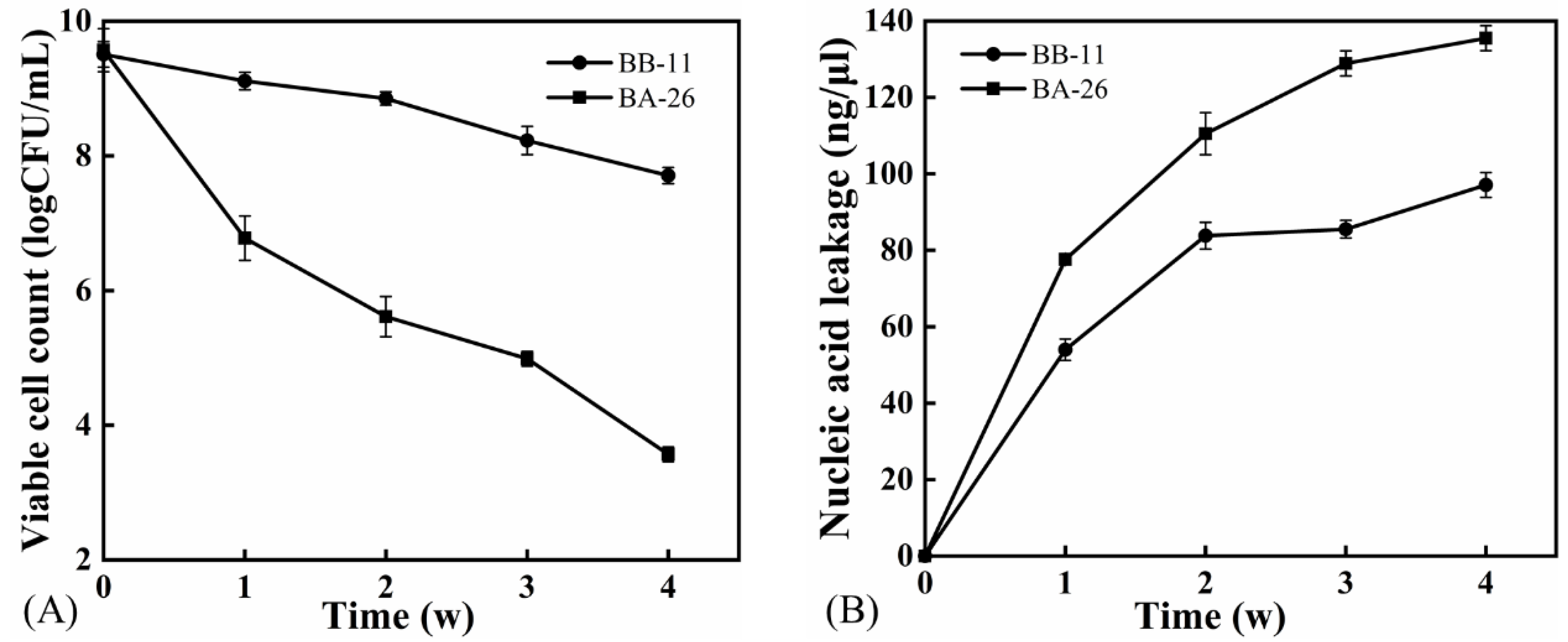
2.3. Viable Cell Enumeration
2.4. Nucleic Acid Leakage Analysis
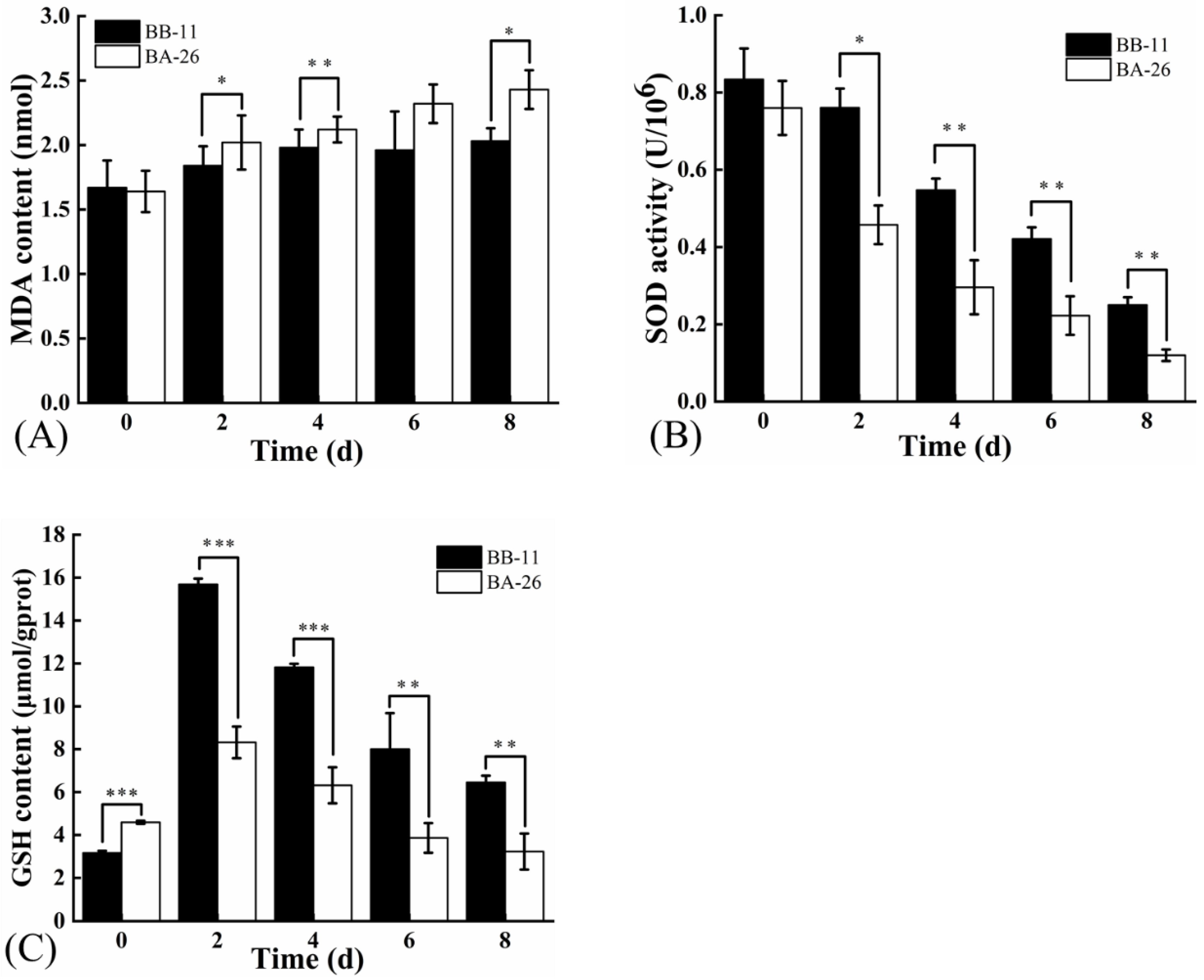
2.5. Determination of Microscale Malondialdehyde (MDA) and Reduced Glutathione (GSH) Contents, and Superoxide Dismutase (SOD) Activity
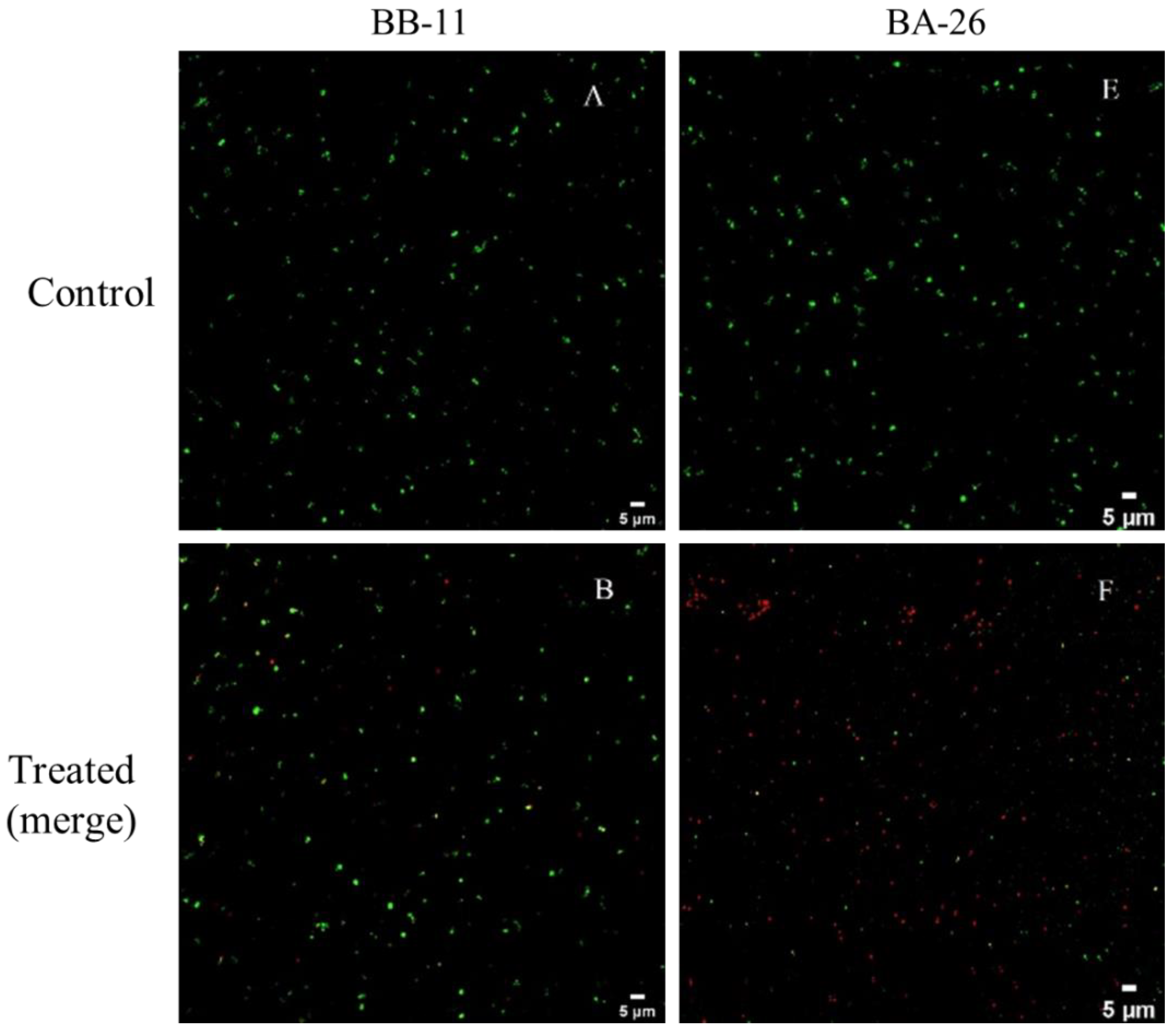
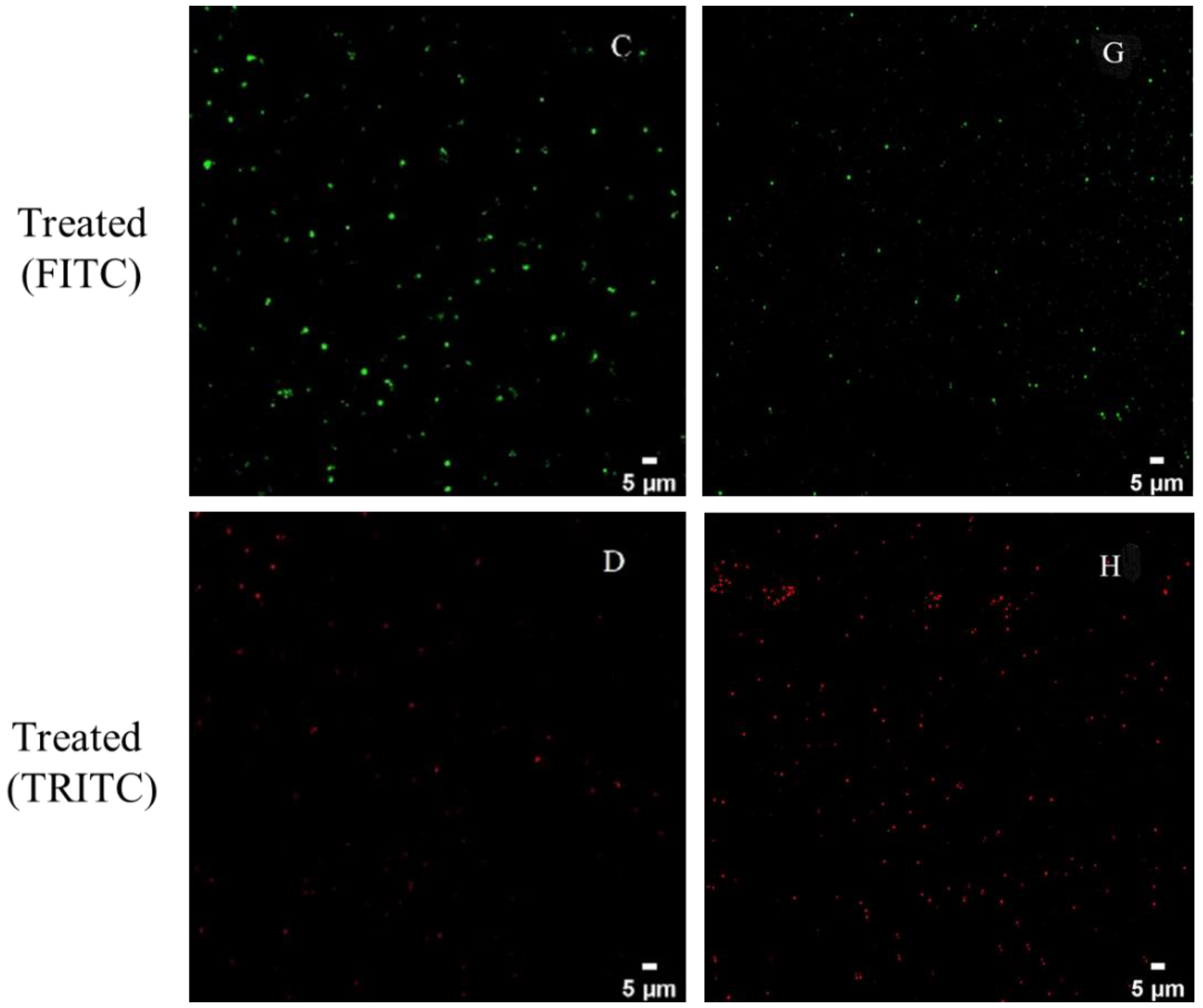
2.6. Confocal Laser Scanning Microscopy (CLSM) Observation
2.7. Transmission Electron Microscopy (TEM) Observation
2.8. RNA-Seq Analysis
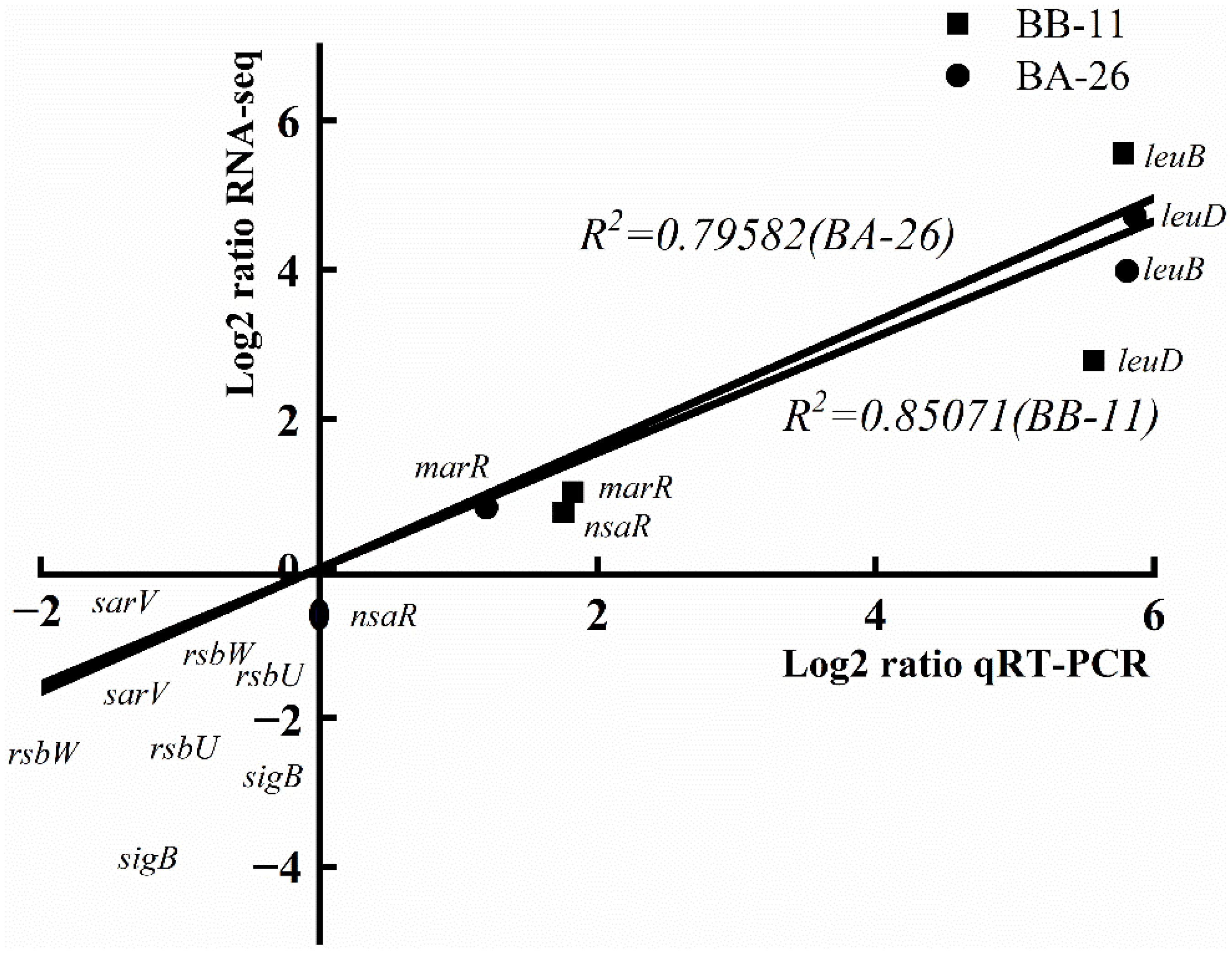
2.9. Validation of RNA-Seq Results Using qRT–PCR
2.10. Statistical Analysis
3. Results and Discussion
3.1. Changes in the Number of Viable Cells and Intracellular Nucleic Acid Leakage of S. aureus
3.2. Oxidative Stress in S. aureus
3.3. CLSM Observations
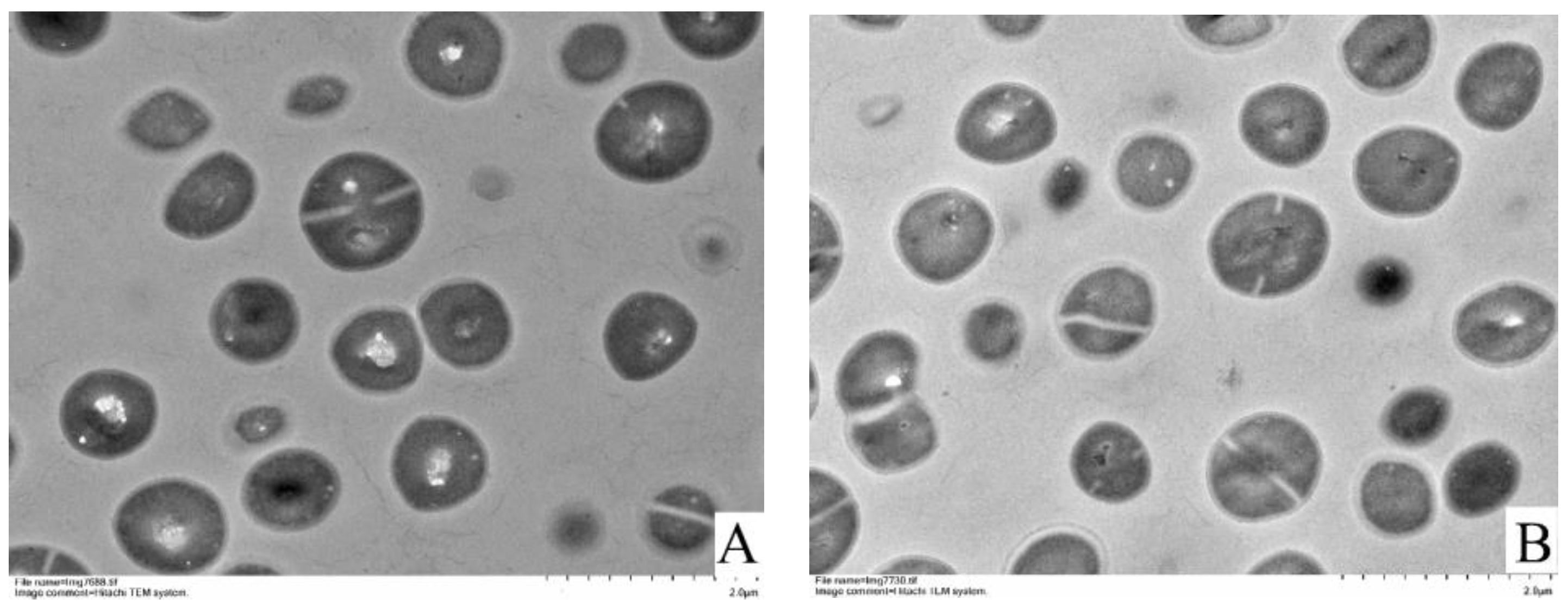
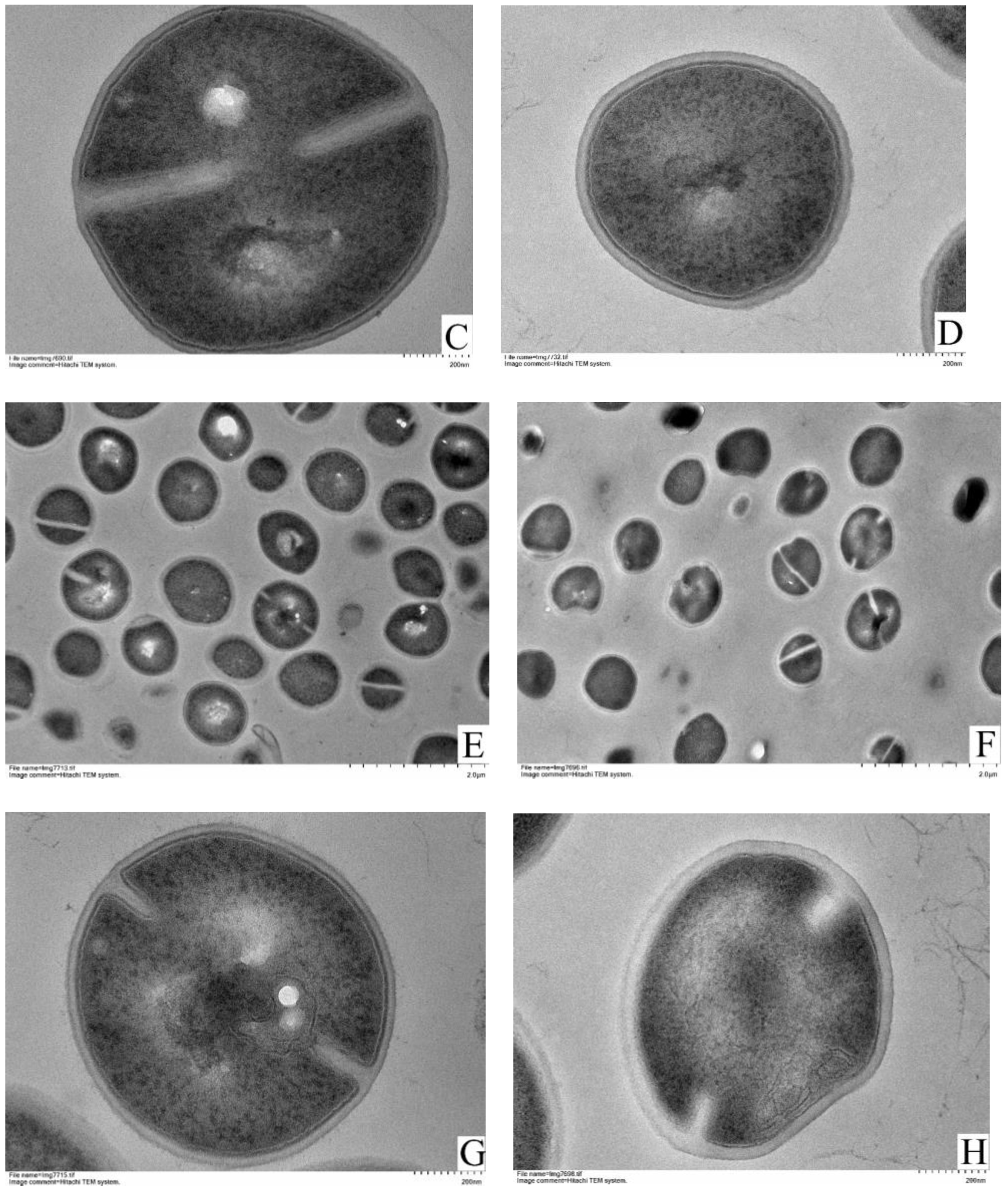
3.4. TEM Observations
3.5. RNA-Seq Data Processing and Analysis
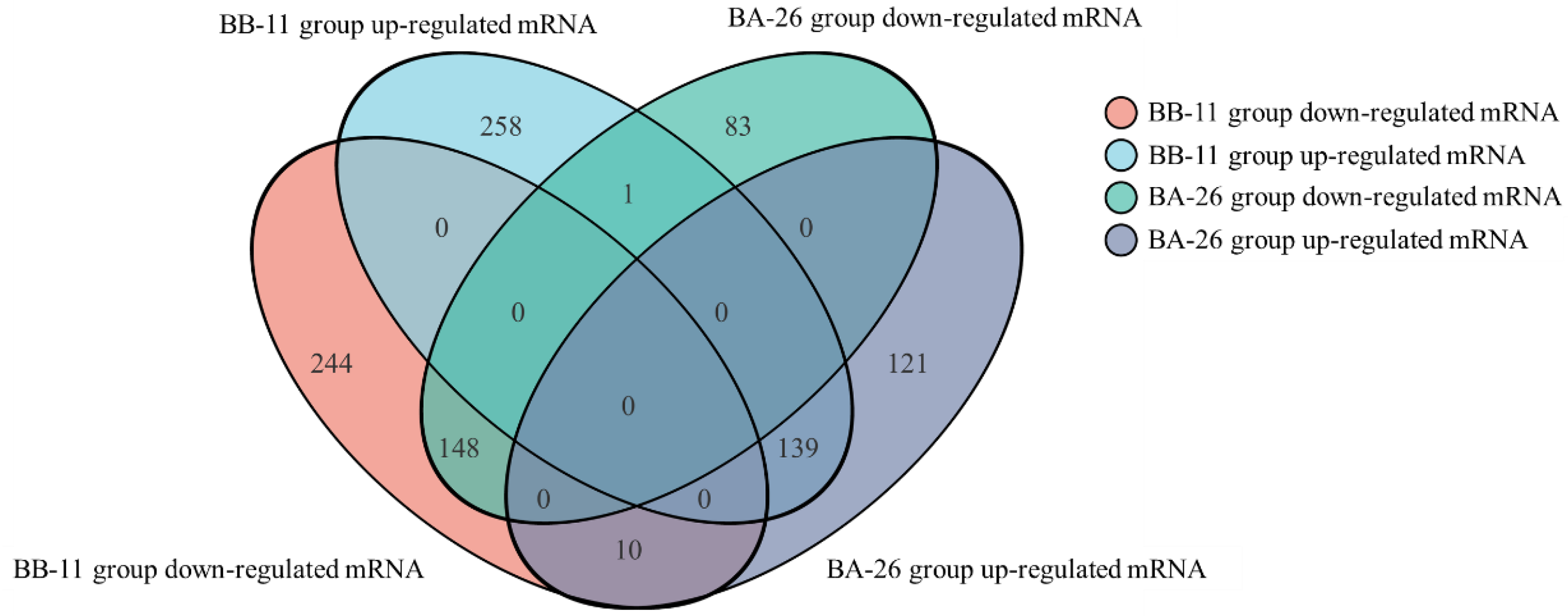
3.6. Differentially Expressed Genes (DGEs) in S. aureus with Different Responses to Low Temperature
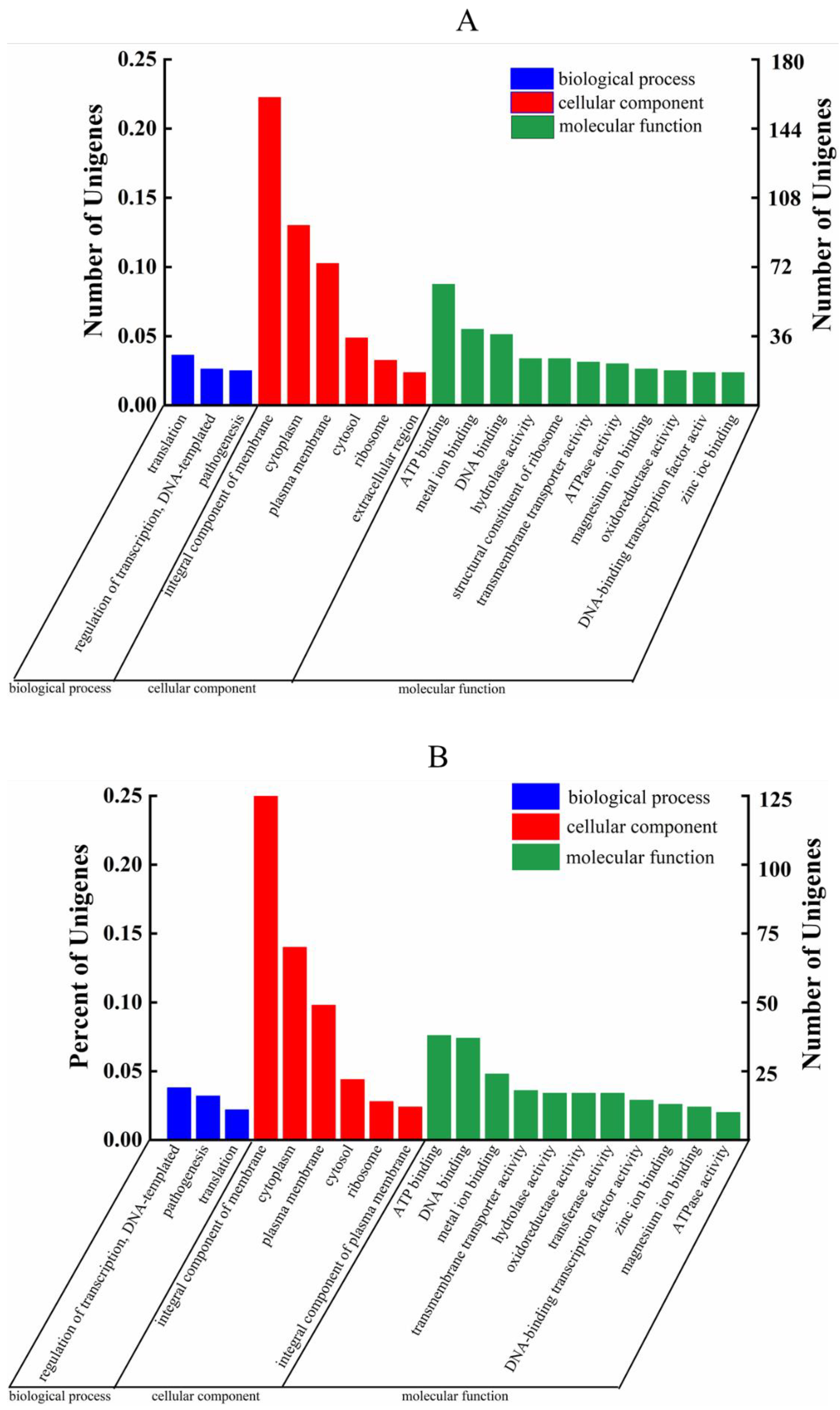
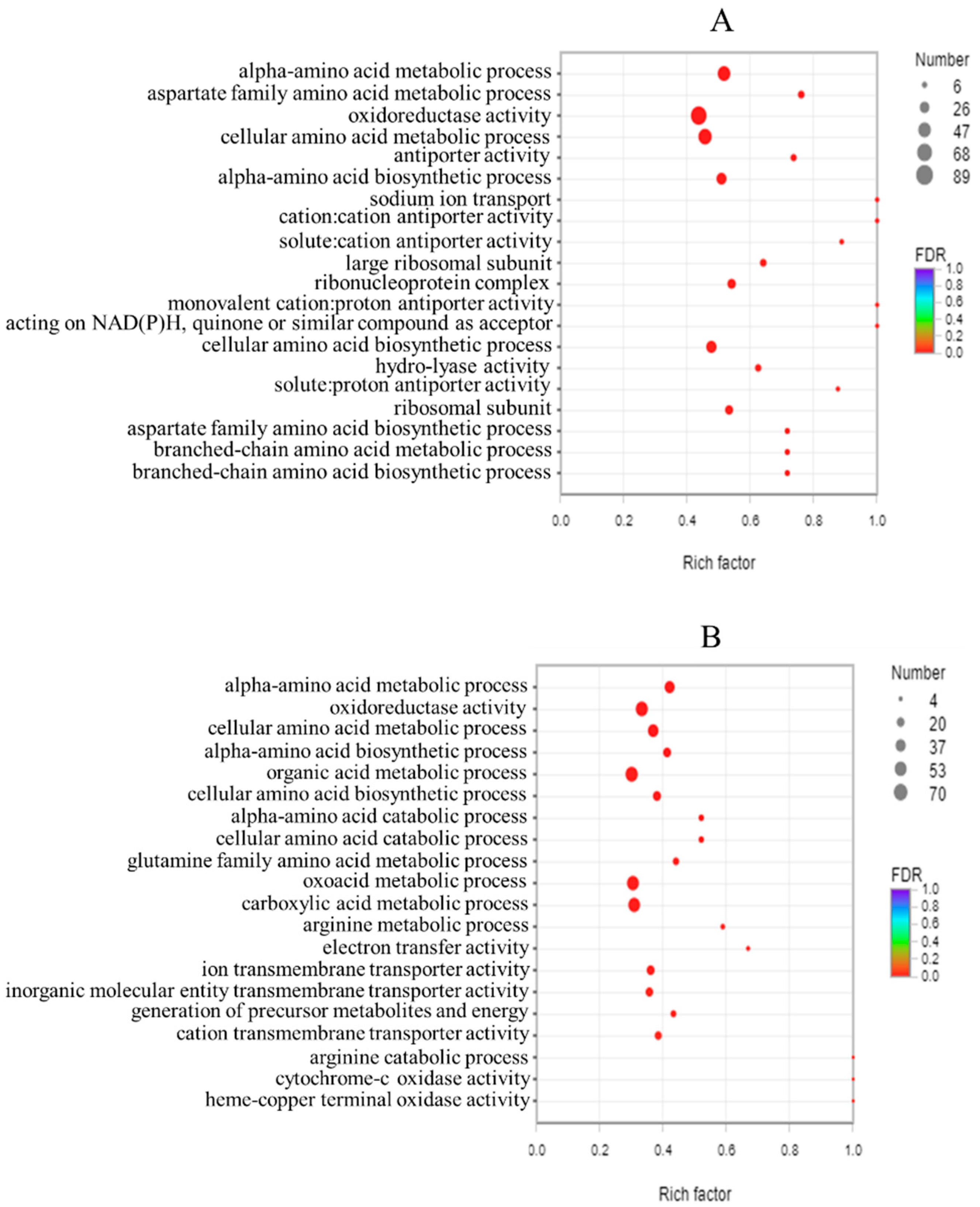
3.7. GO Analysis
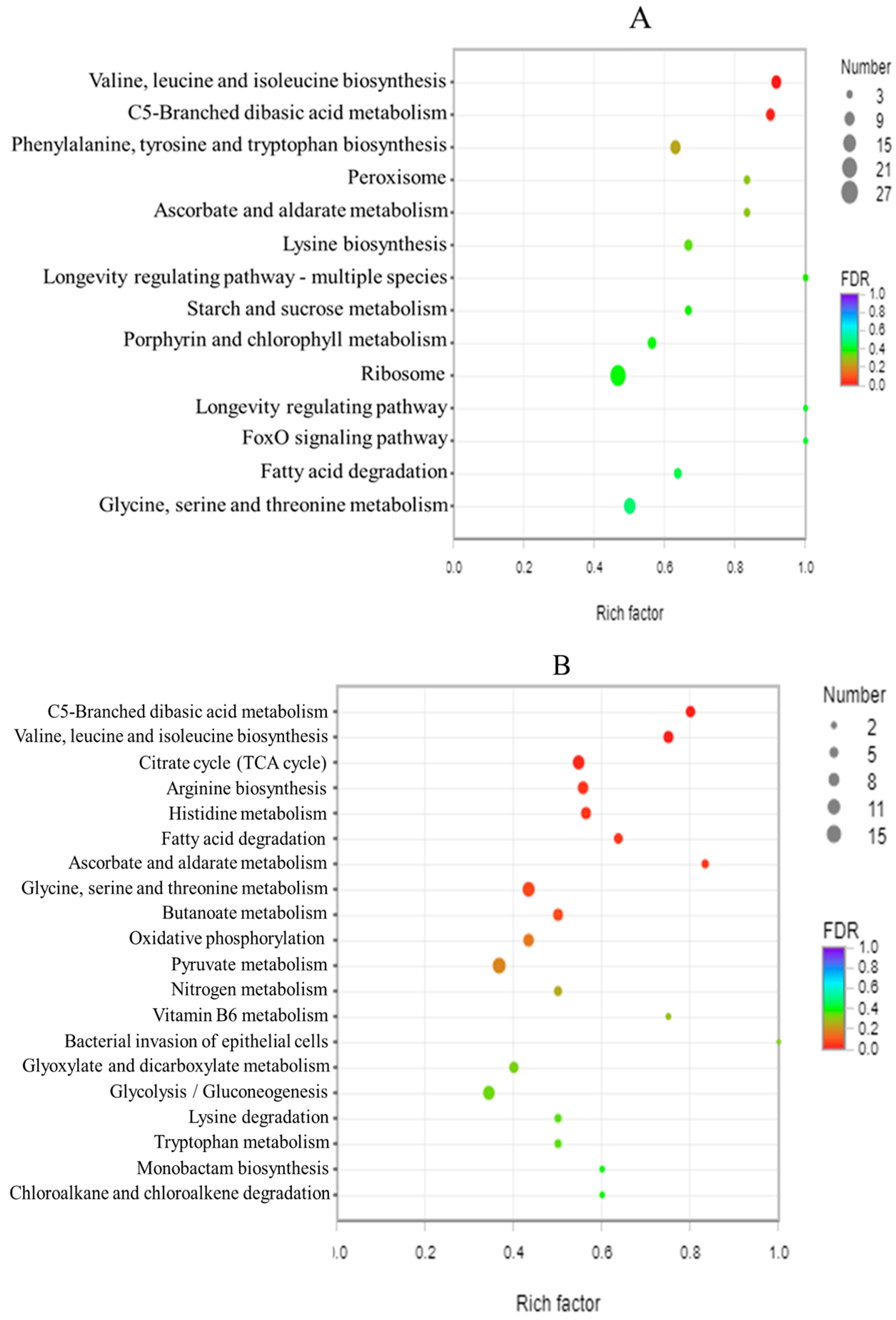
3.8. GO Enrichment Analysis and KEGG Enrichment Analysis
3.9. qRT–PCR Validation
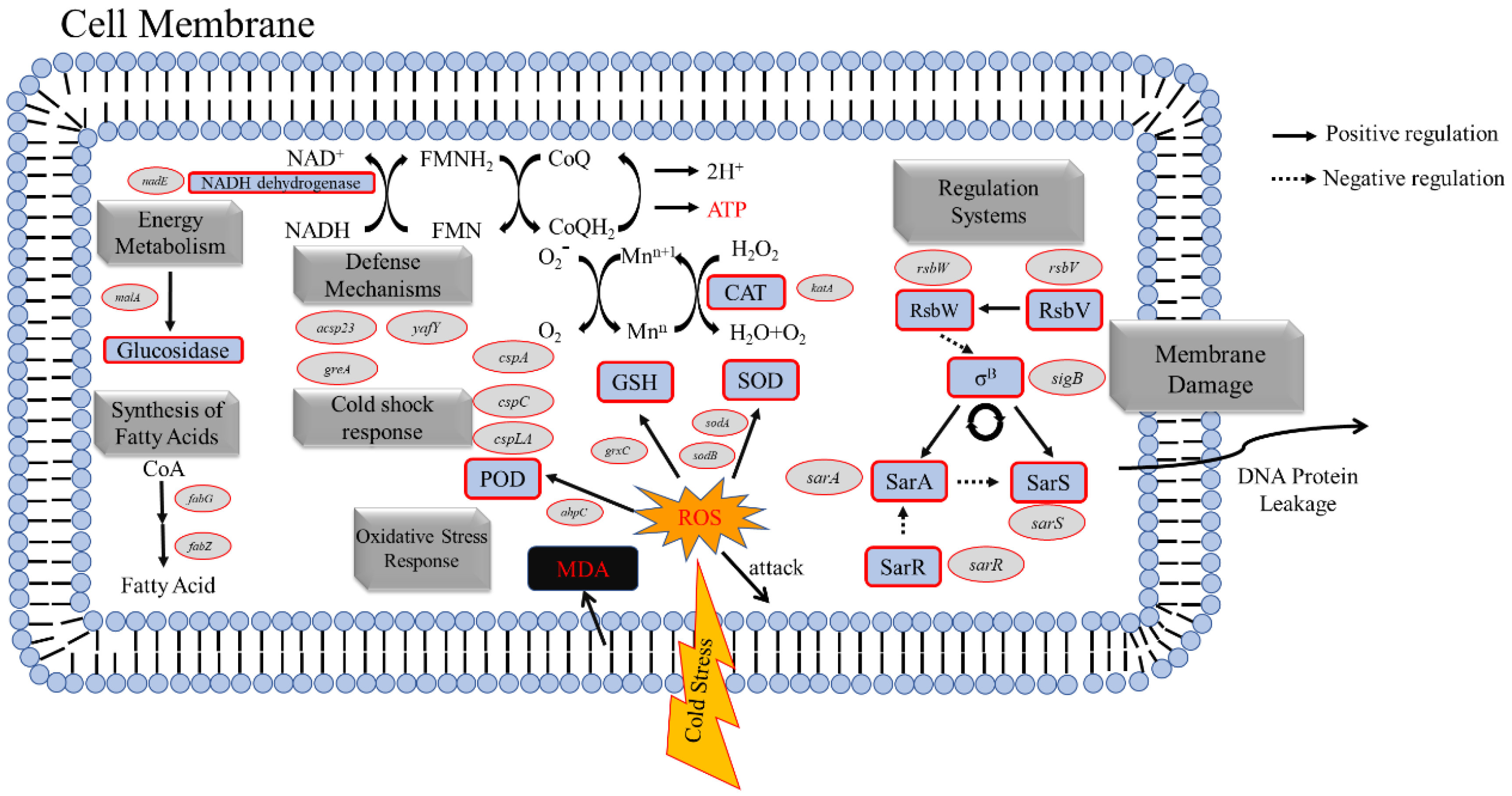
3.10. Differentially Expressed Genes (DEGs) in Response to Low-Temperature Treatment
3.10.1. DEGs Related to Fatty Acid Synthesis in The Cell Membrane of S. aureus
3.10.2. DEGs Related to Oxidative Stress in S. aureus
3.10.3. DEGs Related to Energy Metabolism in S. aureus
3.10.4. DEGs Related to The Regulatory System of S. aureus
3.10.5. DEGs Related to The Defense Systems of S. aureus
3.10.6. DEGs Related to Cold Shock Stress in S. aureus
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zhao, Y.; Xia, D.; Ma, P.; Gao, X.; Kang, W.; Wei, J. Advances in the detection of virulence genes of Staphylococcus aureus originate from food. Food Sci. Hum. Wellness 2020, 9, 40–44. [Google Scholar] [CrossRef]
- Liu, J.; Wu, H.; Ao, X.; Hao, H.; Bi, J.; Hou, H.; Zhang, G. Characterization of the Inclusion Complexes of Isothiocyanates with γ-Cyclodextrin for Improvement of Antibacterial Activities against Staphylococcus aureus. Foods 2022, 11, 60. [Google Scholar] [CrossRef]
- Jiwon, O.; Hoikyung, K.; Beuchat, L.R.; Ryu, J.-H. Inhibition of Staphylococcus aureus on a laboratory medium and black peppercorns by individual and combinations of essential oil vapors. Food Control 2022, 132, 108487. [Google Scholar]
- Zhao, J.; Qian, J.; Zhuang, H.; Luo, J.; Huang, M.; Yan, W.; Zhang, J. Effect of Plasma-Activated Solution Treatment on Cell Biology of Staphylococcus aureus and Quality of Fresh Lettuces. Foods 2021, 10, 2976. [Google Scholar] [CrossRef] [PubMed]
- Rajkovic, A.; Jovanovic, J.; Monteiro, S.; Decleer, M.; Andjelkovic, M.; Foubert, A.; Beloglazova, N.; Tsilla, V.; Sas, B.; Madder, A. Detection of toxins involved in foodborne diseases caused by Gram-positive bacteria. Compr. Rev. Food Sci. Food Saf. 2020, 19, 1605–1657. [Google Scholar] [CrossRef]
- Saklani, P.; Lekshmi, M.; Nayak, B.B.; Kumar, S. Survival of methicillin-resistant Staphylococcus aureus in fish and shrimp under different storage conditions. J. Food Prot. 2020, 83, 844–848. [Google Scholar] [CrossRef]
- Gong, S.; Fei, P.; Sun, Q.; Guo, L.; Jiang, L.; Duo, K.; Bi, X.; Yun, X. Action mode of cranberry anthocyanin on physiological and morphological properties of Staphylococcus aureus and its application in cooked meat. Food Microbiol. 2021, 94, 103632. [Google Scholar] [CrossRef]
- Li, Q.; Yu, S.; Han, J.; Wu, J.; You, L.; Shi, X.; Wang, S. Synergistic antibacterial activity and mechanism of action of nisin/carvacrol combination against Staphylococcus aureus and their application in the infecting pasteurized milk. Food Chem. 2022, 1, 132009. [Google Scholar] [CrossRef]
- Yoo, J.H.; Baek, K.H.; Heo, Y.S.; Yong, H.I.; Jo, C. Synergistic bactericidal effect of clove oil and encapsulated atmospheric pressure plasma against Escherichia coli O157: H7 and Staphylococcus aureus and its mechanism of action. Food Microbiol. 2021, 93, 103611. [Google Scholar] [CrossRef]
- Wang, L.; Zeng, X.; Wang, M.; Brennan, C.S.; Gong, D. Modification of membrane properties and fatty acids biosynthesis-related genes in Escherichia coli and Staphylococcus aureus: Implications for the antibacterial mechanism of naringenin. Biochim. Biophys. Acta (BBA)-Biomembr. 2018, 1860, 481–490. [Google Scholar] [CrossRef]
- Zhang, L.; Zhang, M.; Devahastin, S.; Liu, K. Fabrication of curcumin encapsulated in casein-ethyl cellulose complexes and its antibacterial activity when applied in combination with blue LED irradiation. Food Control 2022, 134, 108702. [Google Scholar] [CrossRef]
- Vaishampayan, A.; Grohmann, E. Antimicrobials Functioning through ROS-Mediated Mechanisms: Current Insights. Microorganisms 2022, 10, 61. [Google Scholar] [CrossRef] [PubMed]
- Shi, Y.; Zhang, R.; Zhu, C.; Xu, M.; Gu, Q.; Ettelaie, R.; Lin, S.; Wang, Y.; Leng, X. Antimicrobial mechanism of alkyl gallates against Escherichia coli and Staphylococcus aureus and its combined effect with electrospun nanofibers on Chinese Taihu icefish preservation. Food Chem. 2021, 346, 128949. [Google Scholar] [CrossRef] [PubMed]
- Tuchscherr, L.; Löffler, B.; Proctor, R.A. Persistence of Staphylococcus aureus: Multiple metabolic pathways impact the expression of virulence factors in Small-Colony Variants (SCVs). Front. Microbiol. 2020, 11, 1028. [Google Scholar] [CrossRef] [PubMed]
- Savijoki, K.; Miettinen, I.; Nyman, T.A.; Kortesoja, M.; Hanski, L.; Varmanen, P.; Fallarero, A. Growth mode and physiological state of cells prior to biofilm formation affect immune evasion and persistence of Staphylococcus aureus. Microorganisms 2020, 8, 106. [Google Scholar] [CrossRef]
- Xiang, Q.; Kang, C.; Zhao, D.; Niu, L.; Liu, X.; Bai, Y. Influence of organic matters on the inactivation efficacy of plasma-activated water against E. coli O157:H7 and S. aureus. Food Control 2019, 99, 28–33. [Google Scholar] [CrossRef]
- Saliba, A.E.; Santos, S.C.; Vogel, J. New RNA-seq approaches for the study of bacterial pathogens. Curr. Opin. Microbiol. 2017, 35, 78–87. [Google Scholar] [CrossRef]
- Park, J.; Jo, S.; Park, K.; Yu, H.; Bai, J.; Ryu, S.; Chang, P. Transcriptomic analysis of Staphylococcus aureus under the stress condition of antibacterial erythorbyl laurate by RNA sequencing. Food Control 2019, 96, 1–8. [Google Scholar] [CrossRef]
- Hingston, P.; Chen, J.; Dhillon, B.K.; Laing, C.; Bertelli, C.; Gannon, V.; Tasara, T.; Allen, K.; Brinkman, F.S.; Truelstrup Hansen, L. Genotypes associated with Listeria monocytogenes isolates displaying impaired or enhanced tolerances to cold, salt, acid, or desiccation stress. Front. Microbiol. 2017, 8, 369. [Google Scholar] [CrossRef]
- Haskell, K.J.; Schriever, S.R.; Fonoimoana, K.D.; Haws, B.; Hair, B.B.; Wienclaw, T.M.; Holmstead, J.G.; Barboza, A.B.; Berges, E.T.; Heaton, M.J. Antibiotic resistance is lower in Staphylococcus aureus isolated from antibiotic-free raw meat as compared to conventional raw meat. PLoS ONE 2018, 13, e0206712. [Google Scholar] [CrossRef]
- Zhang, L.; Hou, L.; Zhang, S.; Kou, X.; Li, R.; Wang, S. Mechanism of S. aureus ATCC 25923 in response to heat stress under different water activity and heating rates. Food Control 2020, 108, 106837. [Google Scholar] [CrossRef]
- Aggarwal, S.; Jena, S.; Panda, S.; Sharma, S.; Dhawan, B.; Nath, G.; Singh, N.; Nayak, K.C.; Singh, D.V. Antibiotic susceptibility, virulence pattern, and typing of Staphylococcus aureus strains isolated from variety of infections in India. Front. Microbiol. 2019, 10, 2763. [Google Scholar] [CrossRef] [PubMed]
- Qiao, J.; Zhu, M.; Lu, Z.; Lv, F.; Zhao, H.; Bie, X. The antibiotics resistance mechanism and pathogenicity of cold stressed Staphylococcus aureus. LWT 2020, 126, 109274. [Google Scholar] [CrossRef]
- Zeng, J.; Hu, Y.; Jia, T.; Zhang, R.; Su, T.; Sun, J.; Gao, H.; Li, G.; Cao, M.; Song, M. Chemoenzymatic synthesis of sialylated lactuloses and their inhibitory effects on Staphylococcus aureus. PLoS ONE 2018, 13, e0199334. [Google Scholar] [CrossRef] [PubMed]
- Bai, J.; Zhong, K.; Wu, Y.; Elena, G.; Gao, H. Antibiofilm activity of shikimic acid against Staphylococcus aureus. Food Control 2019, 95, 327–333. [Google Scholar] [CrossRef]
- Suo, B.; Yang, H.; Wang, Y.; Lv, H.; Zhen, L.; Xu, C.; Ai, Z. Comparative Proteomic and Morphological Change Analyses of Staphylococcus aureus During Resuscitation From Prolonged Freezing. Front. Microbiol. 2018, 9, 866–871. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Ben, L.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Young, M.D.; Matthew, J.W.; Smyth, G.K.; Oshlack, A. Gene ontology analysis for RNA-seq: Accounting for selection bias. Genome Biol. 2010, 11, R14. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Qiu, X.; Choudhry, A.E.; Janson, C.A.; Grooms, M.; Daines, R.A.; Lonsdale, J.T.; Khandekar, S.S. Crystal structure and substrate specificity of the beta-ketoacyl-acyl carrier protein synthase III (FabH) from Staphylococcus aureus. Protein Sci. 2010, 14, 11–20. [Google Scholar]
- Qi, M.; Liu, Y.; Shi, S.; Xian, Y.; Liu, Q.; Yan, H.; Zhang, Y.; Yuan, Y. Inhibition mechanism of high voltage prick electrostatic field (HVPEF) on Staphylococcus aureus through ROS-mediated oxidative stress. LWT 2021, 155, 112990. [Google Scholar] [CrossRef]
- Han, B.; Han, X.; Ren, M.; You, Y.; Zhan, J.; Huang, W. Antimicrobial Effects of Novel H2O2-Ag+ Complex on Membrane Damage to Staphylococcus aureus, Escherichia coli and Salmonella typhimurium The evaluation of novel H2O2-Ag+ complex. J. Food Prot. 2021, 21, 1–12. [Google Scholar]
- Ning, H.; Li, Y.; Tian, Q.; Wang, Z.; Mo, H. The apoptosis of Staphylococcus aureus induced by glycinin basic peptide through ROS oxidative stress response. LWT 2019, 99, 62–68. [Google Scholar] [CrossRef]
- Lalaouna, D.; Baude, J.; Wu, Z.; Tomasini, A.; Chicher, J.; Marzi, S.; Vandenesch, F.; Romby, P.; Caldelari, I.; Moreau, K. RsaC sRNA modulates the oxidative stress response of Staphylococcus aureus during manganese starvation. Nucleic Acids Res. 2019, 47, 9871–9887. [Google Scholar] [CrossRef] [PubMed]
- Christmas, B.A.F.; Rolfe, M.; Rose, M.; Green, J. Staphylococcus aureus adaptation to aerobic low-redox-potential environments: Implications for an intracellular lifestyle. Microbiology 2019, 165, 21–29. [Google Scholar] [CrossRef] [PubMed]
- Queiroux, C.; Bonnet, M.; Saraoui, T.; Delpech, P.; Veisseire, P.; Rifa, E.; Moussard, C.; Gagne, G.; Delbès, C.; Bornes, S. Dialogue between Staphylococcus aureus SA15 and Lactococcus garvieae strains experiencing oxidative stress. BMC Microbiol. 2018, 18, 193. [Google Scholar] [CrossRef]
- Ramón-Sierra, J.; Martínez-Guevara, J.L.; Pool-Yam, L.; Magana-Ortiz, D.; Yam-Puc, A.; Ortiz-Vázquez, E. Effects of phenolic and protein extracts from Melipona beecheii honey on pathogenic strains of Escherichia coli and Staphylococcus aureus. Food Sci. Biotechnol. 2020, 29, 1013–1021. [Google Scholar] [CrossRef]
- Liu, S.; Li, H.; Hassan, M.M.; Ali, S.; Chen, Q. SERS based artificial peroxidase enzyme regulated multiple signal amplified system for quantitative detection of foodborne pathogens. Food Control 2021, 123, 107733. [Google Scholar] [CrossRef]
- Liao, X.; Forghani, F.; Liu, D.; Ding, T. Cumulative damage by nonthermal plasma (NTP) exceeds the defense barrier of multiple antibiotic-resistant Staphylococcus aureus: A key to achieve complete inactivation. Food Qual. Saf. 2021, 5, 127–139. [Google Scholar] [CrossRef]
- Pandey, S.; Sahukhal, G.S.; Elasri, M.O. The msaABCR Operon Regulates Persister Formation by Modulating Energy Metabolism in Staphylococcus aureus. Front. Microbiol. 2021, 12, 881. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.; Kim, M. Leuconostoc mesenteroides MKSR isolated from kimchi possesses α-glucosidase inhibitory activity, antioxidant activity, and cholesterol-lowering effects. LWT 2019, 116, 108570. [Google Scholar] [CrossRef]
- Kang, J.; Jin, W.; Wang, J.; Sun, Y.; Wu, X.; Liu, L. Antibacterial and anti-biofilm activities of peppermint essential oil against Staphylococcus aureus. LWT 2019, 101, 639–645. [Google Scholar] [CrossRef]
- Qiu, Y.; Xu, D.; Xia, X.; Zhang, K.; Wang, J. Five major two components systems of Staphylococcus aureus for adaptation in diverse hostile environment. Microb. Pathog. 2021, 28, 105119. [Google Scholar] [CrossRef]
- Pannu, M.K.; Hudman, D.A.; Sargentini, N.J.; Singh, V.K. Role of SigB and staphyloxanthin in radiation survival of Staphylococcus aureus. Curr. Microbiol. 2019, 76, 70–77. [Google Scholar] [CrossRef] [PubMed]
- Roberts, C.; Anderson, K.L.; Murphy, E.; Projan, S.J. Characterizing the Effect of the Staphylococcus aureus Virulence Factor Regulator, SarA, on Log-Phase mRNA Half-Lives. J. Bacteriol. 2006, 102, 104–118. [Google Scholar] [CrossRef] [PubMed]
- Manna, A.C.; Cheung, A.L. Transcriptional regulation of the agr locus and the identification of DNA binding residues of the global regulatory protein SarR in Staphylococcus aureus. Mol. Microbiol. 2010, 60, 1289–1301. [Google Scholar] [CrossRef]
- Hsieh, H.Y.; Tseng, C.W.; Stewart, G.C. Regulation of Rot expression in Staphylococcus aureus. J. Bacteriol. 2008, 190, 546–554. [Google Scholar] [CrossRef]
- Barros, E.M.; Martin, M.J.; Selleck, E.M.; Lebreton, F.; Sampaio, J.L.M.; Gilmore, M.S. Daptomycin resistance and tolerance due to loss of function in Staphylococcus aureus dsp1 and asp23. Antimicrob. Agents Chemother. 2019, 63, e01542-18. [Google Scholar] [CrossRef]
- Shao, L.; Liu, Y.; Zhao, Y.; Zou, B.; Li, X.; Dai, R. Integrated transcriptomic and metabolomic analysis of the global response of Staphylococcus aureus to ohmic heating. Innov. Food Sci. Emerg. Technol. 2021, 74, 102870. [Google Scholar] [CrossRef]
- Rochat, T.; Delumeau, O.; Figueroa-Bossi, N.; Noirot, P.; Bossi, L.; Dervyn, E.; Bouloc, P. Tracking the elusive function of Bacillus subtilis Hfq. PLoS ONE 2015, 10, e0124977. [Google Scholar] [CrossRef] [PubMed]
- Caballero, C.J.; Menendez-Gil, P.; Catalan-Moreno, A.; Vergara-Irigaray, M.; García, B.; Segura, V.; Irurzun, N.; Villanueva, M.; Ruiz de los Mozos, I.; Solano, C.; et al. The regulon of the RNA chaperone CspA and its auto-regulation in Staphylococcus aureus. Nucleic Acids Res. 2018, 46, 1345–1361. [Google Scholar] [CrossRef] [PubMed]
- Catalan-Moreno, A.; Cela, M.; Menendez-Gil, P.; Irurzun, N.; Caballero, C.J.; Caldelari, I.; Toledo-Arana, A. RNA thermoswitches modulate Staphylococcus aureus adaptation to ambient temperatures. Nucleic Acids Res. 2021, 49, 3409–3426. [Google Scholar] [CrossRef] [PubMed]
- Alreshidi, M.M.; Dunstan, R.H.; Macdonald, M.M.; Smith, N.D.; Gottfries, J.; Roberts, T.K. Metabolomic and proteomic responses of Staphylococcus aureus to prolonged cold stress. J. Proteom. 2015, 121, 44–55. [Google Scholar] [CrossRef]
| Genes | Primer | Sequence (5′-3′) | Product Length (bp) | References |
|---|---|---|---|---|
| leuB | leuB-F | GGTGCAATCGGTGGACCTAA | 124 | this research |
| leuB-R | TAGCGCCTTTGACAACGGTA | |||
| leuD | leuD-F | TGGAAACGCTTGTCTGGTGA | 211 | this research |
| leuD-R | GTCGTGAACATGCTGCTTGG | |||
| sigB | sigB-F | TGGAGTGTACATGTTCCGAGAC | 75 | this research |
| sigB-R | AGCGGTTAGTTCATCGCTCA | |||
| sarV | sarV-F | TCATCCGTTTCAGAACGCAA | 141 | this research |
| sarV-R | TGAAGCTGAAAAGATTAGCGGT | |||
| rsbW | rsbW-F | GCGAAGGTGGCCTAGGTTTA | 125 | this research |
| rsbW-R | GCCATTATTTCGCACCTGCT | |||
| NsaR | NsaR-F | GCGCGTCATGTTAACAGCTA | 273 | this research |
| NsaR-R | ACGATTGCCAAAATTCAAGCAT | |||
| MarR | MarR-F | AATAAGGCCGCAGTAAGCCG | 166 | this research |
| MarR-R | GCGCAATATCTGTCATAATCGCA | |||
| rsbU | rsbU-F | GCTGGTCATGAGCCTGGATA | 171 | this research |
| rsbU-R | AGCTTCAGTCACACCATCCG | |||
| 16s rDNA | 16s rDNA-F | CGGTGAATACGTTCYCGG | 124 | [17] |
| 16s rDNA-R | GGWTACCTTGTTACGACTT |
| Gene ID | Symbol | S. aureus BB-11 | p-Value | S. aureus BA-26 | p-Value | Description |
|---|---|---|---|---|---|---|
| Fold Change/Log2(FC) | Fold Change/Log2(FC) | |||||
| Fatty acid biosynthesis in membrane | ||||||
| E3306_06220 | fabG | 1.056 | 0.001266 | 0.46 | 0.032878 | 3-oxoacyl-[acyl-carrier-protein] reductase |
| E3306_11095 | fabZ | −2.53 | 4.61 × 10−13 | −0.82 | 0.001912 | 3-hydroxyacyl-ACP dehydratase FabZ |
| Oxidative stress | ||||||
| E3306_08020 | sodA | −1.55 | 3.61 × 10−6 | −0.66 | 0.002897 | Superoxide dismutase |
| E3306_00485 | sodB | −1.52 | 1.07 × 10−5 | −0.73 | 0.003111 | [Mn/Fe]-Superoxide dismutase |
| E3306_06825 | katA | 1.19 | 3.74 × 10−4 | 0.71 | 0.004024 | Catalase |
| E3306_13890 | grxC | 1.35 | 3.60 × 10−4 | 0.24 | 0.399629 | Reduced Glutathione |
| E3306_01835 | ahpC | 1.02 | 0.011891 | 0.24 | 0.223756 | Peroxidase |
| Cold shock response | ||||||
| E3306_07190 | cspA | −1.78 | 3.08 × 10−8 | −1.25 | 5.58 × 10−6 | Cold-shock protein (CspA) |
| E3306_14325 | cspLA | −1.21 | 0.063056 | −3.24 | 6.03 × 10−23 | Cold-shock protein (CspLA) |
| E3306_04085 | cspC | −0.66 | 0.284248 | −2.27 | 8.17 × 10−18 | Cold-shock protein (CspC) |
| Energy Metabolism | ||||||
| E3306_02110 | nadE | 5.33 | 4.47 × 10−64 | 2.74 | 1.68 × 10−9 | NADH dehydrogenase |
| E3306_07745 | malA | 3.34 | 2.21 × 10−21 | −1.39 | 1.37 × 10−5 | α-glucosidase |
| Regulation System | ||||||
| E3306_10900 | sigB | −1.57 | 2.20 × 10−8 | −0.65 | 0.001570 | RNA polymerase sigma factor SigB |
| E3306_10910 | rsbV | −1.86 | 2.20 × 10−12 | −0.38 | 0.120798 | STAS domain-containing protein(RsbV) |
| E3306_10905 | rsbW | −1.64 | 2.42 × 10−7 | −1.05 | 0.006128 | anti-sigma B factor RsbW |
| E3306_03140 | sarA | 2.89 | 2.40 × 10−19 | 2.44 | 6.38 × 10−6 | transcriptional regulator(SarA) |
| E3306_00370 | sarS | −2.81 | 2.99 × 10−20 | −1.83 | 9.22 × 10−4 | HTH-type transcriptional regulator SarS |
| E3306_12130 | sarR | 0.24 | 0.675713 | −0.49 | 0.071711 | HTH-type transcriptional regulator SarR |
| Defense Mechanism | ||||||
| E3306_07880 | asp23 | 1.02 | 1.26 × 10−4 | 0.25 | 0.221559 | Asp23/Gls24 family envelope stress response protein |
| E3306_08310 | greA | 1.49 | 1.03 × 10−9 | 0.53 | 0.048271 | transcription elongation factor GreA |
| E3306_12230 | yafY | 1.43 | 6.30 × 10−5 | 0.71 | 9.07 × 10−5 | YafY family transcriptional regulator |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Suo, B.; Guan, P.; Dong, Z.; Zeng, Y.; Fan, S.; Fan, H.; Huang, Z.; Ai, Z. Comparative Transcriptomic Analysis of Staphylococcus aureus Reveals the Genes Involved in Survival at Low Temperature. Foods 2022, 11, 996. https://doi.org/10.3390/foods11070996
Suo B, Guan P, Dong Z, Zeng Y, Fan S, Fan H, Huang Z, Ai Z. Comparative Transcriptomic Analysis of Staphylococcus aureus Reveals the Genes Involved in Survival at Low Temperature. Foods. 2022; 11(7):996. https://doi.org/10.3390/foods11070996
Chicago/Turabian StyleSuo, Biao, Peng Guan, Zijie Dong, Yun Zeng, Shijia Fan, Huiping Fan, Zhongmin Huang, and Zhilu Ai. 2022. "Comparative Transcriptomic Analysis of Staphylococcus aureus Reveals the Genes Involved in Survival at Low Temperature" Foods 11, no. 7: 996. https://doi.org/10.3390/foods11070996
APA StyleSuo, B., Guan, P., Dong, Z., Zeng, Y., Fan, S., Fan, H., Huang, Z., & Ai, Z. (2022). Comparative Transcriptomic Analysis of Staphylococcus aureus Reveals the Genes Involved in Survival at Low Temperature. Foods, 11(7), 996. https://doi.org/10.3390/foods11070996






