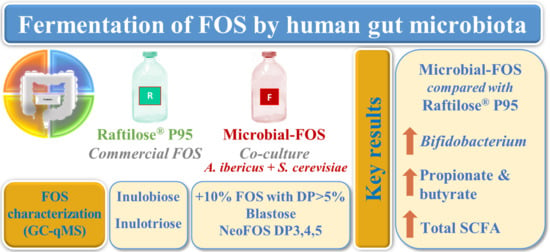
Evaluation of Microbial-Fructo-Oligosaccharides Metabolism by Human Gut Microbiota Fermentation as Compared to Commercial Inulin-Derived Oligosaccharides
Abstract
:1. Introduction
2. Materials and Methods
2.1. FOS Samples
2.2. Microbial-FOS Chemical Characterization
2.3. In Vitro Fecal Fermentation Assays
2.3.1. Fecal Inocula Collection and Preparation
2.3.2. Fermentation Media Preparation
2.3.3. Fecal Fermentations
2.4. Bacterial Population Analysis
2.4.1. DNA Extraction
2.4.2. Quantitative Real-Time Polymerase Chain Reaction
2.5. Carbohydrate and Fermentation Product Analysis
2.6. Statistical Analysis
3. Results
3.1. Microbial-FOS Chemical Characterization
3.2. Fermentation with Human Gut Microbiota
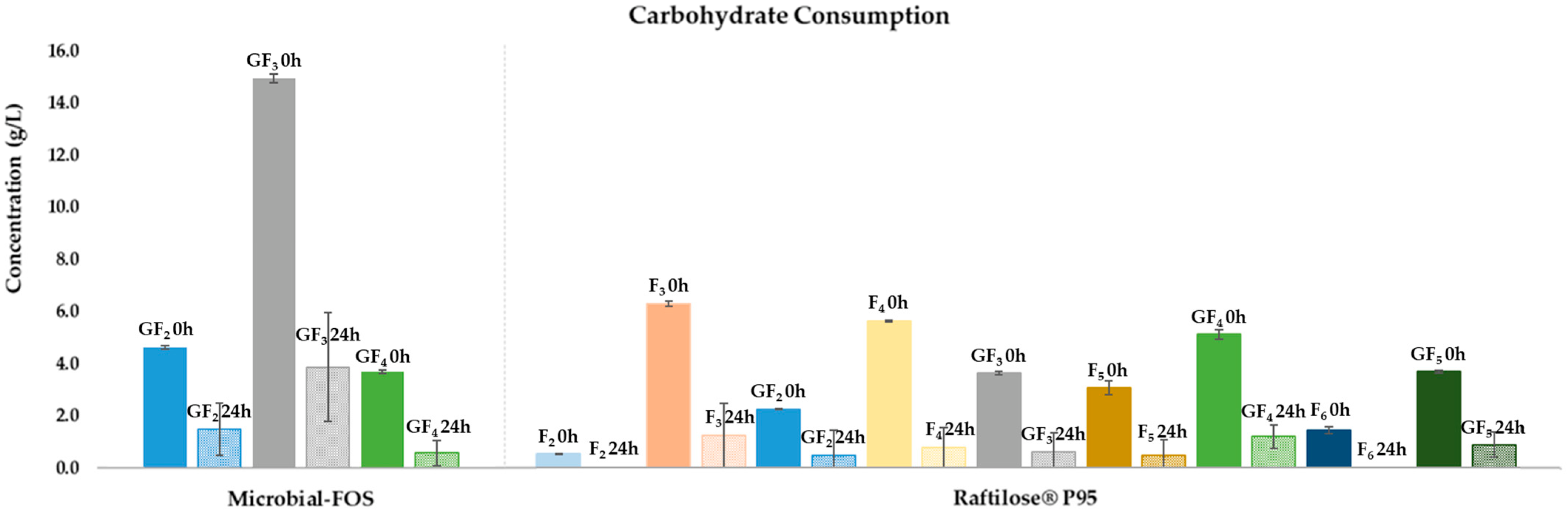
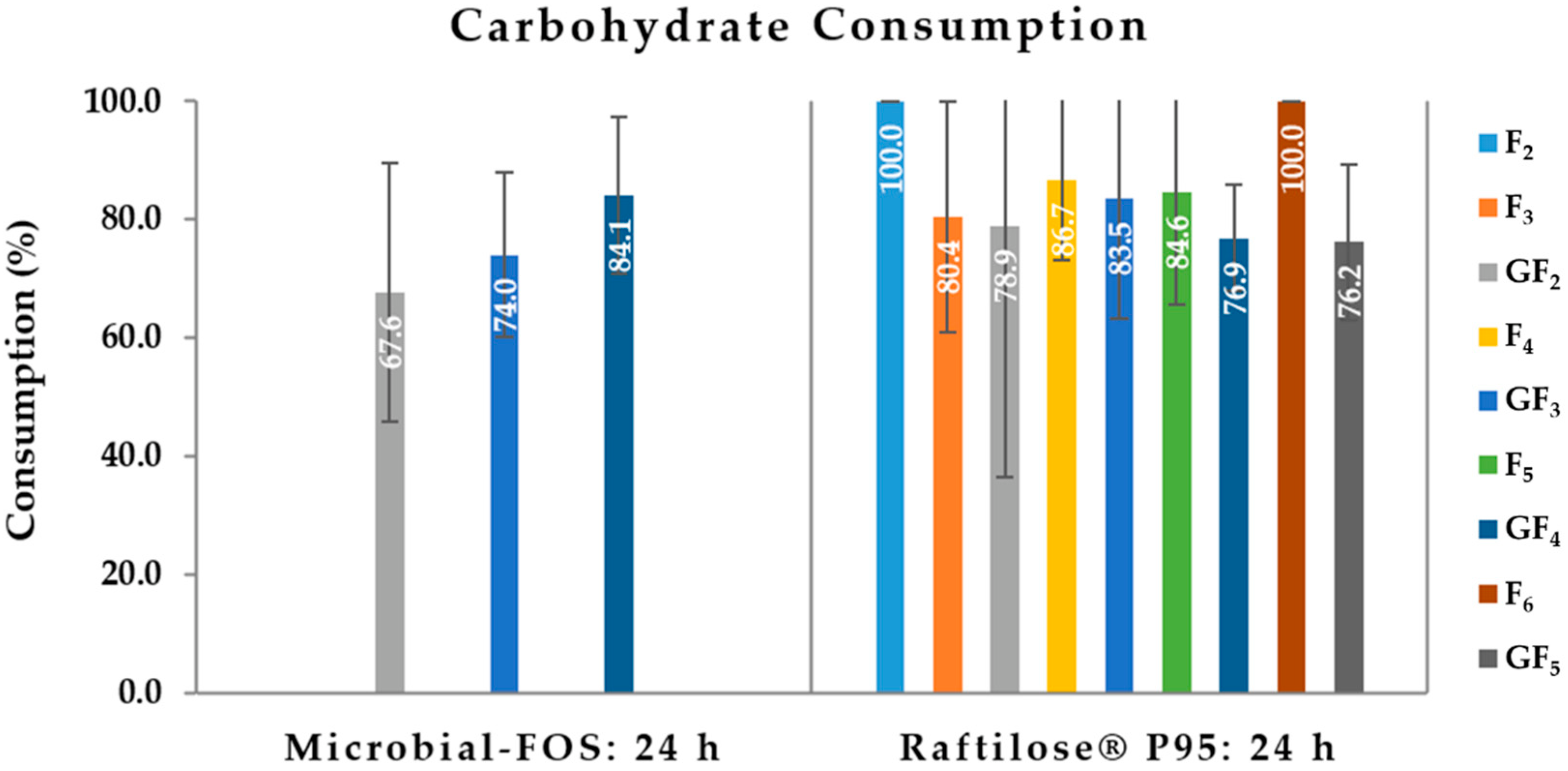
3.2.1. Carbohydrate Consumption
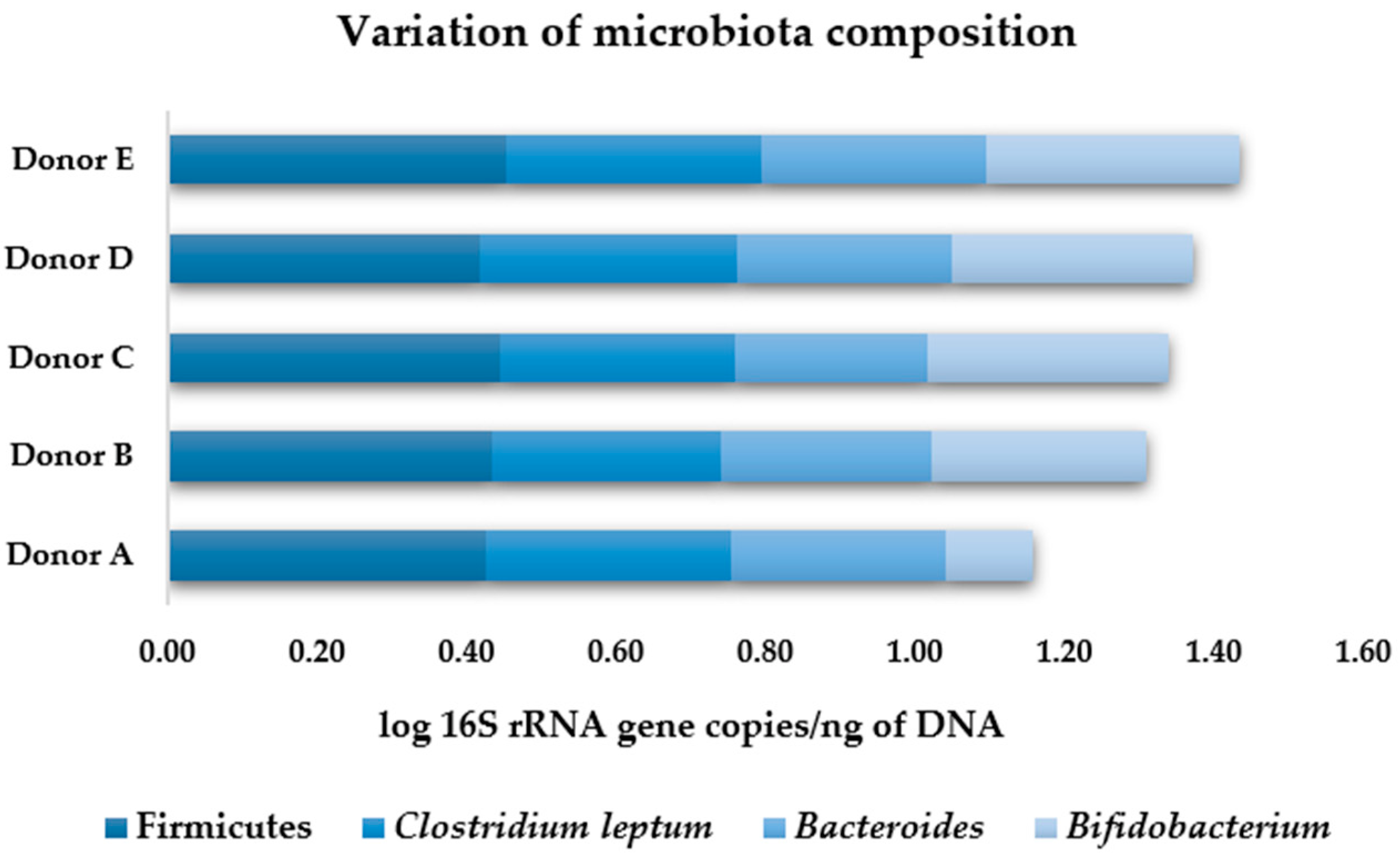
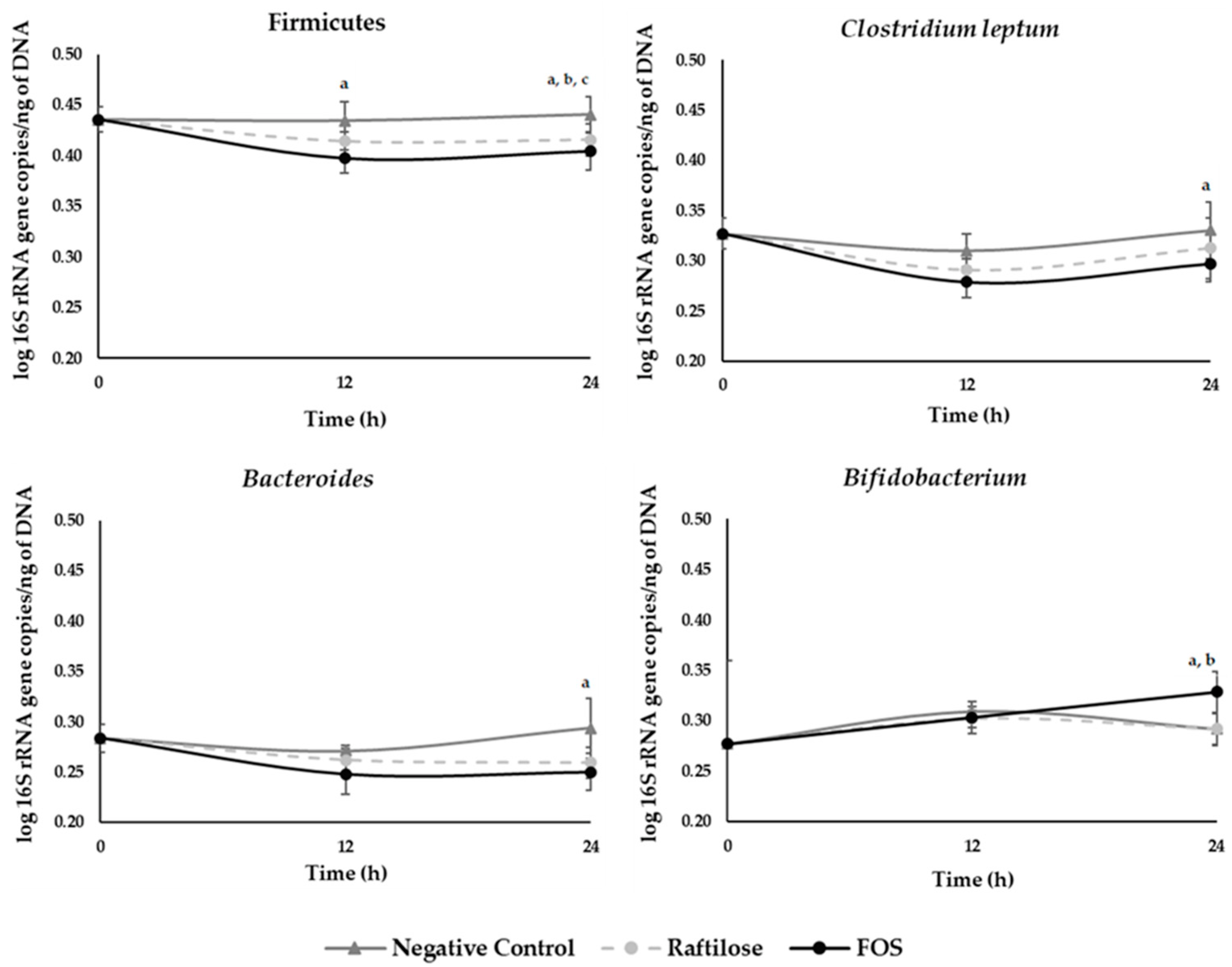
3.2.2. Human Gut Microbiota Composition
3.2.3. Short-Chain Fatty Acid Production
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Gibson, G.R.; Hutkins, R.; Sanders, M.E.; Prescott, S.L.; Reimer, R.A.; Salminen, S.J.; Scott, K.; Stanton, C.; Swanson, K.S.; Cani, P.D.; et al. Expert Consensus Document: The International Scientific Association for Probiotics and Prebiotics (ISAPP) Consensus Statement on the Definition and Scope of Prebiotics. Nat. Rev. Gastroenterol. Hepatol. 2017, 14, 491–502. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Roupar, D.; Berni, P.; Martins, J.T.; Caetano, A.C.; Teixeira, J.A.; Nobre, C. Bioengineering Approaches to Simulate Human Colon Microbiome Ecosystem. Trends Food Sci. Technol. 2021, 112, 808–822. [Google Scholar] [CrossRef]
- Pompei, A.; Cordisco, L.; Raimondi, S.; Amaretti, A.; Pagnoni, U.M.; Matteuzzi, D.; Rossi, M. In Vitro Comparison of the Prebiotic Effects of Two Inulin-Type Fructans. Anaerobe 2008, 14, 280–286. [Google Scholar] [CrossRef] [PubMed]
- Lesmes, U.; Beards, E.J.; Gibson, G.R.; Tuohy, K.M.; Shimoni, E. Effects of Resistant Starch Type III Polymorphs on Human Colon Microbiota and Short Chain Fatty Acids in Human Gut Models. J. Agric. Food Chem. 2008, 56, 5415–5421. [Google Scholar] [CrossRef]
- Li, W.; Wang, K.; Sun, Y.; Ye, H.; Hu, B.; Zeng, X. Influences of Structures of Galactooligosaccharides and Fructooligosaccharides on the Fermentation In Vitro by Human Intestinal Microbiota. J. Funct. Foods 2015, 13, 158–168. [Google Scholar] [CrossRef]
- Reis, S.F.; Gullón, B.; Gullón, P.; Ferreira, S.; Maia, C.J.; Alonso, J.L.; Domingues, F.C.; Abu-Ghannam, N. Evaluation of the Prebiotic Potential of Arabinoxylans from Brewer’s Spent Grain. Appl. Microbiol. Biotechnol. 2014, 98, 9365–9373. [Google Scholar] [CrossRef]
- Macfarlane, G.T.; Macfarlane, S. Models for Intestinal Fermentation: Association between Food Components, Delivery Systems, Bioavailability and Functional Interactions in the Gut. Curr. Opin. Biotechnol. 2007, 18, 156–162. [Google Scholar] [CrossRef]
- Gumienna, M.; Lasik, M.; Czarnecki, Z. Bioconversion of Grape and Chokeberry Wine Polyphenols during Simulated Gastrointestinal In Vitro Digestion. Int. J. Food Sci. Nutr. 2011, 62, 226–233. [Google Scholar] [CrossRef]
- Sarbini, S.R.; Rastall, R. Prebiotics: Metabolism, Structure, and Function. Funct Food Rev. 2011, 3, 93–106. [Google Scholar] [CrossRef]
- Nobre, C.; Cerqueira, M.Â.; Rodrigues, L.R.; Vicente, A.A.; Teixeira, J.A. Production and Extraction of Polysaccharides and Oligosaccharides and Their Use as New Food Additives. In Industrial Biorefineries and White Biotechnology; Elsevier: Amsterdam, The Netherlands, 2015; pp. 653–679. [Google Scholar] [CrossRef]
- Castro, C.C.; Nobre, C.; De Weireld, G.; Hantson, A.L. Microbial Co-Culturing Strategies for Fructo-Oligosaccharide Production. N. Biotechnol. 2019, 51, 1–7. [Google Scholar] [CrossRef] [Green Version]
- Nobre, C.; Alves Filho, E.G.; Fernandes, F.A.N.; Brito, E.S.; Rodrigues, S.; Teixeira, J.A.; Rodrigues, L.R. Production of Fructo-Oligosaccharides by Aspergillus Ibericus and Their Chemical Characterization. LWT—Food Sci. Technol. 2018, 89, 58–64. [Google Scholar] [CrossRef] [Green Version]
- Nobre, C.; do Nascimento, A.K.C.; Silva, S.P.; Coelho, E.; Coimbra, M.A.; Cavalcanti, M.T.H.; Teixeira, J.A.; Porto, A.L.F. Process Development for the Production of Prebiotic Fructo-Oligosaccharides by Penicillium Citreonigrum. Bioresour. Technol. 2019, 282, 464–474. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sánchez-Martínez, M.J.; Soto-Jover, S.; Antolinos, V.; Martínez-Hernández, G.B.; López-Gómez, A. Manufacturing of Short-Chain Fructooligosaccharides: From Laboratory to Industrial Scale. Food Eng. Rev. 2020, 12, 149–172. [Google Scholar] [CrossRef]
- Scott, K.P.; Grimaldi, R.; Cunningham, M.; Sarbini, S.R.; Wijeyesekera, A.; Tang, M.L.K.; Lee, J.C.Y.; Yau, Y.F.; Ansell, J.; Theis, S.; et al. Developments in Understanding and Applying Prebiotics in Research and Practice—An ISAPP Conference Paper. J. Appl. Microbiol. 2020, 128, 934–949. [Google Scholar] [CrossRef]
- Ríos-Covián, D.; Ruas-Madiedo, P.; Margolles, A.; Gueimonde, M.; De los Reyes-Gavilán, C.G.; Salazar, N. Intestinal Short Chain Fatty Acids and Their Link with Diet and Human Health. Front. Microbiol. 2016, 7, 1–9. [Google Scholar] [CrossRef] [Green Version]
- Slavin, J. Fiber and Prebiotics: Mechanisms and Health Benefits. Nutrients 2013, 5, 1417–1435. [Google Scholar] [CrossRef] [Green Version]
- Stojanov, S.; Berlec, A.; Štrukelj, B. The Influence of Probiotics on the Firmicutes/Bacteroidetes Ratio in the Treatment of Obesity and Inflammatory Bowel Disease. Microorganisms 2020, 8, 1–16. [Google Scholar] [CrossRef]
- Bull, M.J.; Plummer, N.T. Part 1: The Human Gut Microbiome in Health and Disease. Integr. Med. A Clin. J. 2014, 13, 17–22. [Google Scholar] [CrossRef]
- Roberfroid, M.B. Inulin-Type Fructans: Functional Food Ingredients. J. Nutr. 2007, 137, 2493S–2502S. [Google Scholar] [CrossRef] [Green Version]
- Bajury, D.M.; Nashri, S.M.; King Jie Hung, P.; Sarbini, S.R. Evaluation of Potential Prebiotics: A Review. Food Rev. Int. 2018, 34, 639–664. [Google Scholar] [CrossRef]
- Flint, H.J.; Duncan, S.H.; Scott, K.P.; Louis, P. Links between Diet, Gut Microbiota Composition and Gut Metabolism. Proc. Nutr. Soc. 2015, 74, 13–22. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nobre, C.; Sousa, S.C.; Silva, S.P.; Pinheiro, A.C.; Coelho, E.; Vicente, A.A.; Gomes, A.M.P.; Coimbra, M.A.; Teixeira, J.A.; Rodrigues, L.R. In Vitro Digestibility and Fermentability of Fructo-Oligosaccharides Produced by Aspergillus Ibericus. J. Funct. Foods 2018, 46, 278–287. [Google Scholar] [CrossRef] [Green Version]
- Nobre, C.; Gonçalves, D.A.; Teixeira, J.A.; Rodrigues, L.R. One-Step Co-Culture Fermentation Strategy to Produce High-Content Fructo-Oligosaccharides. Carbohydr. Polym. 2018, 201, 31–38. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nobre, C.; Teixeira, J.A.; Rodrigues, L.R. Fructo-Oligosaccharides Purification from a Fermentative Broth Using an Activated Charcoal Column. N. Biotechnol. 2012, 29, 395–401. [Google Scholar] [CrossRef] [Green Version]
- Silva, S.P.; Moreira, A.S.P.; Domingues, M.D.R.M.; Evtuguin, D.V.; Coelho, E.; Coimbra, M.A. Contribution of Non-Enzymatic Transglycosylation Reactions to the Honey Oligosaccharides Origin and Diversity. Pure Appl. Chem. 2019, 91, 1231–1242. [Google Scholar] [CrossRef]
- Hartemink, R.; Schoustra, S.E.; Rombouts, F.M. Degradation of Guar Gum by Intestinal Bacteria. Biosci. Microflora 1999, 18, 17–25. [Google Scholar] [CrossRef] [Green Version]
- Madureira, A.R.; Campos, D.; Gullon, B.; Marques, C.; Rodríguez-Alcalá, L.M.; Calhau, C.; Alonso, J.L.; Sarmento, B.; Gomes, A.M.; Pintado, M. Fermentation of Bioactive Solid Lipid Nanoparticles by Human Gut Microflora. Food Funct. 2016, 7, 516–529. [Google Scholar] [CrossRef]
- Marques, C.; Meireles, M.; Norberto, S.; Leite, J.; Freitas, J.; Pestana, D.; Faria, A.; Calhau, C. High-Fat Diet-Induced Obesity Rat Model: A Comparison between Wistar and Sprague-Dawley Rat. Adipocyte 2016, 5, 11–21. [Google Scholar] [CrossRef] [Green Version]
- Magalhães, C.P.; Ribeiro, J.A.; Guedes, A.P.; Arantes, A.L.; Sousa, D.Z.; Stams, A.J.M.; Alves, M.M.; Cavaleiro, A.J. Co-Cultivation of Thermoanaerobacter Strains with a Methanogenic Partner Enhances Glycerol Conversion. Microb. Biotechnol. 2020, 13, 962–973. [Google Scholar] [CrossRef]
- Huebner, J.; Wehling, R.L.; Hutkins, R.W. Functional Activity of Commercial Prebiotics. Int. Dairy J. 2007, 17, 770–775. [Google Scholar] [CrossRef]
- Durieux, A.; Fougnies, C.; Jacobs, H.; Simon, J.P. Metabolism of Chicory Fructooligosaccharides by Bifidobacteria. Biotechnol. Lett. 2001, 23, 1523–1527. [Google Scholar] [CrossRef]
- Park, M.C.; Lim, J.S.; Kim, J.C.; Park, S.W.; Kim, S.W. Continuous Production of Neo-Fructooligosaccharides by Immobilization of Whole Cells of Penicillium Citrinum. Biotechnol. Lett. 2005, 27, 127–130. [Google Scholar] [CrossRef] [PubMed]
- Ning, Y.; Wang, J.; Chen, J.; Yang, N.; Jin, Z.; Xu, X. Production of Neo-Fructooligosaccharides Using Free-Whole-Cell Biotransformation by Xanthophyllomyces Dendrorhous. Bioresour. Technol. 2010, 101, 7472–7478. [Google Scholar] [CrossRef] [PubMed]
- Gallego-Lobillo, P.; Ferreira-Lazarte, A.; Hernández-Hernández, O.; Montilla, A.; Villamiel, M. Evaluation of the Impact of a Rat Small Intestinal Extract on the Digestion of Four Different Functional Fibers. Food Funct. 2020, 11, 4081–4089. [Google Scholar] [CrossRef]
- Kaplan, H.; Hutkins, R.W. Fermentation of Fructooligosaccharides by Lactic Acid Bacteria and Bifidobacteria. Appl. Environ. Microbiol. 2000, 66, 2682–2684. [Google Scholar] [CrossRef] [Green Version]
- Endo, H.; Tamura, K.; Fukasawa, T.; Kanegae, M.; Koga, J. Comparison of Fructooligosaccharide Utilization by Lactobacillus and Bacteroides Species. Biosci. Biotechnol. Biochem. 2012, 76, 176–179. [Google Scholar] [CrossRef] [Green Version]
- Holscher, H.D. Dietary Fiber and Prebiotics and the Gastrointestinal Microbiota. Gut Microbes 2017, 8, 172–184. [Google Scholar] [CrossRef]
- Rossi, M.; Corradini, C.; Amaretti, A.; Nicolini, M.; Pompei, A.; Zanoni, S.; Matteuzzi, D. Fermentation of Fructooligosaccharides and Inulin by Bifidobacteria: A Comparative Study of Pure and Fecal Cultures. Appl. Environ. Microbiol. 2005, 71, 6150–6158. [Google Scholar] [CrossRef] [Green Version]
- Biedrzycka, E.; Bielecka, M. Prebiotic Effectiveness of Fructans of Different Degrees of Polymerization. Trends Food Sci. Technol. 2004, 15, 170–175. [Google Scholar] [CrossRef]
- Hernot, D.C.; Boileau, T.W.; Bauer, L.L.; Middelbos, I.S.; Murphy, M.R.; Swanson, K.S.; Fahey, G.C. In Vitro Fermentation Profiles, Gas Production Rates, and Microbiota Modulation as Affected by Certain Fructans, Galactooligosaccharides, and Polydextrose. J. Agric. Food Chem. 2009, 57, 1354–1361. [Google Scholar] [CrossRef]
- Moon, J.S.; Li, L.; Bang, J.; Han, N.S. Application of In Vitro Gut Fermentation Models to Food Components: A Review. Food Sci. Biotechnol. 2016, 25, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Hasan, N.; Yang, H. Factors Affecting the Composition of the Gut Microbiota, and Its Modulation. PeerJ 2019, 2019, 1–31. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Arboleya, S.; Watkins, C.; Stanton, C.; Ross, R.P. Gut Bifidobacteria Populations in Human Health and Aging. Front. Microbiol. 2016, 7, 1–9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bertranda, R.L. Lag Phase Is a Dynamic, Organized, Adaptive, and Evolvable Period That Prepares Bacteria for Cell Division. J. Bacteriol. 2019, 201, e00697-00718. [Google Scholar] [CrossRef]
- Kilian, S.; Kritzinger, S.; Rycroft, C.; Gibson, G.; Du Preez, J. The Effects of the Novel Bifidogenic Trisaccharide, Neokestose, on the Human Colonic Microbiota. World J. Microbiol. Biotechnol. 2002, 18, 637–644. [Google Scholar] [CrossRef]
- Grizard, D.; Barthomeuf, C. Enzymatic Synthesis and Structure Determination of Neo-FOS. Food Biotechnol. 1999, 13, 93–105. [Google Scholar] [CrossRef]
- Marx, S.P.; Winkler, S.; Hartmeier, W. Metabolization of β-(2,6)-Linked Fructose-Oligosaccharides by Different Bifidobacteria. FEMS Microbiol Lett 2000, 182, 163–169. [Google Scholar] [CrossRef]
- Porras-Domínguez, J.R.; Ávila-Fernández, Á.; Rodríguez-Alegría, M.E.; Miranda-Molina, A.; Escalante, A.; González-Cervantes, R.; Olvera, C.; López Munguía, A. Levan-Type FOS Production Using a Bacillus Licheniformis Endolevanase. Process Biochem. 2014, 49, 783–790. [Google Scholar] [CrossRef]
- Zambelli, P.; Tamborini, L.; Cazzamalli, S.; Pinto, A.; Arioli, S.; Balzaretti, S.; Plou, F.J.; Fernandez-Arrojo, L.; Molinari, F.; Conti, P.; et al. An Efficient Continuous Flow Process for the Synthesis of a Non-Conventional Mixture of Fructooligosaccharides. Food Chem. 2016, 190, 607–613. [Google Scholar] [CrossRef]
- Lim, J.S.; Lee, J.H.; Kang, S.W.; Park, S.W.; Kim, S.W. Studies on Production and Physical Properties of Neo-FOS Produced by Co-Immobilized Penicillium Citrinum and Neo-Fructosyltransferase. Eur. Food Res. Technol. 2007, 225, 457–462. [Google Scholar] [CrossRef]
- Liu, C.; Kolida, S.; Charalampopoulos, D.; Rastall, R.A. An Evaluation of the Prebiotic Potential of Microbial Levans from Erwinia Sp. 10119. J. Funct. Foods 2020, 64, 103668. [Google Scholar] [CrossRef]
- Rivière, A.; Selak, M.; Lantin, D.; Leroy, F.; De Vuyst, L. Bifidobacteria and Butyrate-Producing Colon Bacteria: Importance and Strategies for Their Stimulation in the Human Gut. Front. Microbiol. 2016, 7, 1–21. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jang, K.-H.; Kang, S.-A.; Cho, Y.-H.; Kim, Y.-Y.; Lee, Y.-J.; Hong, K.-H.; Seong, K.-H.; Kim, S.-H.; Kim, C.-H.; Rhee, S.-K.; et al. Prebiotic Properties of Levan in Rats. J. Microbiol. Biotechnol. 2002, 13, 348–353. [Google Scholar]
- Moens, F.; Weckx, S.; De Vuyst, L. Bifidobacterial Inulin-Type Fructan Degradation Capacity Determines Cross-Feeding Interactions between Bifidobacteria and Faecalibacterium Prausnitzii. Int. J. Food Microbiol. 2016, 231, 76–85. [Google Scholar] [CrossRef] [PubMed]
- Adamberg, K.; Tomson, K.; Talve, T.; Pudova, K.; Puurand, M.; Visnapuu, T.; Alamäe, T.; Adamberg, S. Levan Enhances Associated Growth of Bacteroides, Escherichia, Streptococcus and Faecalibacterium in Fecal Microbiota. PLoS ONE 2015, 10, 1–18. [Google Scholar] [CrossRef] [Green Version]
| Assignment | Microbial-FOS Mixture (mg/g) | RSD (%) | |
|---|---|---|---|
| DP 1 | Fructose | 27.4 | 1 |
| DP 2 | Sucrose | 32.2 | 16 |
| DP 3 | 1-Kestose | 204.6 | 13 |
| Neokestose | 8.3 | 8 | |
| DP 4 | Nystose | 495.8 | 18 |
| 1F,6G-Di-β-d-fructofuranosylsucrose or 6G(1-β-d-fructofuranosyl)2-sucrose | 27.0 | 17 | |
| DP 5 | 1F-β-d-Fructofuranosylnystose | 76.2 | 17 |
| 1F-(1-β-d-fructofuranosyl)2-6G-β-d-fructofuranosylsucrose or 6G(1-β-d-fructofuranosyl)3-sucrose or 1F-1-β-d-fructofuranosyl-6G-(1-β-d-fructofuranosyl)2-sucrose | <LOQ | ||
| 1F-(1-β-d-fructofuranosyl)2-6G-β-d-fructofuranosylsucrose or 6G(1-β-d-fructofuranosyl)3-sucrose or 1F-1-β-d-fructofuranosyl-6G-(1-β-d-fructofuranosyl)2-sucrose | <LOQ | ||
| TOTAL | 871.5 | ||
| SCFA (g/L) | Time (h) | Negative Control | Raftilose® P95 | Microbial-FOS | Statistical Significance |
|---|---|---|---|---|---|
| Acetate | 0 | 0.05 ± 0.02 | 0.05 ± 0.02 | 0.05 ± 0.02 | |
| 12 | 0.54 ± 0.16 | 0.83 ± 0.27 | 0.85 ± 0.29 | a | |
| 24 | 0.90 ± 0.10 | 1.48 ± 0.49 | 1.25 ± 0.32 | a, c | |
| Propionate | 0 | 0.90 ± 0.19 | 0.90 ± 0.19 | 0.90 ± 0.19 | |
| 12 | 0.30 ± 0.09 | 1.03 ± 0.13 | 1.25 ± 0.10 | a, b, c | |
| 24 | 0.44 ± 0.12 | 1.03 ± 0.13 | 1.30 ± 0.18 | a, c | |
| Butyrate | 0 | 0.98 ± 0.04 | 0.98 ± 0.04 | 0.98 ± 0.04 | |
| 12 | 0.88 ± 0.31 | 0.89 ± 0.44 | 1.18 ± 0.55 | b | |
| 24 | 0.73 ± 0.19 | 0.73 ± 0.12 | 1.22 ± 0.35 | a, b | |
| Lactate | 0 | 0.25 ± 0.06 | 0.25 ± 0.06 | 0.25 ± 0.06 | |
| 12 | 0.18 ± 0.04 | 0.87 ± 0.30 | 1.08 ± 0.12 | a, c | |
| 24 | 0.13 ± 0.05 | 1.33 ± 0.39 | 1.20 ± 0.42 | a, c | |
| Succinate | 0 | 1.57 ± 0.27 | 1.57 ± 0.27 | 1.57 ± 0.27 | |
| 12 | 0.53 ± 0.41 | 0.98 ± 0.40 | 1.37 ± 0.52 | ||
| 24 | 0.37 ± 0.26 | 0.66 ± 0.41 | 1.17 ± 0.51 | a, b | |
| Formate | 0 | 0.42 ± 0.09 | 0.42 ± 0.09 | 0.42 ± 0.09 | |
| 12 | 0.25 ± 0.06 | 0.31 ± 0.03 | 0.35 ± 0.05 | ||
| 24 | 0.18 ± 0.09 | 0.28 ± 0.08 | 0.35 ± 0.03 | a | |
| Total SCFAs | 0 | 3.92 ± 0.18 | 3.92 ± 0.18 | 3.92 ± 0.18 | |
| 12 | 2.50 ± 0.68 | 4.04 ± 0.48 | 5.00 ± 0.37 | a, b, c | |
| 24 | 2.62 ± 0.50 | 4.18 ± 0.76 | 5.29 ± 0.48 | a, c |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Roupar, D.; Coelho, M.C.; Gonçalves, D.A.; Silva, S.P.; Coelho, E.; Silva, S.; Coimbra, M.A.; Pintado, M.; Teixeira, J.A.; Nobre, C. Evaluation of Microbial-Fructo-Oligosaccharides Metabolism by Human Gut Microbiota Fermentation as Compared to Commercial Inulin-Derived Oligosaccharides. Foods 2022, 11, 954. https://doi.org/10.3390/foods11070954
Roupar D, Coelho MC, Gonçalves DA, Silva SP, Coelho E, Silva S, Coimbra MA, Pintado M, Teixeira JA, Nobre C. Evaluation of Microbial-Fructo-Oligosaccharides Metabolism by Human Gut Microbiota Fermentation as Compared to Commercial Inulin-Derived Oligosaccharides. Foods. 2022; 11(7):954. https://doi.org/10.3390/foods11070954
Chicago/Turabian StyleRoupar, Dalila, Marta C. Coelho, Daniela A. Gonçalves, Soraia P. Silva, Elisabete Coelho, Sara Silva, Manuel A. Coimbra, Manuela Pintado, José A. Teixeira, and Clarisse Nobre. 2022. "Evaluation of Microbial-Fructo-Oligosaccharides Metabolism by Human Gut Microbiota Fermentation as Compared to Commercial Inulin-Derived Oligosaccharides" Foods 11, no. 7: 954. https://doi.org/10.3390/foods11070954
APA StyleRoupar, D., Coelho, M. C., Gonçalves, D. A., Silva, S. P., Coelho, E., Silva, S., Coimbra, M. A., Pintado, M., Teixeira, J. A., & Nobre, C. (2022). Evaluation of Microbial-Fructo-Oligosaccharides Metabolism by Human Gut Microbiota Fermentation as Compared to Commercial Inulin-Derived Oligosaccharides. Foods, 11(7), 954. https://doi.org/10.3390/foods11070954