Supplementation of Enriched Polyunsaturated Fatty Acids and CLA Cheese on High Fat Diet: Effects on Lipid Metabolism and Fat Profile
Abstract
:1. Introduction
2. Materials and Methods
2.1. Experimental Design
2.2. Lipid Analysis
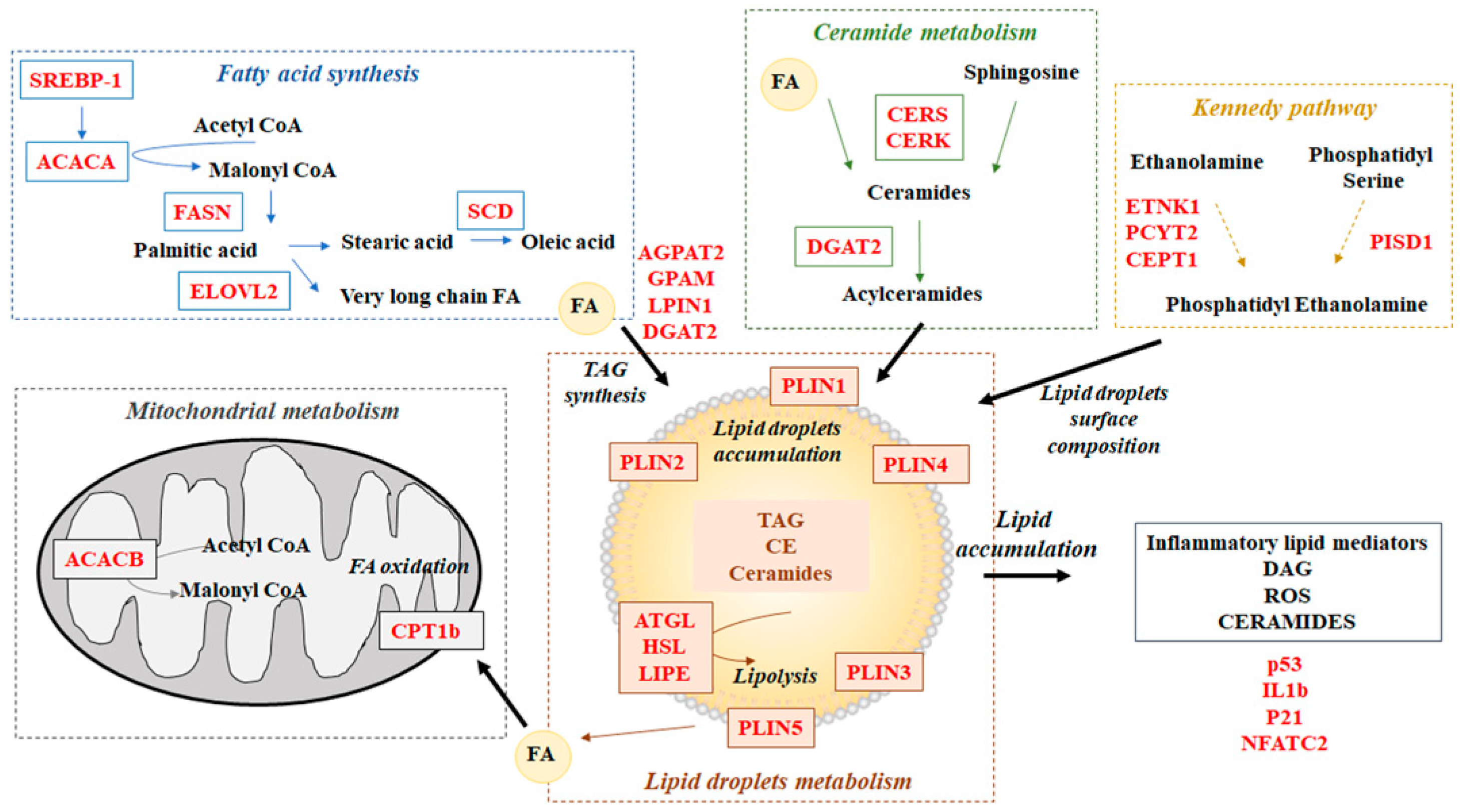
2.3. Gene Expression Analysis
2.4. Statistical Analysis
3. Results
3.1. Effect of ENR Cheese Supplementation on Lipid Fraction Composition
3.1.1. Liver
3.1.2. Brain
3.1.3. Skeletal Muscle
3.1.4. Adipose Tissue
3.2. Effect of Diet on Gene Expression
3.2.1. Liver
3.2.2. Brain
3.2.3. Skeletal Muscle
3.2.4. Adipose Tissue
4. Discussion
4.1. Lipid Profile
4.2. Gene Expression
4.2.1. Skeletal Muscle
4.2.2. Liver
4.2.3. Brain
4.2.4. Adipose Tissue
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ferrucci, L.; Cherubini, A.; Bandinelli, S.; Bartali, B.; Corsi, A.; Lauretani, F.; Martin, A.; Andres-Lacueva, C.; Senin, U.; Guralnik, J.M. Relationship of Plasma Polyunsaturated Fatty Acids to Circulating Inflammatory Markers. J. Clin. Endocrinol. Metab. 2006, 91, 439–446. [Google Scholar] [CrossRef] [PubMed]
- Conte, G.; Serra, A.; Mele, M. Dairy cow breeding and feeding on the milk fatty acid pattern. In Nutrients in Dairy and Their Implications for Health and Disease; Watson, R.R., Collier, R.J., Preedy, V.R., Eds.; Academic Press: San Diego, CA, USA, 2017; pp. 19–41. [Google Scholar]
- Pintus, S.; Murru, E.; Carta, G.; Cordeddu, L.; Batetta, B.; Accossu, S.; Pistis, D.; Uda, S.; Ghiani, M.E.; Mele, M.; et al. Sheep cheese naturally enriched in α-linolenic, conjugated linoleic and vaccenic acids improves the lipid profile and reduces anandamide in the plasma of hypercholesterolaemic subjects. Br. J. Nutr. 2013, 109, 1453–1462. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mele, M.; Contarini, G.; Cercaci, L.; Serra, A.; Buccioni, A.; Povolo, M.; Conte, G.; Funaro, A.; Banni, S.; Lercker, G.; et al. Enrichment of Pecorino cheese with conjugated linoleic acid by feeding dairy ewes with extruded linseed: Effect on fatty acid and triglycerides composition and on oxidative stability. Int. Dairy J. 2011, 21, 365–372. [Google Scholar] [CrossRef]
- Chilliard, Y.; Ferlay, A.; Mansbridge, R.M.; Doreau, M. Ruminant milk fat plasticity: Nutritional control of saturated, polyunsaturated, trans and conjugated fatty acids. Ann. Zootech. 2000, 49, 181–205. [Google Scholar] [CrossRef] [Green Version]
- Franceschi, C.; Campisi, J. Chronic Inflammation (Inflammaging) and Its Potential Contribution to Age-Associated Diseases. J. Gerontol. A Biol. Sci. Med. Sci. 2014, 69 (Suppl. S1), S4–S9. [Google Scholar] [CrossRef] [PubMed]
- Conte, M.; Martucci, M.; Sandri, M.; Franceschi, C.; Salvioli, S. The Dual Role of the Pervasive “Fattish” Tissue Remodeling with Age. Front. Endocrinol. 2019, 10, 114. [Google Scholar] [CrossRef] [Green Version]
- Gastaldelli, A.; Basta, G. Ectopic fat and cardiovascular disease: What is the link? Nutr. Metab. Cardiovasc. Dis. 2010, 20, 481–490. [Google Scholar] [CrossRef]
- Conte, M.; Franceschi, C.; Sandri, M.; Salvioli, S. Perilipin 2 and Age-Related Metabolic Diseases: A New Perspective. Trends Endocrinol. Metab. 2016, 27, 893–903. [Google Scholar] [CrossRef]
- Kuk, J.L.; Saunders, T.; Davidson, L.; Ross, R. Age-related changes in total and regional fat distribution. Ageing Res. Rev. 2009, 8, 339–348. [Google Scholar] [CrossRef]
- Hauck, A.K.; Bernlohr, D.A. Oxidative stress and lipotoxicity. J. Lipid Res. 2016, 57, 1976–1986. [Google Scholar] [CrossRef] [Green Version]
- Unger, R.H. Lipid overload and overflow: Metabolic trauma and the metabolic syndrome. Trends Endocrinol. Metab. 2003, 14, 398–403. [Google Scholar] [CrossRef] [PubMed]
- Argilés, J.M.; Campos, N.; Lopez-Pedrosa, J.M.; Rueda, R.; Rodríguez-Mañas, L. Skeletal Muscle Regulates Metabolism via Interorgan Crosstalk: Roles in Health and Disease. J. Am. Med. Dir. Assoc. 2016, 17, 789–796. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Weickert, M.O.; Pfeiffer, A.F.H. Signalling mechanisms linking hepatic glucose and lipid metabolism. Diabetologia 2006, 49, 1732–1741. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bruce, K.D.; Zsombok, A.; Eckel, R.H. Lipid Processing in the Brain: A Key Regulator of Systemic Metabolism. Front. Endocrinol. 2017, 8, 60. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rodriguez-Estrada, M.; Penazzi, G.; Caboni, M.F.; Bertacco, G.; Lercker, G. Effect of different cooking methods on some lipid and protein components of hamburgers. Meat Sci. 1997, 45, 365–375. [Google Scholar] [CrossRef]
- Christie, W.W. (Ed.) Preparation of ester derivatives of fatty acids for chromatographic analysis. In Advances in Lipid Methodology—Two; Oily Press: Dundee, UK, 1993; pp. 69–111. [Google Scholar]
- Sander, B.D.; Addis, P.B.; Park, S.W.; Smith, D.E. Quantification of Cholesterol Oxidation Products in a Variety of Foods. J. Food Prot. 1989, 52, 109–114. [Google Scholar] [CrossRef] [PubMed]
- Serra, A.; Buccioni, A.; Rodriguez-Estrada, M.; Conte, G.; Cappucci, A.; Mele, M. Fatty acid composition, oxidation status and volatile organic compounds in “Colonnata” lard from Large White or Cinta Senese pigs as affected by curing time. Meat Sci. 2014, 97, 504–512. [Google Scholar] [CrossRef] [PubMed]
- Laugerette, F.; Furet, J.-P.; Debard, C.; Daira, P.; Loizon, E.; Géloën, A.; Soulage, C.O.; Simonet, C.; Lefils-Lacourtablaise, J.; Bernoud-Hubac, N.; et al. Oil composition of high-fat diet affects metabolic inflammation differently in connection with endotoxin receptors in mice. Am. J. Physiol. Endocrinol. Metab. 2012, 302, E374–E386. [Google Scholar] [CrossRef] [Green Version]
- Kinsella, J.E.; Lokesh, B.; Stone, R.A. Dietary n-3 polyunsaturated fatty acids and amelioration of cardiovascular disease: Possible mechanisms. Am. J. Clin. Nutr. 1990, 52, 1–28. [Google Scholar] [CrossRef] [PubMed]
- Marik, P.E.; Varon, J. Omega-3 Dietary Supplements and the Risk of Cardiovascular Events: A Systematic Review. Clin. Cardiol. 2009, 32, 365–372. [Google Scholar] [CrossRef] [PubMed]
- Backes, J.; Anzalone, D.; Hilleman, D.; Catini, J. The clinical relevance of omega-3 fatty acids in the management of hypertriglyceridemia. Lipids Health Dis. 2016, 15, 118. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ibeagha-Awemu, E.M.; Li, R.; Ammah, A.A.; Dudemaine, P.-L.; Bissonnette, N.; Benchaar, C.; Zhao, X. Transcriptome adaptation of the bovine mammary gland to diets rich in unsaturated fatty acids shows greater impact of linseed oil over safflower oil on gene expression and metabolic pathways. BMC Genom. 2016, 17, 104. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yan, Y.; Jiang, W.; Spinetti, T.; Tardivel, A.; Castillo, R.; Bourquin, C.; Guarda, G.; Tian, Z.; Tschopp, J.; Zhou, R. Omega-3 Fatty Acids Prevent Inflammation and Metabolic Disorder through Inhibition of NLRP3 Inflammasome Activation. Immunity 2013, 38, 1154–1163. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pramfalk, C.; Eriksson, M.; Parini, P. Cholesteryl esters and ACAT. Eur. J. Lipid Sci. Technol. 2012, 114, 624–633. [Google Scholar] [CrossRef]
- Tomita, K.; Teratani, T.; Suzuki, T.; Shimizu, M.; Sato, H.; Narimatsu, K.; Usui, S.; Furuhashi, H.; Kimura, A.; Nishiyama, K.; et al. Acyl-CoA:cholesterol acyltransferase 1 mediates liver fibrosisby regulating free cholesterol accumulation in hepatic stellate cells. J. Hepatol. 2014, 61, 98–106. [Google Scholar] [CrossRef]
- Nicolosi, R.J.; Rogers, E.J.; Kritchevsky, D.; Scimeca, J.A.; Huth, P.J. Dietary conjugated linoleic acid reduces plasma lipoproteins and early aortic atherosclerosis in hypercholesterolemic hamsters. Artery 1997, 22, 266–277. [Google Scholar]
- Houseknecht, K.L.; Vanden Heuvel, J.P.; Moya-Camarena, S.Y.; Portocarrero, C.P.; Peck, L.W.; Nickel, K.P.; Belury, M.A. Dietary conjugated linoleic acid normalizes impaired glucose tolerance in the Zucker diabetic fatty fa/fa rat. Biochem. Biophys. Res. Commun. 1998, 244, 678–682. [Google Scholar] [CrossRef]
- Sergeant, S.; Rahbar, E.; Chilton, F.H. Gamma-linolenic acid, Dihommo-gamma linolenic, Eicosanoids and Inflammatory Processes. Eur. J. Pharmacol. 2016, 785, 77–86. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zurlo, F.; Larson, K.; Bogardus, C.; Ravussin, E. Skeletal muscle metabolism is a major determinant of resting energy expenditure. J. Clin. Investig. 1990, 86, 1423–1427. [Google Scholar] [CrossRef] [Green Version]
- Ruderman, N.B.; Saha, A.K.; Vavvas, D.; Witters, L.A. Malonyl-CoA, fuel sensing, and insulin resistance. Am. J. Physiol. Endocrinol. Metab. 1999, 39, E1–E18. [Google Scholar] [CrossRef]
- Sidossis, L.S.; Wolfe, R.R. Glucose and insulin-induced inhibition of fatty acid oxidation: The glucose-fatty acid cycle reversed. Am. J. Physiol. Endocrinol. Metab. 1996, 270, 733–738. [Google Scholar] [CrossRef] [PubMed]
- Schrauwen, P.; van Marken Lichtenbelt, W.D.; Saris, W.H.; Westerterp, K.R. Changes in fat oxidation in response to a high-fat diet. Am. J. Clin. Nutr. 1997, 66, 276–282. [Google Scholar] [CrossRef] [Green Version]
- Duplus, E.; Glorian, M.; Forest, C. Fatty Acid Regulation of Gene Transcription. J. Biol. Chem. 2000, 275, 30749–30752. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vitale, G.; Salvioli, S.; Franceschi, C. Oxidative stress and the ageing endocrine system. Nat. Rev. Endocrinol. 2013, 9, 228–240. [Google Scholar] [CrossRef] [PubMed]
- Conte, M.; Armani, A.; Conte, G.; Serra, A.; Franceschi, C.; Mele, M.; Sandri, M.; Salvioli, S. Muscle-specific Perilipin2 down-regulation affects lipid metabolism and induces myofiber hypertrophy. J. Cachexia Sarcopenia Muscle 2019, 10, 95–110. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bosma, M.; Sparks, L.M.; Hooiveld, G.J.; Jorgensen, J.A.; Houten, S.M.; Schrauwen, P.; Kersten, S.; Hesselink, M.K.C. Overexpression of PLIN5 in skeletal muscle promotes oxidative gene expression and intramyocellualar lipid content without compromising insulin sensitivity. Biochim. Biophys. Acta 2013, 1831, 844–852. [Google Scholar] [CrossRef] [PubMed]
- Covington, J.D.; Noland, R.C.; Hebert, R.C.; Masinter, B.S.; Smith, S.R.; Rustan, A.C.; Ravussin, E.; Bajpeyi, S. Perilipin 3 Differentially Regulates Skeletal Muscle Lipid Oxidation in Active, Sedentary, and Type 2 Diabetic Males. J. Clin. Endocrinol. Metab. 2015, 100, 3683–3692. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Funai, K.; Lodhi, I.J.; Spears, L.D.; Yin, L.; Song, H.; Klein, S.; Semenkovich, C.F. Skeletal Muscle Phospholipid Metabolism Regulates Insulin Sensitivity and Contractile Function. Diabetes 2016, 65, 358–370. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Smith, S.J.; Cases, S.; Jensen, D.R.; Chen, H.C.; Sande, E.; Tow, B.; Sanan, D.A.; Raber, J.; Eckel, R.H.; Farese, R.V., Jr. Obesity resistance and multiple mechanisms of triglyceride synthesis in mice lacking Dgat. Nat. Genet. 2000, 25, 87–90. [Google Scholar] [CrossRef] [PubMed]
- Listenberger, L.L.; Han, X.; Lewis, S.E.; Cases, S.; Farese, R.V., Jr.; Ory, D.S.; Schaffer, J.E. Triglyceride accumulation protects against fatty acid-induced lipotoxicity. Proc. Natl. Acad. Sci. USA 2003, 100, 3077–3082. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jump, D.B.; Clarke, S.D. Regulation of gene expression by dietary fat. Annu. Rev. Nutr. 1999, 19, 63–90. [Google Scholar] [CrossRef] [PubMed]
- Goodpaster, B.H.; He, J.; Watkins, S.; Kelley, D.E. Skeletal muscle lipid content and insulin resistance: Evidence for a paradox in endurance-trained athletes. J. Clin. Endocrinol. Metab. 2001, 86, 5755–5761. [Google Scholar] [CrossRef] [PubMed]
- Patti, M.-E.; Corvera, S. The Role of Mitochondria in the Pathogenesis of Type 2 Diabetes. Endocr. Rev. 2010, 31, 364–395. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Al-Rashed, F.; Ahmad, Z.; Snider, A.J.; Thomas, R.; Kochumon, S.; Melhem, M.; Sindhu, S.; Obeid, L.M.; Al-Mulla, F.; Hannun, Y.A.; et al. Ceramide kinase regulates TNF-α-induced immune responses in human monocytic cells. Sci. Rep. 2021, 11, 8259. [Google Scholar] [CrossRef] [PubMed]
- Mietla, J.A.; Wijesinghe, D.S.; Hoeferlin, L.A.; Shultz, M.D.; Natarajan, R.; Fowler, A.A.; Chalfant, C.E. Characterization of eicosanoid synthesis in a genetic ablation model of ceramide kinase. J. Lipid Res. 2013, 54, 1834–1847. [Google Scholar] [CrossRef] [Green Version]
- Li, W.; Zhao, X.; Yu, T.-T.; Hao, W.; Wang, G.-G. Knockout of PKC θ gene attenuates oleic acid-induced acute lung injury via reduction of inflammation and oxidative stress. Iran J. Basic Med. Sci. 2021, 24, 986–991. [Google Scholar]
- Tyner, S.D.; Venkatachalam, S.; Choi, J.; Jones, S.; Ghebranious, N.; Igelmann, H.; Lu, X.; Soron, G.; Cooper, B.; Brayton, C.; et al. p53 mutant mice that display early ageing-associated phenotypes. Nature 2002, 415, 45–53. [Google Scholar] [CrossRef] [PubMed]
- Maier, B.; Gluba, W.; Bernier, B.; Turner, T.; Mohammad, K.; Guise, T.; Sutherland, A.; Thorner, M.; Scrable, H. Modulation of mammalian life span by the short isoform of p53. Genes Dev. 2004, 18, 306–319. [Google Scholar] [CrossRef] [Green Version]
- Strycharz, J.; Drzewoski, J.; Szemraj, J.; Sliwinska, A. Is p53 Involved in Tissue-Specific Insulin Resistance Formation? Oxid. Med. Cell. Longev. 2017, 2017, 9270549. [Google Scholar] [CrossRef]
- Tsai, T.-H.; Chen, E.; Li, L.; Saha, P.; Lee, H.-J.; Huang, L.-S.; Shelness, G.S.; Chan, L.; Chang, B.H.-J. The constitutive lipid droplet protein PLIN2 regulates autophagy in liver. Autophagy 2017, 13, 1130–1144. [Google Scholar] [CrossRef] [Green Version]
- Griffin, J.D.; Salter, D.M.; Bowman, T.; Greenberg, A. Role of Hepatic PLIN2 and PLIN4 in the Development of Western Type Diet Induced Hepatosteatosis. FASEB J. 2018, 31, 458.3. [Google Scholar] [CrossRef]
- Zhou, J.; Yu, Y.; Ding, L.; Xu, P.; Wang, Y. Matcha Green Tea Alleviates Non-Alcoholic Fatty Liver Disease in High-Fat Diet-Induced Obese Mice by Regulating Lipid Metabolism and Inflammatory Responses. Nutrients 2021, 13, 1950. [Google Scholar] [CrossRef] [PubMed]
- Lally, J.S.V.; Ghoshal, S.; DePeralta, D.K.; Moaven, O.; Wei, L.; Masia, S.; Erstad, D.J.; Fujiwara, N.; Leong, V.; Houde, V.P.; et al. Inhibition of Acetyl-CoA Carboxylase by phosphorylation or the inhibitor ND-654 suppresses lipogenesis and hepatocellular carcinoma. Cell Metab. 2019, 29, 174–182. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nelson, M.E.; Lahiri, S.; Chow, J.D.Y.; Byrne, F.L.; Hargett, S.R.; Breen, D.S.; Olzomer, E.M.; Wu, L.E.; Cooney, C.J.; Turner, R.; et al. Inhibitionof hepatic lipogenesis enhances liver tumorigenesis by increasing antioxidant defence and promoting cell survival. Nat. Commun. 2017, 8, 14689. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, M.T.; Cheon, Y.; Li, Y.; Nara, T.Y. Mechanisms of regulation of gene expression by fatty acids. Lipids 2004, 39, 1077–1083. [Google Scholar] [CrossRef] [PubMed]
- Nakatani, T.; Kim, H.-J.; Kaburagi, Y.; Yasuda, K.; Ezaki, O. A low fish oil inhibits SREBP-1 proteolytic cascade, while a high-fish-oil feeding decreases SREBP-1 mRNA in mice liver: Relationship to anti-obesity. J. Lipid Res. 2003, 44, 369–379. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vieira, P.; Cameron, J.; Rahikkala, E.; Keski-Filppula, R.; Zhang, L.-H.; Santra, S.; Matthews, A.; Myllynen, P.; Nuutinen, M.; Moilanen, J.; et al. Novel homozygous PCK1 mutation causing cytosolic phosphoenolpyruvate carboxykinase deficiency presenting as childhood hypoglycemia, an abnormal pattern of urine metabolites and liver dysfunction. Mol. Genet. Metab. 2017, 120, 337–341. [Google Scholar] [CrossRef]
- Tuo, L.; Xiang, J.; Pan, X.; Hu, J.; Tang, H.; Liang, L.; Xia, J.; Hu, Y.; Zhang, W.; Huang, A.; et al. PCK1 negatively regulates cell cycle progression and hepatoma cell proliferation via the AMPK/p27Kip1 axis. J. Exp. Clin. Cancer Res. 2019, 38, 50. [Google Scholar] [CrossRef] [Green Version]
- Morris, E.M.; Meers, G.M.E.; Booth, F.W.; Fritsche, K.L.; Hardin, C.D.; Thyfault, J.P.; Ibdah, J.A. PGC-1α overexpression results in increased hepatic fatty acid oxidation with reduced triacylglycerol accumulation and secretion. Am. J. Physiol. Gastrointest. Liver Physiol. 2012, 303, G979–G992. [Google Scholar] [CrossRef] [Green Version]
- Adela, R.; Banerjee, S.K. GDF-15 as a Target and Biomarker for Diabetes and Cardiovascular Diseases: A Translational Prospective. J. Diabetes Res. 2015, 2015, 490842. [Google Scholar] [CrossRef]
- Conte, M.; Ostan, R.; Fabbri, C.; Santoro, A.; Guidarelli, G.; Vitale, G.; Mari, D.; Sevini, F.; Capri, M.; Sandri, M.; et al. Human Aging and Longevity Are Characterized by High Levels of Mitokines. J. Gerontol. Ser. A Biol. Sci. Med. Sci. 2019, 74, 600–607. [Google Scholar] [CrossRef] [PubMed]
- Nagao, K.; Yanagita, T. Bioactive lipids in metabolic syndrome. Prog. Lipid Res. 2008, 47, 127–146. [Google Scholar] [CrossRef] [PubMed]
- Uysal, K.T.; Wiesbrock, S.M.; Marino, M.W.; Hotamisligil, G.S. Protection from obesity-induced insulin resistancein mice lacking TNF-α function. Nature 1997, 389, 610–614. [Google Scholar] [CrossRef]
- Pavlovic, Z.; Bakovic, M. Regulation of Phosphatidylethanolamine Homeostasis—The Critical Role of CTP:Phosphoethanolamine Cytidylyltransferase (Pcyt2). Int. J. Mol. Sci. 2013, 14, 2529–2550. [Google Scholar] [CrossRef] [Green Version]
- Aleshin, S.; Reiser, G. Peroxisome proliferator-activated receptor β/δ (PPARβ/δ) protects against ceramide-induced cellular toxicity in rat brain astrocytes and neurons by activation of ceramide kinase. Mol. Cell. Neurosci. 2014, 59, 127–134. [Google Scholar] [CrossRef] [PubMed]
- Conte, M.; Medici, V.; Malagoli, D.; Chiariello, A.; Cirrincione, A.; Davin, A.; Chikhladze, M.; Vasuri, F.; Legname, G.; Ferrer, I.; et al. Expression pattern of perilipins in human brain during aging and in Alzheimer’s disease. Neuropathol. Appl. Neurobiol. 2021, 48, e12756. [Google Scholar] [CrossRef] [PubMed]
- Shimazawa, M.; Inokuchi, Y.; Ito, Y.; Murata, H.; Aihara, M.; Miura, M.; Araie, M.; Hara, H. Involvement of ER stress in retinal cell death. Mol. Vis. 2007, 13, 578–587. [Google Scholar] [PubMed]
- Astarita, G.; Jung, K.-M.; Vasilevko, V.; DiPatrizio, N.V.; Martin, S.K.; Cribbs, D.H.; Head, E.; Cotman, C.W.; Piomelli, D. Elevated Stearoyl-CoA Desaturase in Brains of Patients with Alzheimer’s Disease. PLoS ONE 2011, 6, e24777. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Beckner, M.; Agostino, N.; Fellows-Mayle, W.; Kant, J.; Pollack, I. ATP citrate lyase is a potential therapeutic target for suppression of aberrant metabolic adaptation in glycolytic brain tumors. Cancer Res. 2008, 68. [Google Scholar] [CrossRef] [Green Version]
- Knobloch, M.; Braun, S.; Zurkirchen, L.; Von Schoultz, C.; Zamboni, N.; Araúzo-Bravo, M.J.; Kovacs, W.; Karalay, Ö.; Suter, U.; Machado, R.A.M.; et al. Metabolic control of adult neural stem cell activity by Fasn-dependent lipogenesis. Nature 2013, 493, 226–230. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Bilbao, F.; Arsenijevic, D.; Vallet, P.; Hjelle, O.P.; Ottersen, O.P.; Bouras, C.; Raffin, Y.; Abou, K.; Langhans, W.; Collins, S.; et al. Resistance to cerebral ischemic injury in UCP2 knockout mice: Evidence for a role of UCP2 as a regulator of mitochondrial glutathione levels. J. Neurochem. 2004, 89, 1283–1292. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.C.; Farese, R.V., Jr. Inhibition of Triglyceride Synthesis as a Treatment Strategy for Obesity. Arterioscler. Thromb. Vasc. Biol. 2005, 25, 482–486. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oh, W.K.; Abu-Elheiga, L.; Kordari, P.; Gu, Z.; Shaikenov, T.; Chirala, S.S.; Wakil, S.J. Glucose and fat metabolism in adipose tissue of acetyl-CoA carboxylase 2 knockout mice. Proc. Natl. Acad. Sci. USA 2005, 102, 1384–1389. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Latasa, M.-J.; Moon, Y.S.; Kim, K.-H.; Sul, H.S. Nutritional regulation of the fatty acid synthase promoter in vivo: Sterol regulatory element binding protein functions through an upstream region containing a sterol regulatory element. Proc. Natl. Acad. Sci. USA 2000, 97, 10619–10624. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Horton, J.D.; Shimomur, I.; Bashmakov, Y.; Hammer, R.E. Overexpression of SREBP-1a in Mouse Adipose Tissue Produces Adipocyte Hypertrophy, Increased Fatty Acid Secretion, and Fatty Liver. J. Biol. Chem. 2003, 278, 36652–36660. [Google Scholar] [CrossRef] [PubMed] [Green Version]
| CON | CHE | ENR | |
|---|---|---|---|
| Protein (%) | 14.30 | 21.81 | 19.95 |
| Lipid (%) | 4.00 | 23.07 | 21.89 |
| Carbohydrates (%) | 48.00 | 19.38 | 19.38 |
| Ash (%) | 4.70 | 3.54 | 2.77 |
| Energy (cal/g) | 278.23 | 377.62 | 359.02 |
| C16:0 | 0.50 | 3.03 | 2.41 |
| C18:0 | 0.10 | 1.49 | 1.49 |
| C18:1c9 | 0.70 | 2.85 | 3.17 |
| C18:1c9c12 | 2.00 | 1.10 | 1.11 |
| C18:2c9t11 | 0.00 | 0.13 | 0.73 |
| C18:1c9c12c15 | 0.10 | 0.12 | 0.33 |
| CON | CHE | ENR | SEM | p-Value | |
|---|---|---|---|---|---|
| Initial body weight (g) | 26.00 | 25.00 | 28.00 | 1.50 | 0.450 |
| Final body weight (g) | 26.50 B | 26.00 B | 32.00 A | 1.45 | 0.009 |
| Total Cholesterol (mg/dL) | 78.00 B | 100.00 A | 78.00 A | 3.27 | <0.001 |
| Cholesterol HDL (mg/dL) | 25.50 C | 72.00 A | 40.00 B | 2.50 | <0.001 |
| Cholesterol LDL (mg/dL) | 25.00 A | 15.00 B | 12.50 B | 2.00 | 0.004 |
| Cardiovascular risk (Total Chol./Chol. HDL) | 2.80 A | 1.40 B | 1.50 B | 0.10 | 0.032 |
| CON | CHE | ENR | SEM | p-Value | |
|---|---|---|---|---|---|
| Total Lipids (g/100 g of liver) | 9.47 | 8.20 | 8.26 | 0.98 | 0.762 |
| Phospholipids (g/100 g TL) | 11.12 | 5.59 | 8.40 | 2.75 | 0.237 |
| Triglycerides (g/100 g TL) | 50.47 | 55.56 | 49.99 | 4.15 | 0.321 |
| Free Fatty Acids (g/100 g TL) | 37.70 | 38.09 | 40.73 | 4.93 | 0.550 |
| Free Cholesterol (mg/100 g TL) | 193.01 a | 94.88 b | 109.88 b | 37.37 | 0.025 |
| Esterified Cholesterol (mg/100 g TL) | 4.38 b | 4.50 b | 28.28 a | 0.68 | <0.001 |
| Saturated Fatty Acids | 23.19 B | 30.28 A | 30.38 A | 1.62 | <0.001 |
| Monounsaturated Fatty Acids | 41.99 C | 48.26 B | 55.05 A | 1.75 | <0.001 |
| Polyunsaturated Fatty Acids | 33.36 A | 13.14 B | 14.89 B | 1.20 | <0.001 |
| Polyunsaturated Fatty Acids n-6 | 31.79 A | 10.73 B | 10.02 B | 1.07 | <0.001 |
| Polyunsaturated Fatty Acids n-3 | 1.20 | 0.89 | 1.03 | 0.40 | 0.483 |
| n-6/n-3 | 26.53 A | 12.12 B | 9.75 C | 0.33 | 0.005 |
| Conjugated Linoleic Acid | 0.04 C | 1.22 B | 3.40 A | 0.12 | <0.001 |
| CON | CHE | ENR | SEM | p-Value | |
|---|---|---|---|---|---|
| Total Lipids (g/100 g of brain) | 8.88 | 10.65 | 11.33 | 1.28 | 0.156 |
| Phospholipids (g/100 g TL) | 63.27 | 62.87 | 70.14 | 3.11 | 0.347 |
| Triglycerides (g/100 g TL) | 25.37 | 25.95 | 22.64 | 3.23 | 0.537 |
| Free Fatty Acids (g/100 g TL) | 10.30 | 10.65 | 6.81 | 2.04 | 0.103 |
| Free Cholesterol (mg/100 g TL) | 897.78 | 720.15 | 1217.02 | 258.95 | 0.201 |
| Esterified Cholesterol (mg/100 g TL) | - | - | - | - | NE |
| Saturated Fatty Acids | 43.63 | 50.33 | 44.32 | 3.21 | 0.503 |
| Monounsaturated Fatty Acids | 28.47 | 30.39 | 27.76 | 2.06 | 0.303 |
| Polyunsaturated Fatty Acids | 17.96 | 20.39 | 20.19 | 1.96 | 0.203 |
| Polyunsaturated Fatty Acids n-6 | 13.64 | 13.27 | 13.22 | 1.21 | 0.364 |
| Polyunsaturated Fatty Acids n-3 | 4.24 | 6.54 | 6.60 | 1.16 | 0.540 |
| n-6/n-3 | 3.52 | 2.27 | 2.18 | 0.45 | 0.222 |
| Conjugated Linoleic Acid | 0.40 | 1.05 | 1.07 | 0.33 | 0.401 |
| CON | CHE | ENR | SEM | p-Value | |
|---|---|---|---|---|---|
| Total Lipids (g/100 g of muscle) | 19.25 | 26.84 | 17.15 | 4.73 | 0.252 |
| Phospholipids (g/100 g TL) | 11.25 b | 9.72 b | 20.32 a | 3.36 | 0.013 |
| Triglycerides (g/100 g TL) | 87.58 a | 88.11 a | 75.55 b | 3.37 | 0.011 |
| Free Fatty Acids (g/100 g TL) | 0.45 b | 3.75 a | 3.30 a | 1.37 | 0.033 |
| Free Cholesterol (mg/100 g TL) | 15.60 A | 7.84 B | 7.53 B | 1.36 | <0.001 |
| Esterified Cholesterol (mg/100 g TL) | 2.50 | 4.73 | 3.05 | 1.22 | 0.456 |
| Saturated Fatty Acids | 41.19 C | 63.52 A | 55.46 B | 3.15 | 0.006 |
| Monounsaturated Fatty Acids | 38.27 A | 26.03 C | 29.75 B | 1.35 | <0.001 |
| Polyunsaturated Fatty Acids | 19.96 a | 11.48 b | 17.35 a | 2,22 | 0.019 |
| Polyunsaturated Fatty Acids n-6 | 18.39 a | 9.47 c | 11.48 b | 1,39 | 0.003 |
| Polyunsaturated Fatty Acids n-3 | 1.60 c | 1.99 b | 5.34 a | 1.18 | 0.016 |
| n-6/n-3 | 11.69 A | 5.22 B | 2.78 C | 1.03 | <0.001 |
| Conjugated Linoleic Acid | 0.07 C | 0.42 B | 1.37 A | 0.09 | <0.001 |
| CON | CHE | ENR | SEM | p-Value | |
|---|---|---|---|---|---|
| Total Lipids (g/100 g of ad. tissue) | 64.17 | 51.62 | 57.76 | 4.10 | 0.175 |
| Phospholipids (g/100 g TL) | 1.11 | 1.12 | 1.02 | 0.25 | 0.641 |
| Triglycerides (g/100 g TL) | 95.03 | 95.03 | 95.00 | 0.23 | 0.953 |
| Free Fatty Acids (g/100 g TL) | 2.27 | 1.61 | 1.81 | 0.27 | 0.611 |
| Free Cholesterol (mg/100 g TL) | 5.60 | 7.84 | 7.53 | 1.36 | 0.584 |
| Esterified Cholesterol (mg/100 g TL) | 2.50 | 4.73 | 3.05 | 1.22 | 0.430 |
| Saturated Fatty Acids | 48.92 B | 55.16 A | 53.08 A | 1.45 | <0.001 |
| Monounsaturated Fatty Acids | 27.91 B | 34.54 A | 36.35 A | 0.58 | <0.001 |
| Polyunsaturated Fatty Acids | 22.17 A | 9.30 B | 9.57 B | 0.61 | <0.001 |
| Polyunsaturated Fatty Acids n-6 | 21.14 A | 7.67 B | 6.57 B | 0.56 | <0.001 |
| Polyunsaturated Fatty Acids n-3 | 0.78 | 0.62 | 0.61 | 0.07 | 0.766 |
| n-6/n-3 | 27.10 A | 12.17 B | 10.77 B | 0.65 | <0.001 |
| Conjugated Fatty Acids | 0.02 C | 0.89 B | 2.31 A | 0.09 | <0.001 |
| LIVER | BRAIN | SKELETAL MUSCLE | ADIPOSE TISSUE | |||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CON | CHE | ENR | SEM | p-Value | CON | CHE | ENR | SEM | p-Value | CON | CHE | ENR | SEM | p-Value | CON | CHE | ENR | SEM | p-Value | |
| Lipid metabolism | ||||||||||||||||||||
| PLIN1 | - | - | - | - | NE | - | - | - | - | NE | - | - | - | - | NE | 0.55 | 0.67 | 0.32 | 0.11 | 0.572 |
| PLIN2 | 1.27 a | 0.40 b | 0.57 b | 0.20 | 0.002 | 1.13 A | 1.13 A | 0.59 B | 0.1 | 0.003 | 1.1 | 0.82 | 0.83 | 0.12 | 0.123 | 1.29 | 0.87 | 0.75 | 0.17 | 0.975 |
| PLIN3 | 1.29 a | 1.41 a | 0.75 b | 0.15 | 0.024 | 1.14 | 1.33 | 0.65 | 0.2 | 0.435 | 0.93 a | 0.45 b | 0.75 a | 0.1 | 0.045 | - | - | - | - | NE |
| PLIN4 | 1.09 c | 12.34 b | 18.67 a | 2.81 | 0.008 | 2.96 | 2.55 | 2.45 | 0.75 | 0.655 | 1.21 A | 0.49 B | 0.43 B | 0.11 | 0.009 | 1.10 A | 0.37 B | 0.21 B | 0.09 | <0.001 |
| PLIN5 | 0.59 b | 0.36 b | 1.08 a | 0.12 | 0.005 | 2.46 | 1.39 | 1.89 | 0.61 | 0.699 | 0.78 A | 0.22 B | 0.20 B | 0.08 | <0.001 | - | - | - | - | NE |
| SREBP1 | 2.25 b | 3.38 a | 1.26 c | 0.43 | 0.025 | 1.20 A | 0.57 C | 0.89 B | 0.08 | 0.007 | 0.69 | 1.16 | 0.88 | 0.17 | 0.968 | 1.23 b | 5.39 a | 2.46 b | 0.87 | 0.029 |
| ACACA | 1.38 a | 0.74 b | 0.76 b | 0.16 | 0.037 | 1.52 | 1.31 | 1.03 | 0.24 | 0.213 | 0.78 A | 0.74 A | 0.29 B | 0.09 | 0.004 | 2.51 | 1.94 | 2.94 | 0.5 | 0.144 |
| ACACB | 1.73 A | 0.37 B | 0.55 B | 0.23 | <0.001 | - | - | - | - | NE | 0.76 | 0.65 | 0.70 | 0.10 | 0.650 | 1.50 A | 0.42 B | 0.56 B | 0.13 | 0.002 |
| FASN | 1.08 A | 0.24 B | 0.09 C | 0.06 | <0.001 | 1.54 b | 1.38 b | 3.54 a | 0.43 | 0.034 | 0.84 | 0.91 | 0.91 | 0.08 | 0.411 | 1.40 C | 6.56 B | 16.65 A | 0.9 | <0.001 |
| SCD | 1.09 A | 0.37 C | 0.65 B | 0.07 | <0.001 | 0.92 a | 1.13 a | 0.34 b | 0.15 | 0.021 | 1.69 A | 0.44 B | 0.39 B | 0.2 | 0.004 | 1.1 | 0.86 | 0.97 | 0.18 | 0.167 |
| GPAM | 1.14 A | 1.31 | 1.59 | 0.29 | 0.419 | 1.50 | 1.21 | 1.07 | 0.21 | 0.517 | 1.50 | 1.21 | 1.07 | 0.21 | 0.517 | 2.07 A | 0.33 B | 0.33 B | 0.23 | 0.003 |
| AGPAT2 | 1.48 | 0.93 | 0.99 | 0.31 | 0.839 | 1.43 | 1.62 | 2.31 | 0.54 | 0.321 | 0.86 | 0.93 | 1.05 | 0.2 | 0.635 | 2.13 | 4.68 | 2.73 | 1.00 | 0.383 |
| DGAT1 | 0.84 | 1.00 | 0.78 | 0.16 | 0.408 | 1.61 A | 1.01 B | 0.33 C | 0.16 | <0.001 | 0.63 a | 0.63 a | 0.34 b | 0.11 | 0.034 | 0.99 | 1.10 | 0.91 | 0.21 | 0.901 |
| DGAT2 | 1.14 | 1.49 | 1.56 | 0.26 | 0.496 | 1.17 A | 0.62 B | 0.32 B | 0.15 | 0.007 | 0.95 A | 0.30 B | 0.45 B | 0.06 | <0.001 | 2.36 B | 6.92 A | 0.94 B | 0.88 | 0.002 |
| LPIN1 | 2.10 c | 10.17 b | 22.27 a | 3.78 | 0.021 | 1.13 a | 0.96 a | 0.43 b | 0.17 | 0.016 | 1.13 A | 0.65 B | 1.36 A | 0.11 | 0.006 | 1.83 A | 0.56 B | 0.48 B | 0.13 | <0.001 |
| ELOVL2 | 1.16 | 0.75 | 1.18 | 0.2 | 0.658 | 1.62 a | 1.50 a | 0.33 b | 0.32 | 0.020 | - | - | - | - | NE | - | - | - | - | NE |
| ACLY | 1.63 A | 0.58 B | 0.24 C | 0.11 | <0.001 | 1.11 a | 0.96 a | 0.63 b | 0.12 | 0.016 | 0.28 | 0.26 | 0.13 | 0.06 | 0.863 | 1.63 B | 3.64 A | 2.27 B | 0.21 | <0.001 |
| ETNK1 | 0.80 | 0.68 | 0.61 | 0.08 | 0.881 | 1.48 A | 0.59 B | 0.51 B | 0.15 | 0.008 | 0.95 A | 0.43 B | 0.31 B | 0.06 | <0.001 | 2.58 | 2.48 | 1.26 | 0.37 | 0.688 |
| PCYT2 | 1.00 A | 0.83 B | 0.34 C | 0.07 | <0.001 | 1.10 A | 0.67 B | 0.32 C | 0.1 | 0.007 | 1.28 B | 2.31 A | 1.15 B | 0.17 | 0.001 | 1.49 | 2.7 | 2.85 | 0.43 | 0.975 |
| CEPT-1 | 0.57 | 0.35 | 0.54 | 0.09 | 0.754 | 1.40 | 1.44 | 1.35 | 0.35 | 0.445 | 0.84 A | 1.06 A | 0.31 B | 0.09 | <0.001 | 1.59 | 1.51 | 1.06 | 0.2 | 0.916 |
| PISD | 0.98 b | 1.03 b | 1.56 b | 0.13 | 0.035 | 1.36 | 1.64 | 1.54 | 0.23 | 0.644 | 0.79 a | 0.45 b | 0.76 a | 0.09 | 0.045 | 2.77 | 1.03 | 1.32 | 0.57 | 0.732 |
| CERK | 1.71 | 1.63 | 1.17 | 0.32 | 0.761 | 1.23 | 0.99 | 0.6 | 0.14 | 0.029 | 0.82 A | 0.41 B | 0.25 B | 0.06 | 0.002 | 1.94 A | 1.15 B | 0.67 C | 0.14 | 0.001 |
| CERS6 | 2.13 | 1.88 | 2.54 | 0.52 | 0.384 | 1.37 | 0.91 | 1.14 | 0.23 | 0.714 | - | - | - | - | NE | 2.02 | 1.60 | 1.12 | 0.21 | 0.202 |
| PCK1 | 0.92 b | 0.66 c | 1.29 a | 0.17 | 0.029 | - | - | - | - | NE | - | - | - | - | NE | 0.92 A | 0.22 B | 0.16 B | 0.1 | 0.002 |
| PRKAA1 | 0.76 b | 1.10 a | 0.82 b | 0.05 | 0.006 | 1.38 | 0.91 | 1.46 | 0.24 | 0.816 | 0.44 | 0.40 | 0.41 | 0.05 | 0.444 | 2.67 a | 1.29 b | 0.98 b | 0.32 | 0.027 |
| ATGL | 1.33 | 1.78 | 1.65 | 0.27 | 0.385 | 1.78 | 1.78 | 1.5 | 0.35 | 0.885 | 0.79 | 0.43 | 0.58 | 0.11 | 0.738 | 1.61 A | 0.24 B | 0.26 B | 0.15 | <0.001 |
| CPT1 b | 1.03 b | 1.26 b | 2.45 a | 0.41 | 0.036 | - | - | - | - | NE | 0.94 | 0.68 | 0.99 | 0.10 | 0.489 | 2.37 A | 1.97 A | 0.00 B | 0.6 | 0.007 |
| LIPE | 1.58 | 2.44 | 1.93 | 0.28 | 0.843 | 1.54 | 1.32 | 0.91 | 0.28 | 0.421 | 0.45 | 0.42 | 0.31 | 0.07 | 0.521 | 1.86 | 0.94 | 0.91 | 0.31 | 0.641 |
| Inflammation and cell cycle | ||||||||||||||||||||
| PRKCQ | 0.52 A | 0.45 A | 0.28 A | 0.14 | 0.005 | 1.42 A | 1.11 B | 0.32 C | 0.14 | <0.001 | 0.98 A | 1.24 A | 0.36 B | 0.13 | 0.009 | - | - | - | - | NE |
| NFATC2 | - | - | - | - | NE | - | - | - | - | NE | 1.11 B | 7.26 A | 2.46 B | 0.52 | <0.001 | 1.56 | 1.36 | 1.39 | 0.29 | 0.669 |
| TRP53 | 1.12 a | 0.51 b | 0.24 c | 0.12 | 0.002 | 1.46 A | 0.77 B | 0.25 C | 0.15 | 0.006 | 1.18 b | 2.01 a | 1.13 b | 0.2 | 0.011 | 1.08 a | 1.12 a | 0.29 b | 0.18 | 0.012 |
| P21 | 1.91 A | 1.59 A | 1.84 A | 0.27 | 0.194 | 1.05 | 0.88 | 0.9 | 0.16 | 0.589 | 0.71 A | 0.23 B | 0.28 B | 0.07 | 0.002 | 1.54 A | 0.29 B | 0.32 B | 0.14 | <0.001 |
| FGF21 | 0.28 b | 0.08 c | 0.43 a | 0.1 | 0.024 | - | - | - | - | NE | - | - | - | - | NE | 0.82 A | 0.10 B | 0.15 B | 0.04 | <0.001 |
| IL1 B | 1.11 A | 0.33 B | 0.41 B | 0.10 | <0.001 | - | - | - | - | NE | - | - | - | - | NE | 0.25 | 0.43 | 0.00 | 0.06 | 0.530 |
| Mitochondrial metabolism | ||||||||||||||||||||
| PGC1 A | 1.21 c | 1.90 b | 2.20 a | 0.18 | 0.002 | 1.36 | 0.96 | 1.62 | 0.23 | 0.662 | 1.36 | 0.96 | 1.62 | 0.23 | 0.662 | 3.15 | 2.33 | 2.05 | 0.28 | 0.535 |
| OPA-1 | 0.77 c | 1.22 b | 1.60 a | 0.19 | 0.022 | 1.14 | 0.83 | 0.85 | 0.11 | 0.435 | 1.14 | 0.83 | 0.85 | 0.11 | 0.435 | 1.69 | 1.92 | 1.42 | 0.23 | 0.622 |
| MFN1 | 0.83 a | 0.40 b | 0.35 b | 0.10 | 0.043 | - | - | - | - | NE | - | - | - | - | NE | 2.67 | 7.98 | 4.69 | 1.88 | 0.789 |
| DRP1 | 1.16 | 0.95 | 0.84 | 0.12 | 0.654 | - | - | - | - | NE | - | - | - | - | NE | 1.99 | 2.26 | 1.94 | 0.26 | 0.964 |
| UCP2 | 1.71 a | 0.74 c | 1.14 b | 0.26 | 0.014 | 1.38 A | 0.55 B | 0.38 B | 0.13 | <0.001 | 1.38 A | 0.55 B | 0.38 B | 0.13 | <0.001 | 1.80 A | 0.60 B | 0.69 B | 0.15 | <0.001 |
| UCP3 | - | - | - | - | NE | - | - | - | - | NE | - | - | - | - | NE | 1.27 | 0.82 | 1.00 | 0.15 | 0.720 |
| NDUFS3 | 0.97 A | 0.99 | 1.71 | 0.36 | 0.791 | 0.99 | 1.45 | 1.24 | 0.18 | 0.954 | 0.99 | 1.45 | 1.24 | 0.18 | 0.954 | 1.51 A | 1.06 A | 0.34 B | 0.17 | 0.001 |
| UQCR10 | 0.67 A | 0.55 | 0.66 | 0.13 | 0.756 | 2.09 a | 1.10 b | 1.03 b | 0.27 | 0.013 | 2.09 a | 1.10 b | 1.03 b | 0.27 | 0.019 | 3.72 A | 2.31 B | 0.17 C | 0.55 | <0.001 |
| COX10 | 1.77 a | 1.34 b | 1.04 c | 0.16 | 0.034 | 1.54 | 1.78 | 1.14 | 0.31 | 0.484 | 1.54 | 1.78 | 1.14 | 0.31 | 0.484 | 1.74 A | 1.79 A | 0.81 B | 0.15 | 0.007 |
| ATP5 G1 | 0.81 a | 0.68 b | 0.46 c | 0.10 | 0.016 | 1.56 a | 0.72 b | 0.92 b | 0.24 | 0.022 | 1.56 a | 0.72 b | 0.92 b | 0.24 | 0.026 | 2.26 | 4.67 | 3.75 | 0.87 | 0.675 |
| KLOTOB | - | - | - | - | NE | - | - | - | - | NE | - | - | - | - | NE | 0.96 A | 0.09 B | 0.19 B | 0.08 | <0.001 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tognocchi, M.; Conte, M.; Testai, L.; Martucci, M.; Serra, A.; Salvioli, S.; Calderone, V.; Mele, M.; Conte, G. Supplementation of Enriched Polyunsaturated Fatty Acids and CLA Cheese on High Fat Diet: Effects on Lipid Metabolism and Fat Profile. Foods 2022, 11, 398. https://doi.org/10.3390/foods11030398
Tognocchi M, Conte M, Testai L, Martucci M, Serra A, Salvioli S, Calderone V, Mele M, Conte G. Supplementation of Enriched Polyunsaturated Fatty Acids and CLA Cheese on High Fat Diet: Effects on Lipid Metabolism and Fat Profile. Foods. 2022; 11(3):398. https://doi.org/10.3390/foods11030398
Chicago/Turabian StyleTognocchi, Monica, Maria Conte, Lara Testai, Morena Martucci, Andrea Serra, Stefano Salvioli, Vincenzo Calderone, Marcello Mele, and Giuseppe Conte. 2022. "Supplementation of Enriched Polyunsaturated Fatty Acids and CLA Cheese on High Fat Diet: Effects on Lipid Metabolism and Fat Profile" Foods 11, no. 3: 398. https://doi.org/10.3390/foods11030398
APA StyleTognocchi, M., Conte, M., Testai, L., Martucci, M., Serra, A., Salvioli, S., Calderone, V., Mele, M., & Conte, G. (2022). Supplementation of Enriched Polyunsaturated Fatty Acids and CLA Cheese on High Fat Diet: Effects on Lipid Metabolism and Fat Profile. Foods, 11(3), 398. https://doi.org/10.3390/foods11030398









